Placenta-Specific Genes, Their Regulation During Villous Trophoblast Differentiation and Dysregulation in Preterm Preeclampsia
Abstract
:1. Introduction
2. Results
2.1. Identification of Predominantly Placenta-Expressed Genes
2.2. Predominantly Placenta-Expressed Genes are Key to Villous Trophoblastic Functions
2.3. Predominantly Placenta-Expressed Genes Are Primarily Expressed in Villous Trophoblasts
2.4. Most Predominantly Placenta-Expressed Genes are Differentially Expressed During Villous Trophoblast Differentiation
2.5. Microarray Analysis Reveals Global Transcriptomic and Functional Changes During Villous Trophoblast Differentiation
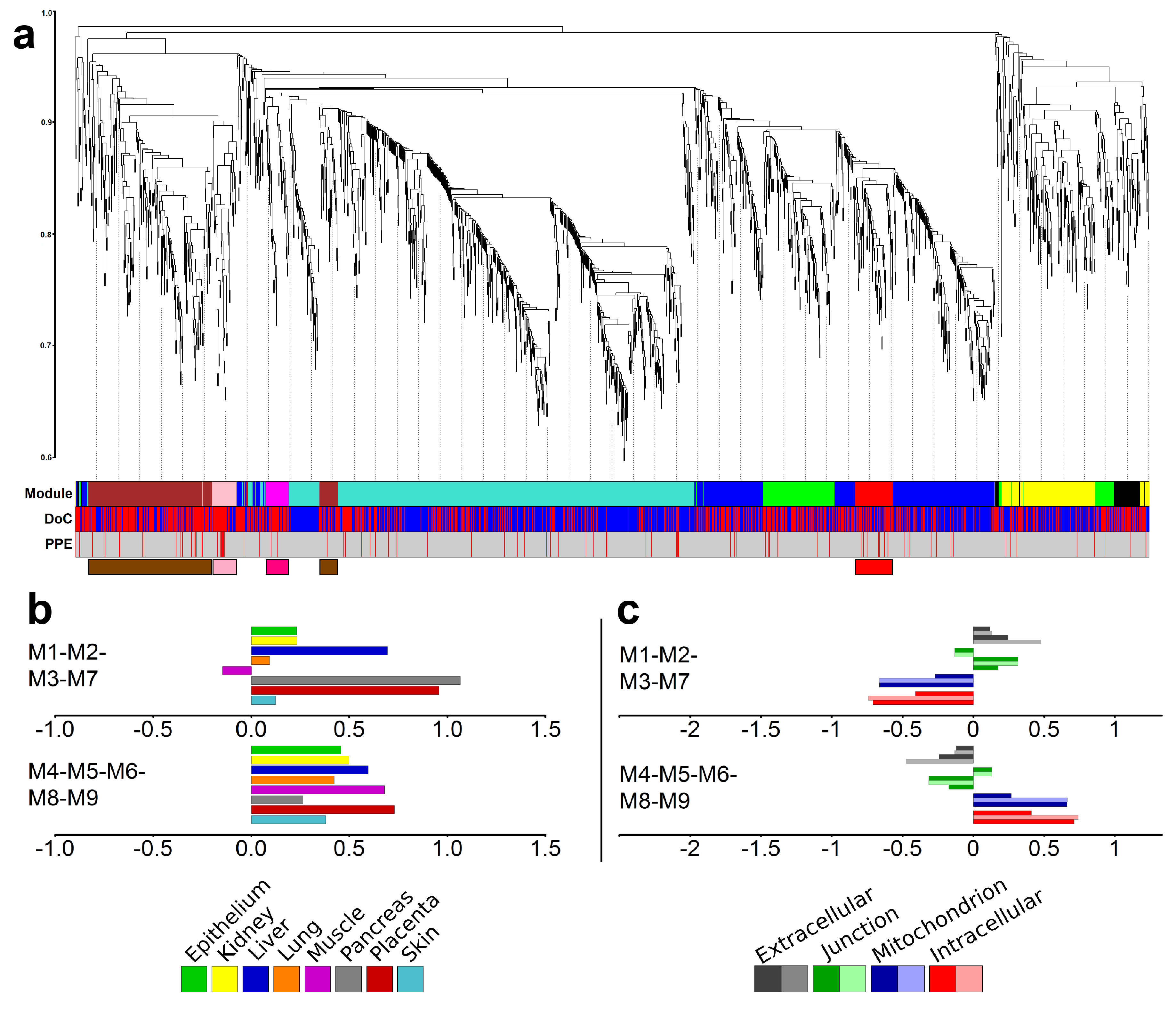
2.6. Identification of Gene Modules and Module Groups From the Co-Expression Network During Villous Trophoblast Differentiation
2.7. Major Differences in Pathway Perturbation of Placental and Non-Placental Modules
2.8. Major Differences in the Time Course of Gene Expression in Placental and Non-Placental Modules
2.9. Identification of Genes with High Expression Change
2.10. High Expression Change Gene Clusters Point to Temporal Changes in Structural and Functional Rearrangements of Differentiating Villous Trophoblasts
2.11. Identification of Upstream Regulators of High Expression Change Gene Clusters
2.12. Identification of the Transcription Regulatory Network During Villous Trophoblast Differentiation
2.13. Different Transcription Factors Regulate Placental and Non-Placental Modules
2.14. Identification of Transcription Regulatory Genes in the Trophoblast Differentiation Co-Expression Network that are Key to Preterm Preeclampsia Development
3. Discussion
4. Materials and Methods
4.1. Experimental Procedures
4.1.1. Collection and Culturing of Cytotrophoblasts
4.1.2. RNA Microarray Analysis
4.1.3. qRT-PCR Validation
4.2. Bioinformatics Analyses
4.2.1. Identification and Analysis of Predominantly Placenta-Expressed Genes
4.2.2. Identification and Analysis of Differentially Expressed Genes During Villous Trophoblast Differentiation
4.2.3. Functional Annotation and Pathway Analysis
4.2.4. Co-Expression Network Analysis
4.2.5. Genes with High Expression Change
4.2.6. Determining Transcriptional Networks from DNase Footprinting Data
4.3. Data Availability
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Burton, G.J.; Jauniaux, E. What is the placenta? Am. J. Obstet. Gynecol. 2015, 213, S6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, M.S.; Aleksunes, L.M.; Boeuf, P.; Chung, M.K.; Daoud, G.; Desoye, G.; Díaz, P.; Golos, T.G.; Illsley, N.P.; Kikuchi, K.; et al. IFPA Meeting 2012 Workshop Report II: epigenetics and imprinting in the placenta, growth factors and villous trophoblast differentiation, role of the placenta in regulating fetal exposure to xenobiotics during pregnancy, infection and the placenta. Placenta 2013, 34, S6–S10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bischof, P.; Irminger-Finger, I. The human cytotrophoblastic cell, a mononuclear chameleon. Int. J. Biochem. Cell Biol. 2005, 37, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Mayhew, T.M. Turnover of human villous trophoblast in normal pregnancy: What do we know and what do we need to know? Placenta 2014, 35, 229–240. [Google Scholar] [CrossRef] [PubMed]
