Beneficial Effects of SREBP Decoy Oligodeoxynucleotide in an Animal Model of Hyperlipidemia
Abstract
:1. Introduction
2. Results
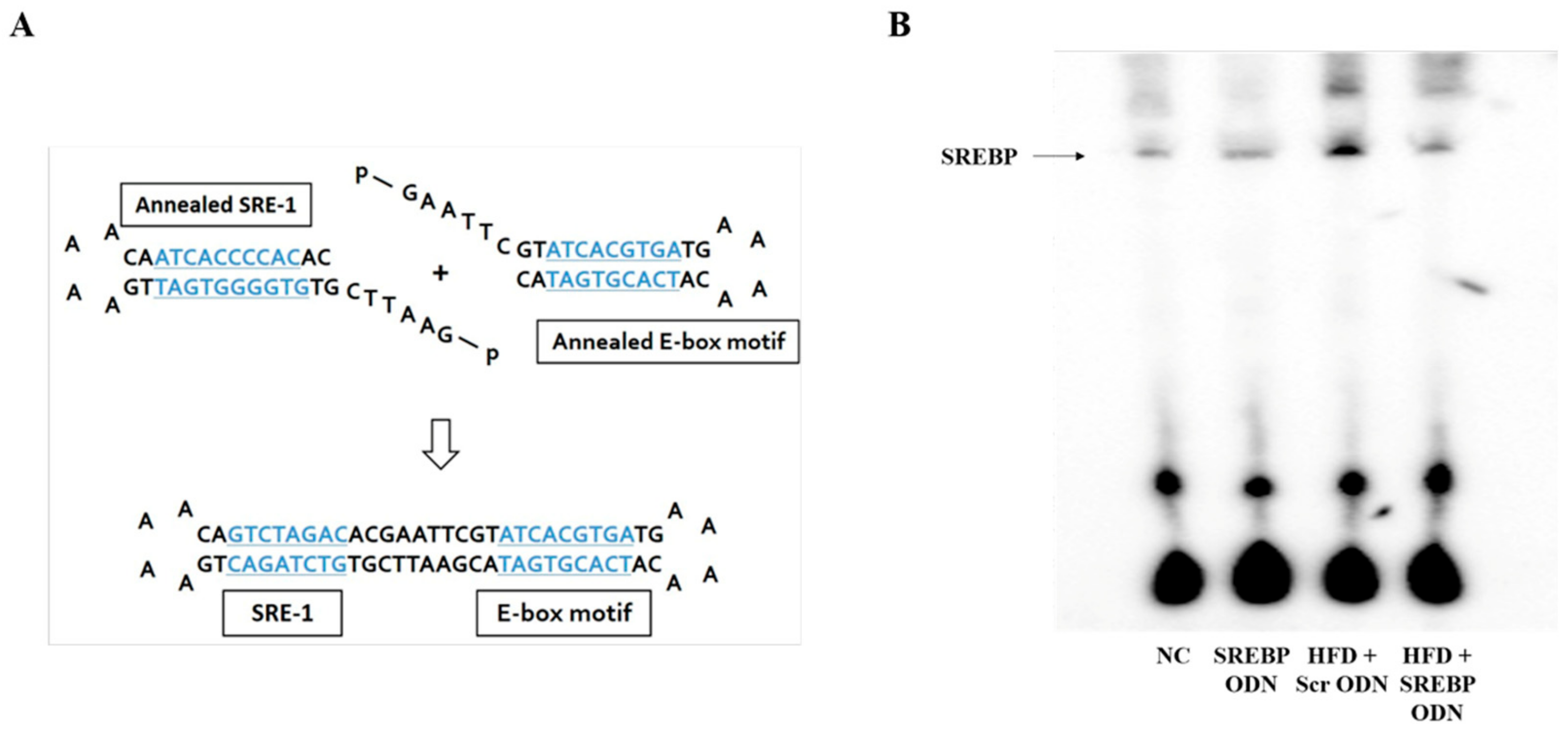
2.1. Construction of Decoy ODNs
2.2. SREBP Decoy ODN Attenuated Morphological Change on Hyperlipidemic Mice Liver
2.3. Effects of SREBP Decoy ODN on the Inflammation-Related Factors in Hyperlipidemic Mice Livers
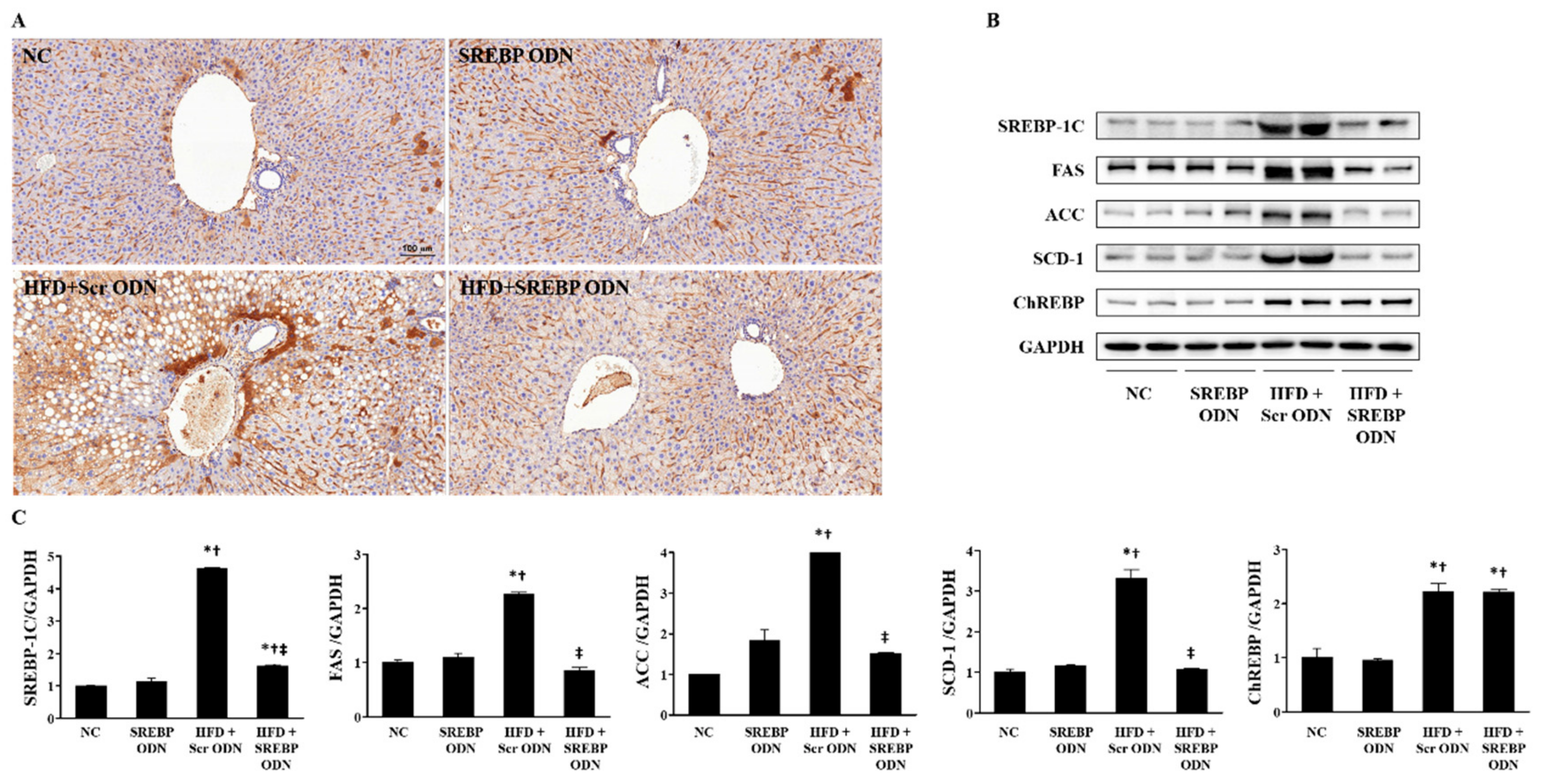
2.4. Effects of SREBP Decoy ODN on the Expression of Cholesterol Metabolism-Related Factors and Lipid Metabolism-Related Factors in the Liver
2.5. Effects of SREBP Decoy ODN in Hyperlipidemic Mice Aorta
3. Discussion
4. Materials and Methods
4.1. Construction of Decoy ODNs
4.2. Animal Model
4.3. Histological Analysis
4.4. Immunohistochemical Staining
4.5. Immunofluorescence Staining and Confocal Microscopy
4.6. Serum Biochemical Analysis
4.7. Enzyme-Linked Immunosorbent Assay (ELISA)
4.8. Real-Time PCR
4.9. Western Blot Analysis
4.10. Electrophoretic Mobility Shift Assay (EMSA)
4.11. Statistical Analysis
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| NAFLD | Non-alcoholic fatty liver disease |
| SREBP | Sterol regulatory element binding protein |
| ODN | Oligodeoxynucleotide |
| HFD | High-fat diet |
| SRE-1 | Sterol regulated element-1 |
| ALT | Alanine aminotransferase |
| AST | Aspartate aminotransferase |
| FAS | Fatty acid synthase |
| ACC | Acetyl-CoA carboxylase |
| SCD | Stearoyl-CoA desaturase |
| HMGCR | HMG-CoA reductase |
| ChREBP | Carbohydrate response element binding protein |
References
- Moon, Y.A. The SCAP/SREBP Pathway: A Mediator of Hepatic Steatosis. Endocrinol. Metab. (Seoul) 2017, 32, 6–10. [Google Scholar] [CrossRef] [PubMed]
- Tevar, A.D.; Clarke, C.; Wang, J.; Rudich, S.M.; Woodle, E.S.; Lentsch, A.B.; Edwards, M.L. Clinical review of nonalcoholic steatohepatitis in liver surgery and transplantation. J. Am. Coll. Surg. 2010, 210, 515–526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaggini, M.; Morelli, M.; Buzzigoli, E.; DeFronzo, R.A.; Bugianesi, E.; Gastaldelli, A. Non-alcoholic fatty liver disease (NAFLD) and its connection with insulin resistance, dyslipidemia, atherosclerosis and coronary heart disease. Nutrients 2013, 5, 1544–1560. [Google Scholar] [CrossRef] [PubMed]
