Identification of 56 Proteins Involved in Embryo–Maternal Interactions in the Bovine Oviduct
Abstract
1. Introduction
2. Results
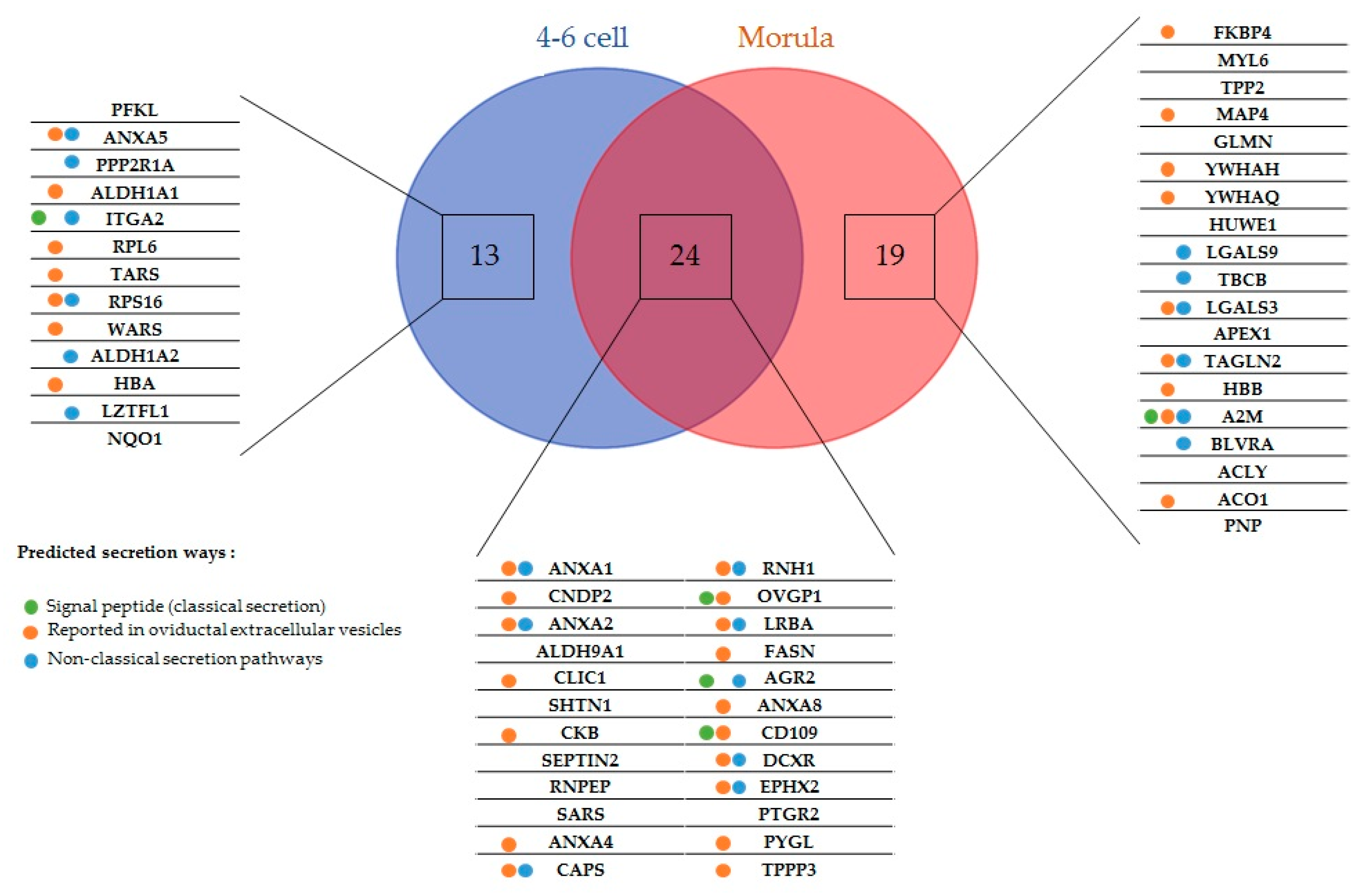
2.1. New Embryo-Interacting Proteins Were Identified by NanoLC-MS/MS and Changed According to the Embryonic Stage
2.2. Most Embryo-Interacting Proteins Were Presumed to be Exosomal and Secreted via Non-Conventional Pathways
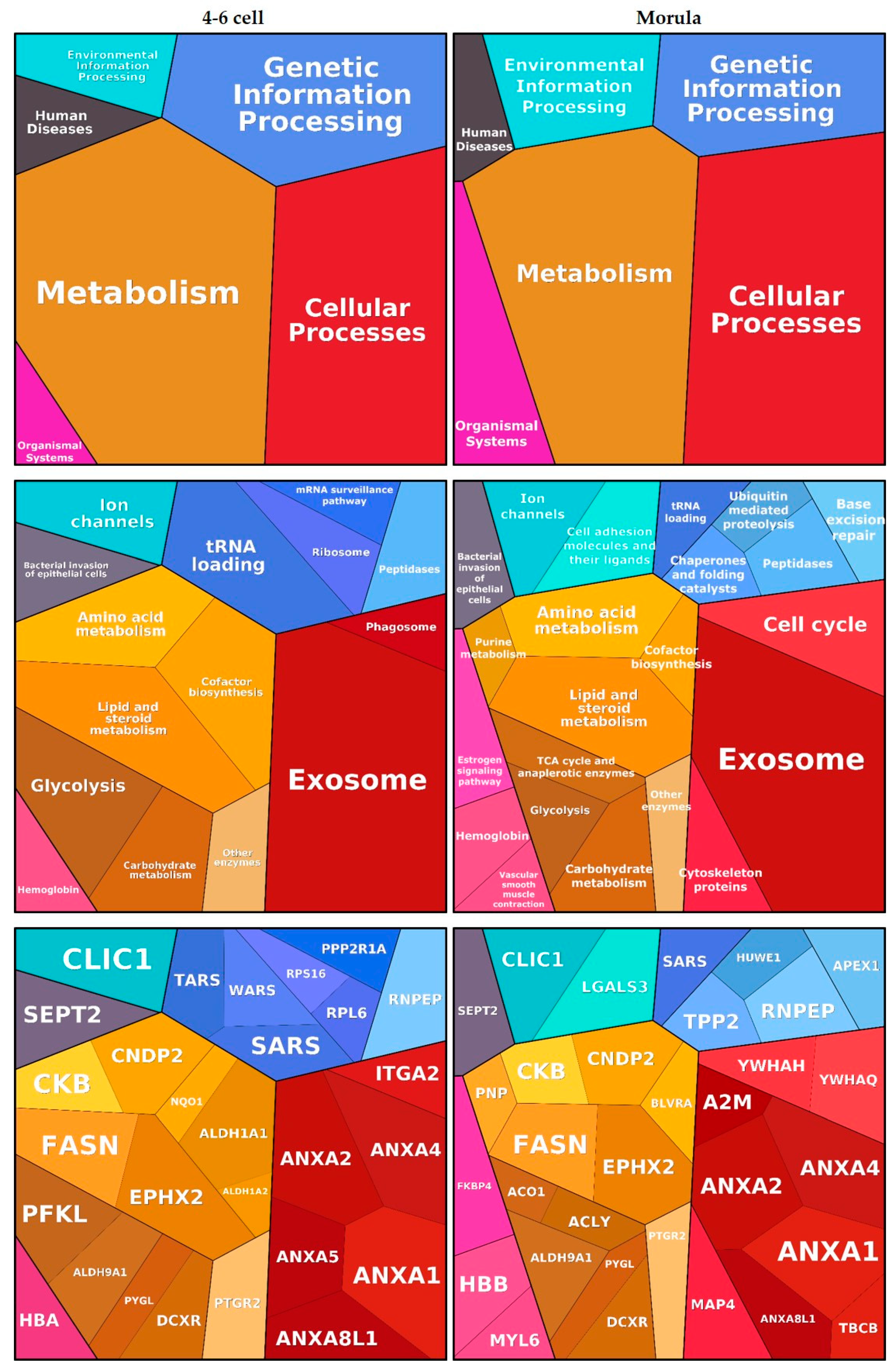
2.3. Embryo-Interacting Proteins Were Mainly Involved in Metabolism and Cellular Processes
2.4. Protein Interactions Were Localized in Different Embryo Subcompartments
3. Discussion
4. Materials and Methods
4.1. Bovine Oviductal Fluid (OF) Collection
4.2. Embryo In-Vitro Production and Incubation with OF
4.3. Nanoliquid Chromatography Coupled with Tandem Mass Spectrometry (NanoLC-MS/MS)
4.4. Quantification of Proteins, Identification of Embryo-Interacting Proteins and Statistical Analysis
4.5. Functional Analyses of Interacting Proteins
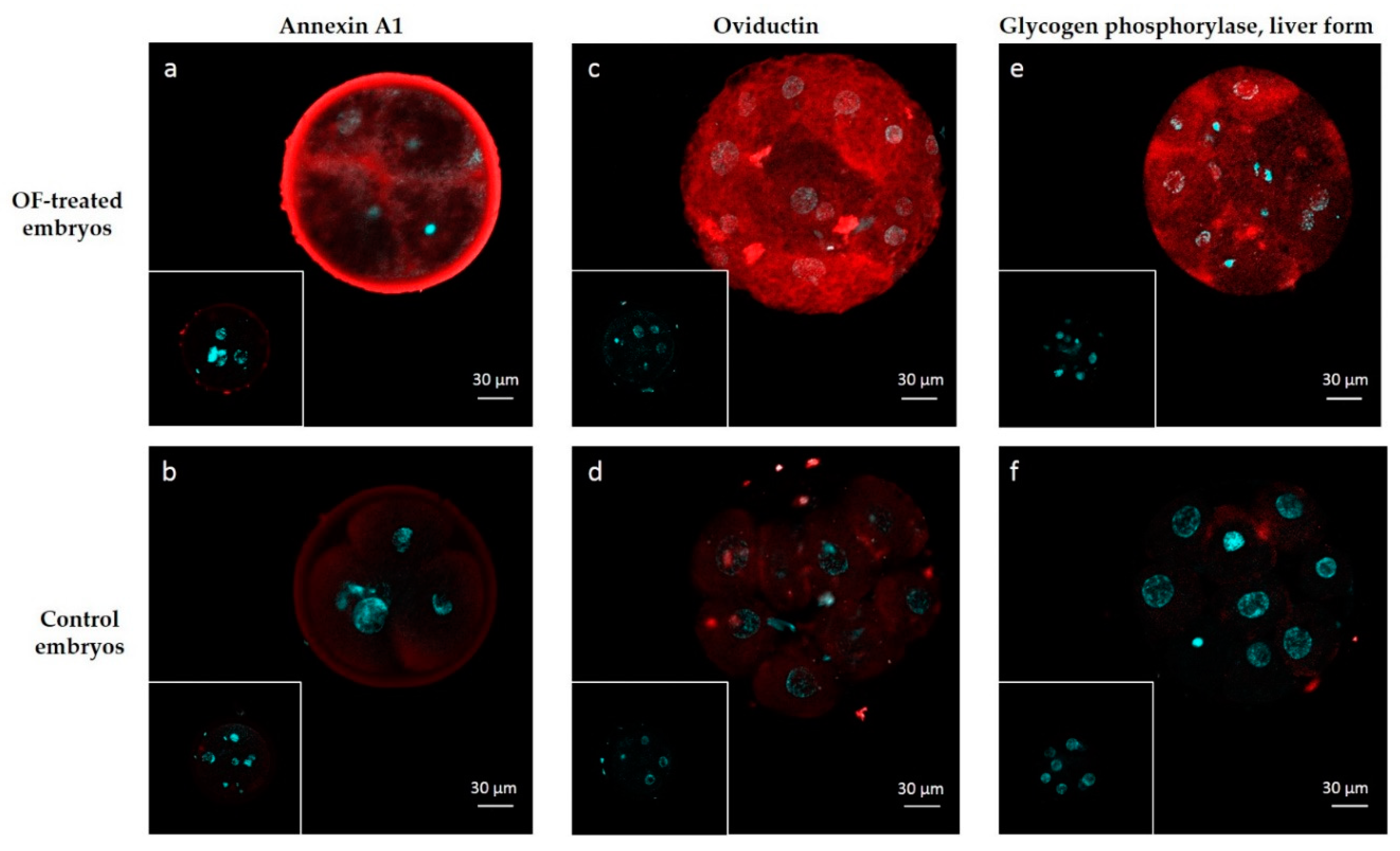
4.6. Immunolocalization of ANXA1, OVGP1 and PYGL
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| OF | Oviductal fluid |
| EV | Extracellular vesicles |
| MS | Mass spectrometry |
| nanoLC-MS/MS | Nanoliquid chromatography coupled with tandem mass spectrometry |
| NWS | Normalized weighted spectra |
References
- Kolle, S.; Dubielzig, S.; Reese, S.; Wehrend, A.; Konig, P.; Kummer, W. Ciliary transport, gamete interaction, and effects of the early embryo in the oviduct: Ex vivo analyses using a new digital videomicroscopic system in the cow. Biol. Reprod. 2009, 81, 267–274. [Google Scholar] [CrossRef]
- Lonergan, P.; Forde, N. Maternal-embryo interaction leading up to the initiation of implantation of pregnancy in cattle. Animal 2014, 8 (Suppl. 1), 64–69. [Google Scholar] [CrossRef]
- Graf, A.; Krebs, S.; Heininen-Brown, M.; Zakhartchenko, V.; Blum, H.; Wolf, E. Genome activation in bovine embryos: Review of the literature and new insights from RNA sequencing experiments. Anim. Reprod. Sci. 2014, 149, 46–58. [Google Scholar] [CrossRef]
- Leese, H.J.; Hugentobler, S.A.; Gray, S.M.; Morris, D.G.; Sturmey, R.G.; Whitear, S.L.; Sreenan, J.M. Female reproductive tract fluids: Composition, mechanism of formation and potential role in the developmental origins of health and disease. Reprod. Fertil. Dev. 2008, 20, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Lamy, J.; Labas, V.; Harichaux, G.; Tsikis, G.; Mermillod, P.; Saint-Dizier, M. Regulation of the bovine oviductal fluid proteome. Reproduction 2016, 152, 629–644. [Google Scholar] [CrossRef]