- Aronow, B.J.; Richardson, B.D.; Handwerger, S. Microarray analysis of trophoblast differentiation: Gene expression reprogramming in key gene function categories. Physiol. Genom. 2001, 6, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Handwerger, S.; Aronow, B. Dynamic changes in gene expression during human trophoblast differentiation. Recent Prog. Horm. Res. 2003, 58, 263–281. [Google Scholar] [CrossRef]
- Knöfler, M.; Saleh, L.; Bauer, S.; Vasicek, R.; Griesinger, G.; Strohmer, H.; Helmer, H.; Husslein, P. Promoter elements and transcription factors involved in differentiation-dependent human chorionic gonadotrophin-alpha messenger ribonucleic acid expression of term villous trophoblasts. Endocrinology 2000, 141, 3737–3748. [Google Scholar] [CrossRef]
- Knöfler, M.; Saleh, L.; Bauer, S.; Galos, B.; Rotheneder, H.; Husslein, P.; Helmer, H. Transcriptional regulation of the human chorionic gonadotropin beta gene during villous trophoblast differentiation. Endocrinology 2004, 145, 1685–1694. [Google Scholar] [CrossRef] [Green Version]
- Loregger, T.; Pollheimer, J.; Knöfler, M. Regulatory transcription factors controlling function and differentiation of human trophoblast—A review. Placenta 2003, 24, S104–S110. [Google Scholar] [CrossRef]
- Kliman, H.J.; Nestler, J.E.; Sermasi, E.; Sanger, J.M.; Strauss, J.F. Purification, characterization, and in vitro differentiation of cytotrophoblasts from human term placentae. Endocrinology 1986, 118, 1567–1582. [Google Scholar] [CrossRef] [Green Version]
- Feinman, M.A.; Kliman, H.J.; Caltabiano, S.; Strauss, J.F. 8-Bromo-3’,5’-adenosine monophosphate stimulates the endocrine activity of human cytotrophoblasts in culture. J. Clin. Endocrinol. Metab. 1986, 63, 1211–1217. [Google Scholar] [CrossRef] [PubMed]
- Kao, L.C.; Caltabiano, S.; Wu, S.; Strauss, J.F.; Kliman, H.J. The human villous cytotrophoblast: interactions with extracellular matrix proteins, endocrine function, and cytoplasmic differentiation in the absence of syncytium formation. Dev. Biol. 1988, 130, 693–702. [Google Scholar] [CrossRef]
- Rejniak, K.A.; Kliman, H.J.; Fauci, L.J. A computational model of the mechanics of growth of the villous trophoblast bilayer. Bull. Math. Biol. 2004, 66, 199–232. [Google Scholar] [CrossRef]
- Than, G.N.; Bohn, H.; Szabó, D.G. Advances in Pregnancy-Related Protein Research: Functional and Clinical Applications; CRC Press Inc.: Boca Raton, FL, USA, 1993; ISBN 0-8493-4962-1. [Google Scholar]