- Zhong, H.; Chen, K.; Feng, M.; Shao, W.; Wu, J.; Liang, T.; Liu, C. Genipin alleviates high-fat diet-induced hyperlipidemia and hepatic lipid accumulation in mice via miR-142a-5p/SREBP-1c axis. FEBS J. 2018, 285, 501–517. [Google Scholar] [CrossRef] [PubMed]
- Plutzky, J. Emerging concepts in metabolic abnormalities associated with coronary artery disease. Curr. Opin. Cardiol. 2000, 15, 416–421. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Mudgal, J.; Parihar, V.K.; Nayak, P.G.; Kutty, N.G.; Rao, C.M. Sesamol treatment reduces plasma cholesterol and triacylglycerol levels in mouse models of acute and chronic hyperlipidemia. Lipids 2013, 48, 633–638. [Google Scholar] [CrossRef]
- Ullrich, I.H. Evaluation of a high-fiber diet in hyperlipidemia: A review. J. Am. Coll. Nutr. 1987, 6, 19–25. [Google Scholar] [CrossRef]
- Hulsmann, W.C.; Kort, W.J. Saturated fat feeding, hyperlipidemia and hyperinsulinemia. Biochim. Biophys. Acta 1983, 754, 231–237. [Google Scholar] [CrossRef]
- Henk, H.J.; Paoli, C.J.; Gandra, S.R. A Retrospective Study to Examine Healthcare Costs Related to Cardiovascular Events in Individuals with Hyperlipidemia. Adv. Ther. 2015, 32, 1104–1116. [Google Scholar] [CrossRef] [Green Version]
- Ertunc, M.E.; Hotamisligil, G.S. Lipid signaling and lipotoxicity in metaflammation: Indications for metabolic disease pathogenesis and treatment. J. Lipid Res. 2016, 57, 2099–2114. [Google Scholar] [CrossRef] [Green Version]
- Eberle, D.; Hegarty, B.; Bossard, P.; Ferre, P.; Foufelle, F. SREBP transcription factors: Master regulators of lipid homeostasis. Biochimie 2004, 86, 839–848. [Google Scholar] [CrossRef] [PubMed]
- Edwards, P.A.; Tabor, D.; Kast, H.R.; Venkateswaran, A. Regulation of gene expression by SREBP and SCAP. Biochim. Biophys. Acta 2000, 1529, 103–113. [Google Scholar] [CrossRef]