- Pillai, V.V.; Weber, D.M.; Phinney, B.S.; Selvaraj, V. Profiling of proteins secreted in the bovine oviduct reveals diverse functions of this luminal microenvironment. PLoS ONE 2017, 12, e0188105. [Google Scholar] [CrossRef] [PubMed]
- Papp, S.M.; Frohlich, T.; Radefeld, K.; Havlicek, V.; Kosters, M.; Yu, H.; Mayrhofer, C.; Brem, G.; Arnold, G.J.; Besenfelder, U. A novel approach to study the bovine oviductal fluid proteome using transvaginal endoscopy. Theriogenology 2019, 132, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Alminana, C.; Bauersachs, S. Extracellular vesicles in the oviduct: Progress, challenges and implications for the reproductive success. Bioengineering 2019, 6, 32. [Google Scholar] [CrossRef] [PubMed]
- Alminana, C.; Corbin, E.; Tsikis, G.; Alcantara-Neto, A.S.; Labas, V.; Reynaud, K.; Galio, L.; Uzbekov, R.; Garanina, A.S.; Druart, X.; et al. Oviduct extracellular vesicles protein content and their role during oviduct-embryo cross-talk. Reproduction 2017, 154, 153–168. [Google Scholar] [CrossRef] [PubMed]
- Alminana, C.; Tsikis, G.; Labas, V.; Uzbekov, R.; da Silveira, J.C.; Bauersachs, S.; Mermillod, P. Deciphering the oviductal extracellular vesicles content across the estrous cycle: Implications for the gametes-oviduct interactions and the environment of the potential embryo. BMC Genom. 2018, 19, 622. [Google Scholar] [CrossRef]
- Fair, T.; Lonergan, P.; Dinnyes, A.; Cottell, D.C.; Hyttel, P.; Ward, F.A.; Boland, M.P. Ultrastructure of bovine blastocysts following cryopreservation: Effect of method of blastocyst production. Mol. Reprod. Dev. 2001, 58, 186–195. [Google Scholar] [CrossRef]
- Rizos, D.; Clemente, M.; Bermejo-Alvarez, P.; de La Fuente, J.; Lonergan, P.; Gutierrez-Adan, A. Consequences of in vitro culture conditions on embryo development and quality. Reprod. Domest. Anim. 2008, 43 (Suppl. 4), 44–50. [Google Scholar] [CrossRef]
- Maillo, V.; Lopera-Vasquez, R.; Hamdi, M.; Gutierrez-Adan, A.; Lonergan, P.; Rizos, D. Maternal-embryo interaction in the bovine oviduct: Evidence from in vivo and in vitro studies. Theriogenology 2016, 86, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Lopera-Vasquez, R.; Hamdi, M.; Maillo, V.; Lloreda, V.; Coy, P.; Gutierrez-Adan, A.; Bermejo-Alvarez, P.; Rizos, D. Effect of bovine oviductal fluid on development and quality of bovine embryos produced in vitro. Reprod. Fertil. Dev. 2015. [Google Scholar] [CrossRef] [PubMed]