- Than, N.G.; Romero, R.; Goodman, M.; Weckle, A.; Xing, J.; Dong, Z.; Xu, Y.; Tarquini, F.; Szilagyi, A.; Gal, P.; et al. A primate subfamily of galectins expressed at the maternal-fetal interface that promote immune cell death. Proc. Natl. Acad. Sci. USA 2009, 106, 9731–9736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Than, N.G.; Romero, R.; Kim, C.J.; McGowen, M.R.; Papp, Z.; Wildman, D.E. Galectins: Guardians of eutherian pregnancy at the maternal-fetal interface. Trends Endocrinol. Metab. 2012, 23, 23–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rawn, S.M.; Cross, J.C. The evolution, regulation, and function of placenta-specific genes. Annu. Rev. Cell Dev. Biol. 2008, 24, 159–181. [Google Scholar] [CrossRef]
- Cross, J.C.; Baczyk, D.; Dobric, N.; Hemberger, M.; Hughes, M.; Simmons, D.G.; Yamamoto, H.; Kingdom, J.C.P. Genes, development and evolution of the placenta. Placenta 2003, 24, 123–130. [Google Scholar] [CrossRef]
- Than, N.G.; Romero, R.; Xu, Y.; Erez, O.; Xu, Z.; Bhatti, G.; Leavitt, R.; Chung, T.H.; El-Azzamy, H.; LaJeunesse, C.; et al. Evolutionary origins of the placental expression of chromosome 19 cluster galectins and their complex dysregulation in preeclampsia. Placenta 2014, 35, 855–865. [Google Scholar] [CrossRef] [Green Version]
- Wildman, D.E. Review: Toward an integrated evolutionary understanding of the mammalian placenta. Placenta 2011, 32, S142–S145. [Google Scholar] [CrossRef] [Green Version]
- Morales-Prieto, D.M.; Ospina-Prieto, S.; Chaiwangyen, W.; Schoenleben, M.; Markert, U.R. Pregnancy-associated miRNA-clusters. J. Reprod. Immunol. 2013, 97, 51–61. [Google Scholar] [CrossRef]
- Ouyang, Y.; Mouillet, J.-F.; Coyne, C.B.; Sadovsky, Y. Review: Placenta-specific microRNAs in exosomes—Good things come in nano-packages. Placenta 2014, 35, S69–S73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hromadnikova, I.; Dvorakova, L.; Kotlabova, K.; Krofta, L. The Prediction of Gestational Hypertension, Preeclampsia and Fetal Growth Restriction via the First Trimester Screening of Plasma Exosomal C19MC microRNAs. Int. J. Mol. Sci. 2019, 20, 2972. [Google Scholar] [CrossRef]
- Hromadnikova, I.; Kotlabova, K.; Ivankova, K.; Krofta, L. First trimester screening of circulating C19MC microRNAs and the evaluation of their potential to predict the onset of preeclampsia and IUGR. PLoS ONE 2017, 12, e0171756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hromadnikova, I.; Kotlabova, K.; Ondrackova, M.; Pirkova, P.; Kestlerova, A.; Novotna, V.; Hympanova, L.; Krofta, L. Expression profile of C19MC microRNAs in placental tissue in pregnancy-related complications. DNA Cell Biol. 2015, 34, 437–457. [Google Scholar] [CrossRef] [PubMed]