- Horton, J.D.; Goldstein, J.L.; Brown, M.S. SREBPs: Activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Investig. 2002, 109, 1125–1131. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; DeBose-Boyd, R.A. Regulation of cholesterol and fatty acid synthesis. Cold Spring Harb. Perspect. Biol. 2011, 3, 004754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pai, J.T.; Guryev, O.; Brown, M.S.; Goldstein, J.L. Differential stimulation of cholesterol and unsaturated fatty acid biosynthesis in cells expressing individual nuclear sterol regulatory element-binding proteins. J. Biol. Chem. 1998, 273, 26138–26148. [Google Scholar] [CrossRef] [Green Version]
- Horton, J.D.; Shah, N.A.; Warrington, J.A.; Anderson, N.N.; Park, S.W.; Brown, M.S.; Goldstein, J.L. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc. Natl. Acad. Sci. USA 2003, 100, 12027–12032. [Google Scholar] [CrossRef] [Green Version]
- McPherson, R.; Gauthier, A. Molecular regulation of SREBP function: The Insig-SCAP connection and isoform-specific modulation of lipid synthesis. Biochem. cell Biol. Biochim. Biol. Cell. 2004, 82, 201–211. [Google Scholar] [CrossRef]
- Ferre, P.; Foufelle, F. SREBP-1c transcription factor and lipid homeostasis: Clinical perspective. Horm. Res. 2007, 68, 72–82. [Google Scholar] [CrossRef]
- Lin, J.; Yang, R.; Tarr, P.T.; Wu, P.H.; Handschin, C.; Li, S.; Yang, W.; Pei, L.; Uldry, M.; Tontonoz, P.; et al. Hyperlipidemic effects of dietary saturated fats mediated through PGC-1beta coactivation of SREBP. Cell 2005, 120, 261–273. [Google Scholar] [CrossRef] [Green Version]
- Boullart, A.C.; de Graaf, J.; Stalenhoef, A.F. Serum triglycerides and risk of cardiovascular disease. Biochim. Biophys. Acta 2012, 1821, 867–875. [Google Scholar] [CrossRef]
- Nguyen, N.T.; Varela, J.E. Bariatric surgery for obesity and metabolic disorders: State of the art. Nature Rev. Gastroenterol. Hepatol. 2017, 14, 160–169. [Google Scholar] [CrossRef]
- Golomb, B.A.; Evans, M.A. Statin adverse effects: A review of the literature and evidence for a mitochondrial mechanism. Am. J. Cardiovasc. Drugs Drugs Devices Other Interv. 2008, 8, 373–418. [Google Scholar] [CrossRef]
- Sung, W.J.; Kim, K.H.; Kim, Y.J.; Chang, Y.C.; Lee, I.H.; Park, K.K. Antifibrotic effect of synthetic Smad/Sp1 chimeric decoy oligodeoxynucleotide through the regulation of epithelial mesenchymal transition in unilateral ureteral obstruction model of mice. Exp. Mol. Pathol. 2013, 95, 136–143. [Google Scholar] [CrossRef]