- Hamdi, M.; Lopera-Vasquez, R.; Maillo, V.; Sanchez-Calabuig, M.J.; Nunez, C.; Gutierrez-Adan, A.; Rizos, D. Bovine oviductal and uterine fluid support in vitro embryo development. Reprod. Fertil. Dev. 2017. [Google Scholar] [CrossRef]
- Lopera-Vasquez, R.; Hamdi, M.; Maillo, V.; Gutierrez-Adan, A.; Bermejo-Alvarez, P.; Ramirez, M.A.; Yanez-Mo, M.; Rizos, D. Effect of bovine oviductal extracellular vesicles on embryo development and quality in vitro. Reproduction 2017, 153, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Pradeep, M.A.; Jagadeesh, J.; De, A.K.; Kaushik, J.K.; Malakar, D.; Kumar, S.; Dang, A.K.; Das, S.K.; Mohanty, A.K. Purification, sequence characterization and effect of goat oviduct-specific glycoprotein on in vitro embryo development. Theriogenology 2011, 75, 1005–1015. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, S.; Kumaresan, A.; Kumar, M.; Chhillar, S.; Malik, H.; Kumar, S.; Kaushik, J.K.; Datta, T.K.; Mohanty, A.K. Effect of recombinant and native buffalo OVGP1 on sperm functions and in vitro embryo development: A comparative study. J. Anim. Sci. Biotechnol. 2017, 8, 69. [Google Scholar] [CrossRef]
- Algarra, B.; Maillo, V.; Aviles, M.; Gutierrez-Adan, A.; Rizos, D.; Jimenez-Movilla, M. Effects of recombinant OVGP1 protein on in vitro bovine embryo development. J. Reprod. Dev. 2018, 64, 433–443. [Google Scholar] [CrossRef]
- Malette, B.; Bleau, G. Biochemical characterization of hamster oviductin as a sulphated zona pellucida-binding glycoprotein. Biochem. J. 1993, 295, 437–445. [Google Scholar] [CrossRef]
- O’Day-Bowman, M.B.; Mavrogianis, P.A.; Minshall, R.D.; Verhage, H.G. In vivo versus in vitro oviductal glycoprotein (OGP) association with the zona pellucida (ZP) in the hamster and baboon. Mol. Reprod. Dev. 2002, 62, 248–256. [Google Scholar] [CrossRef] [PubMed]
- Boice, M.L.; McCarthy, T.J.; Mavrogianis, P.A.; Fazlebas, A.T.; Verhage, H.G. Localization of oviductal glycoproteins within the zona pellucida and perivitelline space of ovulated ova and early embryos in baboons (Papio anubis). Biol. Reprod. 1990, 43, 340–346. [Google Scholar] [CrossRef] [PubMed]
- Buhi, W.C.; O’Brien, B.; Alvarez, I.M.; Erdos, G.; Dubois, D. Immunogold localization of porcine oviductal secretory proteins within the zona pellucida, perivitelline space, and plasma membrane of oviductal and uterine oocytes and early embryos. Biol. Reprod. 1993, 48, 1274–1283. [Google Scholar] [CrossRef] [PubMed]