- Apicella, C.; Ruano, C.S.M.; Méhats, C.; Miralles, F.; Vaiman, D. The Role of Epigenetics in Placental Development and the Etiology of Preeclampsia. Int. J. Mol. Sci. 2019, 20, 2837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ellery, P.M.; Cindrova-Davies, T.; Jauniaux, E.; Ferguson-Smith, A.C.; Burton, G.J. Evidence for transcriptional activity in the syncytiotrophoblast of the human placenta. Placenta 2009, 30, 329–334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kudo, Y.; Boyd, C.a.R.; Sargent, I.L.; Redman, C.W.G.; Lee, J.M.; Freeman, T.C. An analysis using DNA microarray of the time course of gene expression during syncytialization of a human placental cell line (BeWo). Placenta 2004, 25, 479–488. [Google Scholar] [CrossRef]
- Sawai, K.; Azuma, C.; Koyama, M.; Hashimoto, K.; Kimura, T.; Samejima, Y.; Nobunaga, T.; Takemura, M.; Saji, F. The novel role of 3’,5’-guanosine monophosphate (cGMP) on the differentiation of trophoblasts: comparison with the effects of 3’,5’-adenosine monophosphate (cAMP). Early Pregnancy 1996, 2, 244–252. [Google Scholar]
- Soncin, F.; Natale, D.; Parast, M.M. Signaling pathways in mouse and human trophoblast differentiation: A comparative review. Cell. Mol. Life Sci. 2015, 72, 1291–1302. [Google Scholar] [CrossRef] [Green Version]
- Kusama, K.; Bai, R.; Imakawa, K. Regulation of human trophoblast cell syncytialization by transcription factors STAT5B and NR4A3. J. Cell. Biochem. 2018, 119, 4918–4927. [Google Scholar] [CrossRef]
- Knöfler, M.; Haider, S.; Saleh, L.; Pollheimer, J.; Gamage, T.K.J.B.; James, J. Human placenta and trophoblast development: key molecular mechanisms and model systems. Cell. Mol. Life Sci. 2019, 76, 3479–3496. [Google Scholar] [CrossRef] [Green Version]
- Yoshie, M.; Kaneyama, K.; Kusama, K.; Higuma, C.; Nishi, H.; Isaka, K.; Tamura, K. Possible role of the exchange protein directly activated by cyclic AMP (Epac) in the cyclic AMP-dependent functional differentiation and syncytialization of human placental BeWo cells. Hum. Reprod. 2010, 25, 2229–2238. [Google Scholar] [CrossRef] [Green Version]
- Morrish, D.W.; Bhardwaj, D.; Dabbagh, L.K.; Marusyk, H.; Siy, O. Epidermal growth factor induces differentiation and secretion of human chorionic gonadotropin and placental lactogen in normal human placenta. J. Clin. Endocrinol. Metab. 1987, 65, 1282–1290. [Google Scholar] [CrossRef]
- Mirlesse, V.; Alsat, E.; Fondacci, C.; Evain-Brion, D. Epidermal growth factor receptors in cultured human trophoblast cells from first- and third-trimester placentas. Horm. Res. 1990, 34, 234–239. [Google Scholar] [CrossRef]
- Amemiya, K.; Kurachi, H.; Adachi, H.; Morishige, K.I.; Adachi, K.; Imai, T.; Miyake, A. Involvement of epidermal growth factor (EGF)/EGF receptor autocrine and paracrine mechanism in human trophoblast cells: functional differentiation in vitro. J. Endocrinol. 1994, 143, 291–301. [Google Scholar] [CrossRef]
- Meyer zu Schwabedissen, H.E.; Grube, M.; Dreisbach, A.; Jedlitschky, G.; Meissner, K.; Linnemann, K.; Fusch, C.; Ritter, C.A.; Völker, U.; Kroemer, H.K. Epidermal growth factor-mediated activation of the map kinase cascade results in altered expression and function of ABCG2 (BCRP). Drug Metab. Dispos. 2006, 34, 524–533. [Google Scholar] [CrossRef] [Green Version]