- Morishita, R.; Higaki, J.; Tomita, N.; Ogihara, T. Application of transcription factor "decoy" strategy as means of gene therapy and study of gene expression in cardiovascular disease. Circ. Res. 1998, 82, 1023–1028. [Google Scholar] [CrossRef] [Green Version]
- Tomita, N.; Kashihara, N.; Morishita, R. Transcription factor decoy oligonucleotide-based therapeutic strategy for renal disease. Clin. Exp. Nephrol. 2007, 11, 7–17. [Google Scholar] [CrossRef]
- Kim, K.H.; Lee, W.R.; Kang, Y.N.; Chang, Y.C.; Park, K.K. Inhibitory effect of nuclear factor-kappaB decoy oligodeoxynucleotide on liver fibrosis through regulation of the epithelial-mesenchymal transition. Human Gene Ther. 2014, 25, 721–729. [Google Scholar] [CrossRef] [Green Version]
- Gwon, M.G.; Kim, J.Y.; An, H.J.; Kim, W.H.; Gu, H.; Kim, M.K.; Park, S.C.; Park, K.K. Antifibrotic Effect of Smad Decoy Oligodeoxynucleotide in a CCl(4)-Induced Hepatic Fibrosis Animal Model. Molecules 2018, 23, 1991. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Song, Y.; Feng, M.; Zhou, X.; Lu, Y.; Gao, L.; Yu, C.; Jiang, X.; Zhao, J. Thyroid-stimulating hormone decreases HMG-CoA reductase phosphorylation via AMP-activated protein kinase in the liver. J. Lipid Res. 2015, 56, 963–971. [Google Scholar] [CrossRef] [Green Version]
- Denechaud, P.D.; Bossard, P.; Lobaccaro, J.M.; Millatt, L.; Staels, B.; Girard, J.; Postic, C. ChREBP, but not LXRs, is required for the induction of glucose-regulated genes in mouse liver. J. Clin. Investig. 2008, 118, 956–964. [Google Scholar] [CrossRef]
- Postic, C.; Dentin, R.; Girard, J. Role of the liver in the control of carbohydrate and lipid homeostasis. Diabetes Metab. 2004, 30, 398–408. [Google Scholar] [CrossRef]
- Dentin, R.; Girard, J.; Postic, C. Carbohydrate responsive element binding protein (ChREBP) and sterol regulatory element binding protein-1c (SREBP-1c): Two key regulators of glucose metabolism and lipid synthesis in liver. Biochimie 2005, 87, 81–86. [Google Scholar] [CrossRef]