- Boice, M.L.; Mavrogianis, P.A.; Murphy, C.N.; Prather, R.S.; Day, B.N. Immunocytochemical analysis of the association of bovine oviduct-specific glycoproteins with early embryos. J. Exp. Zool. 1992, 263, 225–229. [Google Scholar] [CrossRef] [PubMed]
- Wegner, C.C.; Killian, G.J. In vitro and in vivo association of an oviduct estrus-associated protein with bovine zona pellucida. Mol. Reprod. Dev. 1991, 29, 77–84. [Google Scholar] [CrossRef]
- Kan, F.W.; Roux, E.; Bleau, G. Immunolocalization of oviductin in endocytic compartments in the blastomeres of developing embryos in the golden hamster. Biol. Reprod. 1993, 48, 77–88. [Google Scholar] [CrossRef]
- Buhi, W.C. Characterization and biological roles of oviduct-specific, oestrogen-dependent glycoprotein. Reproduction 2002, 123, 355–362. [Google Scholar] [CrossRef]
- Staros, A.L.; Killian, G.J. In vitro association of six oviductal fluid proteins with the bovine zona pellucida. J. Reprod. Fertil. 1998, 112, 131–137. [Google Scholar] [CrossRef][Green Version]
- Goncalves, R.F.; Staros, A.L.; Killian, G.J. Oviductal fluid proteins associated with the bovine zona pellucida and the effect on in vitro sperm-egg binding, fertilization and embryo development. Reprod. Domest. Anim. 2008, 43, 720–729. [Google Scholar] [CrossRef]
- Ferraz, M.; Carothers, A.; Dahal, R.; Noonan, M.J.; Songsasen, N. Oviductal extracellular vesicles interact with the spermatozoon’s head and mid-piece and improves its motility and fertilizing ability in the domestic cat. Sci. Rep. 2019, 9, 9484. [Google Scholar] [CrossRef]
- Lamy, J.; Nogues, P.; Combes-Soia, L.; Tsikis, G.; Labas, V.; Mermillod, P.; Druart, X.; Saint-Dizier, M. Identification by proteomics of oviductal sperm-interacting proteins. Reproduction 2018. [Google Scholar] [CrossRef] [PubMed]
- Tse, P.K.; Lee, Y.L.; Chow, W.N.; Luk, J.M.; Lee, K.F.; Yeung, W.S. Preimplantation embryos cooperate with oviductal cells to produce embryotrophic inactivated complement-3b. Endocrinology 2008, 149, 1268–1276. [Google Scholar] [CrossRef] [PubMed]
- Maillo, V.; Gaora, P.O.; Forde, N.; Besenfelder, U.; Havlicek, V.; Burns, G.W.; Spencer, T.E.; Gutierrez-Adan, A.; Lonergan, P.; Rizos, D. Oviduct-embryo interactions in cattle: Two-way traffic or a one-way street? Biol. Reprod. 2015, 92, 144. [Google Scholar] [CrossRef] [PubMed]
- Ghersevich, S.; Massa, E.; Zumoffen, C. Oviductal secretion and gamete interaction. Reproduction 2015, 149, R1–R14. [Google Scholar] [CrossRef]