- Prast, J.; Saleh, L.; Husslein, H.; Sonderegger, S.; Helmer, H.; Knöfler, M. Human chorionic gonadotropin stimulates trophoblast invasion through extracellularly regulated kinase and AKT signaling. Endocrinology 2008, 149, 979–987. [Google Scholar] [CrossRef] [Green Version]
- Daoud, G.; Amyot, M.; Rassart, E.; Masse, A.; Simoneau, L.; Lafond, J. ERK1/2 and p38 regulate trophoblasts differentiation in human term placenta. J. Physiol. 2005, 566, 409–423. [Google Scholar] [CrossRef]
- Johnstone, E.D.; Sibley, C.P.; Lowen, B.; Guilbert, L.J. Epidermal growth factor stimulation of trophoblast differentiation requires MAPK11/14 (p38 MAP kinase) activation. Biol. Reprod. 2005, 73, 1282–1288. [Google Scholar] [CrossRef]
- Daoud, G.; Rassart, E.; Masse, A.; Lafond, J. Src family kinases play multiple roles in differentiation of trophoblasts from human term placenta. J. Physiol. 2006, 571, 537–553. [Google Scholar] [CrossRef]
- Daoud, G.; Le bellego, F.; Lafond, J. PP2 regulates human trophoblast cells differentiation by activating p38 and ERK1/2 and inhibiting FAK activation. Placenta 2008, 29, 862–870. [Google Scholar] [CrossRef]
- Luo, Y.; Kumar, P.; Mendelson, C.R. Estrogen-related receptor γ (ERRγ) regulates oxygen-dependent expression of voltage-gated potassium (K+) channels and tissue kallikrein during human trophoblast differentiation. Mol. Endocrinol. 2013, 27, 940–952. [Google Scholar] [CrossRef]
- Kumar, P.; Mendelson, C.R. Estrogen-related receptor gamma (ERRgamma) mediates oxygen-dependent induction of aromatase (CYP19) gene expression during human trophoblast differentiation. Mol. Endocrinol. 2011, 25, 1513–1526. [Google Scholar] [CrossRef] [Green Version]
- Paul, S.; Home, P.; Bhattacharya, B.; Ray, S. GATA factors: Master regulators of gene expression in trophoblast progenitors. Placenta 2017, 60, S61–S66. [Google Scholar] [CrossRef]
- Bai, H.; Sakurai, T.; Godkin, J.D.; Imakawa, K. Expression and potential role of GATA factors in trophoblast development. J. Reprod. Dev. 2013, 59, 1–6. [Google Scholar] [CrossRef]
- Ray, S.; Dutta, D.; Rumi, M.A.K.; Kent, L.N.; Soares, M.J.; Paul, S. Context-dependent function of regulatory elements and a switch in chromatin occupancy between GATA3 and GATA2 regulate Gata2 transcription during trophoblast differentiation. J. Biol. Chem. 2009, 284, 4978–4988. [Google Scholar] [CrossRef] [Green Version]
- Cross, J.C.; Anson-Cartwright, L.; Scott, I.C. Transcription factors underlying the development and endocrine functions of the placenta. Recent Prog. Horm. Res. 2002, 57, 221–234. [Google Scholar] [CrossRef] [Green Version]