- Ishibashi, H.; Nakagawa, K.; Onimaru, M.; Castellanous, E.J.; Kaneda, Y.; Nakashima, Y.; Shirasuna, K.; Sueishi, K. Sp1 decoy transfected to carcinoma cells suppresses the expression of vascular endothelial growth factor, transforming growth factor beta1, and tissue factor and also cell growth and invasion activities. Cancer Res. 2000, 60, 6531–6536. [Google Scholar]
- Magae, J.; Munemura, K.; Ichikawa, C.; Osada, K.; Hanada, T.; Tsuji, R.F.; Yamashita, M.; Hino, A.; Horiuchi, T.; Uramoto, M.; et al. Effects of microbial products on glucose consumption and morphology of macrophages. Biosci. Biotechnol. Biochem. 1993, 57, 1628–1631. [Google Scholar] [CrossRef]
- Yamashita, J.; Yoshimasa, T.; Arai, H.; Hiraoka, J.; Takaya, K.; Miyamoto, Y.; Ogawa, Y.; Itoh, H.; Nakao, K. Identification of cis-elements of the human endothelin-A receptor gene and inhibition of the gene expression by the decoy strategy. J. Biol. Chem. 1998, 273, 15993–15999. [Google Scholar] [CrossRef] [Green Version]
- Xiao, X.; Song, B.L. SREBP: A novel therapeutic target. Acta Biochim. Biophys. Sin. 2013, 45, 2–10. [Google Scholar] [CrossRef] [Green Version]
- Shimomura, I.; Shimano, H.; Horton, J.D.; Goldstein, J.L.; Brown, M.S. Differential expression of exons 1a and 1c in mRNAs for sterol regulatory element binding protein-1 in human and mouse organs and cultured cells. J. Clin. Investig. 1997, 99, 838–845. [Google Scholar] [CrossRef] [Green Version]
- Shimano, H. Sterol regulatory element-binding proteins (SREBPs): Transcriptional regulators of lipid synthetic genes. Prog. Lipid Res. 2001, 40, 439–452. [Google Scholar] [CrossRef]
- Yang, J.; Goldstein, J.L.; Hammer, R.E.; Moon, Y.A.; Brown, M.S.; Horton, J.D. Decreased lipid synthesis in livers of mice with disrupted Site-1 protease gene. Proc. Natl. Acad. Sci. USA 2001, 98, 13607–13612. [Google Scholar] [CrossRef] [Green Version]
- Sato, R. Sterol metabolism and SREBP activation. Arch. Biochem. Biophys. 2010, 501, 177–181. [Google Scholar] [CrossRef]
- Ipsen, D.H.; Lykkesfeldt, J.; Tveden-Nyborg, P. Molecular mechanisms of hepatic lipid accumulation in non-alcoholic fatty liver disease. Cell. Mol. Life Sci. CMLS 2018, 75, 3313–3327. [Google Scholar] [CrossRef] [Green Version]
- Kuhajda, F.P. Fatty-acid synthase and human cancer: New perspectives on its role in tumor biology. Nutrition 2000, 16, 202–208. [Google Scholar] [CrossRef]