- Vanroose, G.; Nauwynck, H.; Soom, A.V.; Ysebaert, M.T.; Charlier, G.; Oostveldt, P.V.; de Kruif, A. Structural aspects of the zona pellucida of in vitro-produced bovine embryos: A scanning electron and confocal laser scanning microscopic study. Biol. Reprod. 2000, 62, 463–469. [Google Scholar] [CrossRef]
- Turner, K.; Horobin, R.W. Permeability of the mouse zona pellucida: A structure-staining-correlation model using coloured probes. J. Reprod. Fertil. 1997, 111, 259–265. [Google Scholar] [CrossRef]
- Legge, M. Oocyte and zygote zona pellucida permeability to macromolecules. J. Exp. Zool. 1995, 271, 145–150. [Google Scholar] [CrossRef]
- Graf, A.; Krebs, S.; Zakhartchenko, V.; Schwalb, B.; Blum, H.; Wolf, E. Fine mapping of genome activation in bovine embryos by RNA sequencing. Proc. Natl. Acad. Sci. USA 2014, 111, 4139–4144. [Google Scholar] [CrossRef]
- Saadeldin, I.M.; Kim, S.J.; Choi, Y.B.; Lee, B.C. Improvement of cloned embryos development by co-culturing with parthenotes: A possible role of exosomes/microvesicles for embryos paracrine communication. Cell. Reprogram. 2014, 16, 223–234. [Google Scholar] [CrossRef]
- Nancarrow, C.D.; Hill, J.L. Oviduct proteins in fertilization and early embryo development. J. Reprod. Fertil. Suppl. 1995, 49, 3–13. [Google Scholar] [CrossRef]
- Kouba, A.J.; Abeydeera, L.R.; Alvarez, I.M.; Day, B.N.; Buhi, W.C. Effects of the porcine oviduct-specific glycoprotein on fertilization, polyspermy, and embryonic development in vitro. Biol. Reprod. 2000, 63, 242–250. [Google Scholar] [CrossRef] [PubMed]
- Yong, P.; Gu, Z.; Luo, J.P.; Wang, J.R.; Tso, J.K. Antibodies against the C-terminal peptide of rabbit oviductin inhibit mouse early embryo development to pass 2-cell stage. Cell Res. 2002, 12, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Sheikh, M.H.; Solito, E. Annexin A1: Uncovering the many talents of an old protein. Int. J. Mol. Sci. 2018, 19, 1045. [Google Scholar] [CrossRef] [PubMed]
- Smits, K.; Nelis, H.; Van Steendam, K.; Govaere, J.; Roels, K.; Ververs, C.; Leemans, B.; Wydooghe, E.; Deforce, D.; Van Soom, A. Proteome of equine oviducal fluid: Effects of ovulation and pregnancy. Reprod. Fertil. Dev. 2016. [Google Scholar] [CrossRef]
- Smits, K.; Willems, S.; Van Steendam, K.; Van De Velde, M.; De Lange, V.; Ververs, C.; Roels, K.; Govaere, J.; Van Nieuwerburgh, F.; Peelman, L.; et al. Proteins involved in embryo-maternal interaction around the signalling of maternal recognition of pregnancy in the horse. Sci. Rep. 2018, 8, 5249. [Google Scholar] [CrossRef]