- Janatpour, M.J.; Utset, M.F.; Cross, J.C.; Rossant, J.; Dong, J.; Israel, M.A.; Fisher, S.J. A repertoire of differentially expressed transcription factors that offers insight into mechanisms of human cytotrophoblast differentiation. Dev. Genet. 1999, 25, 146–157. [Google Scholar] [CrossRef]
- Jacquemin, P.; Sapin, V.; Alsat, E.; Evain-Brion, D.; Dollé, P.; Davidson, I. Differential expression of the TEF family of transcription factors in the murine placenta and during differentiation of primary human trophoblasts in vitro. Dev. Dyn. 1998, 212, 423–436. [Google Scholar] [CrossRef]
- Morrish, D.W.; Dakour, J.; Li, H. Life and death in the placenta: New peptides and genes regulating human syncytiotrophoblast and extravillous cytotrophoblast lineage formation and renewal. Curr. Protein Pept. Sci. 2001, 2, 245–259. [Google Scholar] [CrossRef]
- Depoix, C.L.; Debiève, F.; Hubinont, C. Inhibin alpha gene expression in human trophoblasts is regulated by interactions between TFAP2 and cAMP signaling pathways. Mol. Reprod. Dev. 2014, 81, 1009–1018. [Google Scholar] [CrossRef]
- Kuckenberg, P.; Kubaczka, C.; Schorle, H. The role of transcription factor Tcfap2c/TFAP2C in trophectoderm development. Reprod. Biomed. Online 2012, 25, 12–20. [Google Scholar] [CrossRef] [Green Version]
- Richardson, B.D.; Cheng, Y.H.; Langland, R.A.; Handwerger, S. Differential expression of AP-2gamma and AP-2alpha during human trophoblast differentiation. Life Sci. 2001, 69, 2157–2165. [Google Scholar] [CrossRef]
- Cheng, Y.-H.; Aronow, B.J.; Hossain, S.; Trapnell, B.; Kong, S.; Handwerger, S. Critical role for transcription factor AP-2alpha in human trophoblast differentiation. Physiol. Genom. 2004, 18, 99–107. [Google Scholar] [CrossRef]
- Vaiman, D.; Miralles, F. An Integrative Analysis of Preeclampsia Based on the Construction of an Extended Composite Network Featuring Protein-Protein Physical Interactions and Transcriptional Relationships. PLoS ONE 2016, 11, e0165849. [Google Scholar] [CrossRef]
- Than, N.G.; Romero, R.; Tarca, A.L.; Kekesi, K.A.; Xu, Y.; Xu, Z.; Juhasz, K.; Bhatti, G.; Leavitt, R.J.; Gelencser, Z.; et al. Integrated Systems Biology Approach Identifies Novel Maternal and Placental Pathways of Preeclampsia. Front. Immunol. 2018, 9, 1661. [Google Scholar] [CrossRef]
- Burton, G.J.; Redman, C.W.; Roberts, J.M.; Moffett, A. Pre-eclampsia: pathophysiology and clinical implications. BMJ 2019, 366, l2381. [Google Scholar] [CrossRef] [Green Version]
- Chaiworapongsa, T.; Chaemsaithong, P.; Yeo, L.; Romero, R. Pre-eclampsia part 1: current understanding of its pathophysiology. Nat. Rev. Nephrol. 2014, 10, 466–480. [Google Scholar] [CrossRef] [Green Version]
- Kalkunte, S.; Boij, R.; Norris, W.; Friedman, J.; Lai, Z.; Kurtis, J.; Lim, K.-H.; Padbury, J.F.; Matthiesen, L.; Sharma, S. Sera from preeclampsia patients elicit symptoms of human disease in mice and provide a basis for an in vitro predictive assay. Am. J. Pathol. 2010, 177, 2387–2398. [Google Scholar] [CrossRef]