- Moon, Y.S.; Latasa, M.J.; Griffin, M.J.; Sul, H.S. Suppression of fatty acid synthase promoter by polyunsaturated fatty acids. J. Lipid Res. 2002, 43, 691–698. [Google Scholar]
- Semenkovich, C.F. Regulation of fatty acid synthase (FAS). Prog. Lipid Res. 1997, 36, 43–53. [Google Scholar] [CrossRef]
- Lounis, M.A.; Bergeron, K.F.; Burhans, M.S.; Ntambi, J.M.; Mounier, C. Oleate activates SREBP-1 signaling activity in SCD1-deficient hepatocytes. Am. J. Physiol. Endocrinol. Metab. 2017, 313, E710–E720. [Google Scholar] [CrossRef]
- Miyazaki, M.; Dobrzyn, A.; Man, W.C.; Chu, K.; Sampath, H.; Kim, H.J.; Ntambi, J.M. Stearoyl-CoA desaturase 1 gene expression is necessary for fructose-mediated induction of lipogenic gene expression by sterol regulatory element-binding protein-1c-dependent and -independent mechanisms. J. Biol. Chem. 2004, 279, 25164–25171. [Google Scholar] [CrossRef] [Green Version]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
An, H.-J.; Kim, J.-Y.; Gwon, M.-G.; Gu, H.; Kim, H.-J.; Leem, J.; Youn, S.W.; Park, K.-K. Beneficial Effects of SREBP Decoy Oligodeoxynucleotide in an Animal Model of Hyperlipidemia. Int. J. Mol. Sci. 2020, 21, 552. https://doi.org/10.3390/ijms21020552
An H-J, Kim J-Y, Gwon M-G, Gu H, Kim H-J, Leem J, Youn SW, Park K-K. Beneficial Effects of SREBP Decoy Oligodeoxynucleotide in an Animal Model of Hyperlipidemia. International Journal of Molecular Sciences. 2020; 21(2):552. https://doi.org/10.3390/ijms21020552
Chicago/Turabian StyleAn, Hyun-Jin, Jung-Yeon Kim, Mi-Gyeong Gwon, Hyemin Gu, Hyun-Ju Kim, Jaechan Leem, Sung Won Youn, and Kwan-Kyu Park. 2020. "Beneficial Effects of SREBP Decoy Oligodeoxynucleotide in an Animal Model of Hyperlipidemia" International Journal of Molecular Sciences 21, no. 2: 552. https://doi.org/10.3390/ijms21020552
APA StyleAn, H.-J., Kim, J.-Y., Gwon, M.-G., Gu, H., Kim, H.-J., Leem, J., Youn, S. W., & Park, K.-K. (2020). Beneficial Effects of SREBP Decoy Oligodeoxynucleotide in an Animal Model of Hyperlipidemia. International Journal of Molecular Sciences, 21(2), 552. https://doi.org/10.3390/ijms21020552