- Hebeda, C.B.; Machado, I.D.; Reif-Silva, I.; Moreli, J.B.; Oliani, S.M.; Nadkarni, S.; Perretti, M.; Bevilacqua, E.; Farsky, S.H.P. Endogenous annexin A1 (AnxA1) modulates early-phase gestation and offspring sex-ratio skewing. J. Cell. Physiol. 2018, 233, 6591–6603. [Google Scholar] [CrossRef]
- Garrido-Gomez, T.; Dominguez, F.; Quinonero, A.; Estella, C.; Vilella, F.; Pellicer, A.; Simon, C. Annexin A2 is critical for embryo adhesiveness to the human endometrium by RhoA activation through F-actin regulation. FASEB J. 2012, 26, 3715–3727. [Google Scholar] [CrossRef]
- Wang, B.; Ye, T.M.; Lee, K.F.; Chiu, P.C.; Pang, R.T.; Ng, E.H.; Yeung, W.S. Annexin A2 acts as an adhesion molecule on the endometrial epithelium during implantation in mice. PLoS ONE 2015, 10, e0139506. [Google Scholar] [CrossRef]
- Leffler, H.; Carlsson, S.; Hedlund, M.; Qian, Y.; Poirier, F. Introduction to galectins. Glycoconj. J. 2002, 19, 433–440. [Google Scholar] [CrossRef]
- Kim, M.; Kim, S.; Kim, H.; Joo, H.G.; Shin, T. Immunohistochemical localization of galectin-3 in the reproductive organs of the cow. Acta Histochem. 2008, 110, 473–480. [Google Scholar] [CrossRef]
- Yang, H.; Lei, C.; Zhang, W. Expression of galectin-3 in mouse endometrium and its effect during embryo implantation. Reprod. Biomed. Online 2012, 24, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Holm, P.; Booth, P.J.; Schmidt, M.H.; Greve, T.; Callesen, H. High bovine blastocyst development in a static in vitro production system using SOFaa medium supplemented with sodium citrate and myo-inositol with or without serum-proteins. Theriogenology 1999, 52, 683–700. [Google Scholar] [CrossRef]
- Keller, A.; Nesvizhskii, A.I.; Kolker, E.; Aebersold, R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002, 74, 5383–5392. [Google Scholar] [CrossRef] [PubMed]
- Nesvizhskii, A.I.; Keller, A.; Kolker, E.; Aebersold, R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003, 75, 4646–4658. [Google Scholar] [CrossRef]

| Gene Name | Protein Name | Accession Number (UniprotKB) | Molecular Weight (kDa) | OF-Treated: Control Ratio |
|---|---|---|---|---|
| LZTFL1 | Leucine zipper transcription factor-like protein 1 | Q3ZBL4 | 35 | T 1 |
| NQO1 | NAD(P)H quinone dehydrogenase 1 | Q3ZBH2 | 31 | T 1 |
| ALDH1A2 | Aldehyde dehydrogenase 1 family member A2 | G3X6U1 | 57 | T 1 |
| HBA | Hemoglobin subunit alpha | P01966 | 15 | T 1 |
| ITGA2 | Integrin alpha-2 (Fragment) | P53710 | 129 | 22 |
| WARS | Tryptophan--tRNA ligase. cytoplasmic | P17248 | 54 | 8.7 |
| RPS16 | 40S ribosomal protein S16 | Q3T0X6 | 16 | 8.3 |
| TARS | Threonine--tRNA ligase. cytoplasmic | Q3ZBV8 | 83 | 8.1 |
| PFKL | ATP-dependent 6-phosphofructokinase. liver type | A1A4J1 | 85 | 4.9 |
| RPL6 | 60S ribosomal protein L6 | Q58DQ3 | 33 | 4.5 |
| ALDH1A1 | Retinal dehydrogenase 1 | P48644 | 55 | 2.9 |
| PPP2R1A | Alpha isoform of regulatory subunit A. protein phosphatase 2 | Q32PI5 | 65 | 2.3 |
| ANXA5 | Annexin A5 | P81287 | 36 | 2.3 |
| Gene Name | Protein Name | Accession Number (UniprotKB) | Molecular Weight (kDa) | OF-Treated: Control Ratio |
|---|---|---|---|---|
| ACLY | ATP-citrate synthase | Q32PF2 | 120 | T 1 |
| ACO1 | Cytoplasmic aconitate hydratase | Q0VCU1 | 98 | T 1 |
| PNP | Purine nucleoside phosphorylase | P55859 | 32 | T 1 |
| BLVRA | Biliverdin reductase A | A5D7K0 | 34 | 9.9 |
| A2M | Alpha-2-macroglobulin | Q7SIH1 | 168 | 7 |
| HBB | Hemoglobin subunit beta | P02070 | 16 | 6.7 |
| APEX1 | DNA-(apurinic or apyrimidinic site) lyase | P23196 | 36 | 5.8 |
| TAGLN2 | Transgelin-2 | Q5E9F5 | 22 | 5.8 |
| LGALS3 | Galectin-3 | A6QLZ0 | 28 | 5.3 |
| LGALS9 | Galectin-9 | Q3MHZ8 | 39 | 4.3 |
| TBCB | Tubulin-folding cofactor B | Q5E951 | 28 | 4.3 |
| YWHAQ | 14-3-3 protein theta | Q3SZI4 | 28 | 3 |
| GLMN | Glomulin. FKBP associated protein | E1BA27 | 68 | 2.9 |
| YWHAH | 14-3-3 protein eta | P68509 | 28 | 2.9 |
| MAP4 | Microtubule-associated protein | P36225 | 112 | 2.6 |
| TPP2 | Tripeptidyl-peptidase 2 | A5PK39 | 138 | 2.3 |
| MYL6 | Myosin light polypeptide 6 | P60661 | 17 | 2.2 |
| FKBP4 | Peptidyl-prolyl cis-trans isomerase | Q9TRY0 | 51 | 2 |
| HUWE1 | HECT. UBA and WWE domain containing 1. E3 ubiquitin protein ligase | E1BNY9 | 482 | 3.4 |
| Gene Name | Protein Name | Accession Number (UniprotKB) | Molecular Weight (kDa) | 4–6 Cell OF-Treated: Control Ratio | Morula OF-Treated: Control Ratio |
|---|---|---|---|---|---|
| ANXA8 | Annexin A8 | Q95L54 | 37 | T 1 | T 1 |
| AGR2 | Anterior gradient 2. protein disulphide isomerase family member | F1N3J3 | 20 | T 1 | T 1 |
| CAPS | Calcyphosin | Q0VCC0 | 21 | T 1 | 61 |
| CD109 | CD109 molecule | F1MPE1 | 161 | T 1 | T 1 |
| CKB | Creatine kinase B-type | Q5EA61 | 43 | T 1 | 4.1 |
| CNDP2 | Cytosolic non-specific dipeptidase | Q3ZC84 | 53 | T 1 | 17 |
| EPHX2 | Epoxide hydrolase 2 | F6QS88 | 63 | T 1 | T 1 |
| PYGL | Glycogen phosphorylase. liver form | Q0VCM4 | 97 | T 1 | T 1 |
| LRBA | LPS responsive beige-like anchor protein | E1BND6 | 316 | T 1 | 8.2 |
| DCXR | l-xylulose reductase | Q1JP75 | 26 | T 1 | T 1 |
| OVGP1 | Oviduct-specific glycoprotein | Q28042 | 60 | T 1 | 71 |
| PTGR2 | Prostaglandin reductase 2 | Q32L99 | 38 | T 1 | T 1 |
| SEPTIN2 | Septin-2 | Q2NKY7 | 41 | T 1 | 4.2 |
| SARS | Serine--tRNA ligase. cytoplasmic | Q9GMB8 | 59 | T 1 | 5 |
| TPPP3 | Tubulin polymerization-promoting protein family member 3 | Q3ZCC8 | 19 | T 1 | T 1 |
| ANXA1 | Annexin A1 | P46193 | 39 | 37 | 15 |
| RNPEP | Uncharacterized protein | G3X743 | 72 | 9.6 | 4.6 |
| FASN | Fatty acid synthase | Q71SP7 | 274 | 8.5 | 9.6 |
| RNH1 | Ribonuclease/angiogenin inhibitor 1 | Q3SZN8 | 49 | 6.9 | 7.5 |
| ANXA4 | Annexin A4 | P13214 | 36 | 5.4 | 6.5 |
| SHTN1 | Shootin 1 | F1MUA7 | 63 | 5.3 | 4 |
| ALDH9A1 | 4-trimethylaminobutyraldehyde dehydrogenase | Q2KJH9 | 54 | 4.2 | 2.9 |
| ANXA2 | Annexin A2 | P04272 | 39 | 3.6 | 2.2 |
| CLIC1 | Chloride intracellular channel protein 1 | Q5E9B7 | 27 | 2.1 | 3 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Banliat, C.; Tsikis, G.; Labas, V.; Teixeira-Gomes, A.-P.; Com, E.; Lavigne, R.; Pineau, C.; Guyonnet, B.; Mermillod, P.; Saint-Dizier, M. Identification of 56 Proteins Involved in Embryo–Maternal Interactions in the Bovine Oviduct. Int. J. Mol. Sci. 2020, 21, 466. https://doi.org/10.3390/ijms21020466
Banliat C, Tsikis G, Labas V, Teixeira-Gomes A-P, Com E, Lavigne R, Pineau C, Guyonnet B, Mermillod P, Saint-Dizier M. Identification of 56 Proteins Involved in Embryo–Maternal Interactions in the Bovine Oviduct. International Journal of Molecular Sciences. 2020; 21(2):466. https://doi.org/10.3390/ijms21020466
Chicago/Turabian StyleBanliat, Charles, Guillaume Tsikis, Valérie Labas, Ana-Paula Teixeira-Gomes, Emmanuelle Com, Régis Lavigne, Charles Pineau, Benoit Guyonnet, Pascal Mermillod, and Marie Saint-Dizier. 2020. "Identification of 56 Proteins Involved in Embryo–Maternal Interactions in the Bovine Oviduct" International Journal of Molecular Sciences 21, no. 2: 466. https://doi.org/10.3390/ijms21020466
APA StyleBanliat, C., Tsikis, G., Labas, V., Teixeira-Gomes, A.-P., Com, E., Lavigne, R., Pineau, C., Guyonnet, B., Mermillod, P., & Saint-Dizier, M. (2020). Identification of 56 Proteins Involved in Embryo–Maternal Interactions in the Bovine Oviduct. International Journal of Molecular Sciences, 21(2), 466. https://doi.org/10.3390/ijms21020466