- Burton, G.J.; Fowden, A.L.; Thornburg, K.L. Placental Origins of Chronic Disease. Physiol. Rev. 2016, 96, 1509–1565. [Google Scholar] [CrossRef]
- Millership, S.J.; Van de Pette, M.; Withers, D.J. Genomic imprinting and its effects on postnatal growth and adult metabolism. Cell. Mol. Life Sci. 2019, 76, 4009–4021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Than, N.G.; Vaisbuch, E.; Kim, C.J.; Mazaki-Tovi, S.; Erez, O.; Yeo, L.; Mittal, P.; Hupuczi, P.; Varkonyi, T.; Hassan, S.S.; et al. Early-Onset Preeclampsia and HELLP Syndrome: An Overview. In Handbook of Growth and Growth Monitoring in Health and Disease; Preedy, V.R., Ed.; Springer: New York, NY, USA, 2012; pp. 1867–1891. ISBN 978-1-4419-1794-2. [Google Scholar]
- Kamity, R.; Sharma, S.; Hanna, N. MicroRNA-Mediated Control of Inflammation and Tolerance in Pregnancy. Front. Immunol. 2019, 10, 718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aouache, R.; Biquard, L.; Vaiman, D.; Miralles, F. Oxidative Stress in Preeclampsia and Placental Diseases. Int. J. Mol. Sci. 2018, 19, 1496. [Google Scholar] [CrossRef] [Green Version]
- Szabo, S.; Mody, M.; Romero, R.; Xu, Y.; Karaszi, K.; Mihalik, N.; Xu, Z.; Bhatti, G.; Fule, T.; Hupuczi, P.; et al. Activation of villous trophoblastic p38 and ERK1/2 signaling pathways in preterm preeclampsia and HELLP syndrome. Pathol. Oncol. Res. 2015, 21, 659–668. [Google Scholar] [CrossRef]
- Balogh, A.; Pozsgay, J.; Matkó, J.; Dong, Z.; Kim, C.J.; Várkonyi, T.; Sammar, M.; Rigó, J.; Meiri, H.; Romero, R.; et al. Placental protein 13 (PP13/galectin-13) undergoes lipid raft-associated subcellular redistribution in the syncytiotrophoblast in preterm preeclampsia and HELLP syndrome. Am. J. Obstet. Gynecol. 2011, 205, 156-e1. [Google Scholar] [CrossRef] [Green Version]
- Than, N.G.; Abdul Rahman, O.; Magenheim, R.; Nagy, B.; Fule, T.; Hargitai, B.; Sammar, M.; Hupuczi, P.; Tarca, A.L.; Szabo, G.; et al. Placental protein 13 (galectin-13) has decreased placental expression but increased shedding and maternal serum concentrations in patients presenting with preterm pre-eclampsia and HELLP syndrome. Virchows Arch. 2008, 453, 387–400. [Google Scholar] [CrossRef] [Green Version]
- Szabo, S.; Xu, Y.; Romero, R.; Fule, T.; Karaszi, K.; Bhatti, G.; Varkonyi, T.; Varkonyi, I.; Krenacs, T.; Dong, Z.; et al. Changes of placental syndecan-1 expression in preeclampsia and HELLP syndrome. Virchows Arch. 2013, 463, 445–458. [Google Scholar] [CrossRef] [Green Version]
- Hannke-Lohmann, A.; Pildner von Steinburg, S.; Dehne, K.; Benard, V.; Kolben, M.; Schmitt, M.; Lengyel, E. Downregulation of a mitogen-activated protein kinase signaling pathway in the placentas of women with preeclampsia. Obstet. Gynecol. 2000, 96, 582–587. [Google Scholar]
- Cindrova-Davies, T. Gabor Than Award Lecture 2008: Pre-eclampsia—From placental oxidative stress to maternal endothelial dysfunction. Placenta 2009, 30, S55–S65. [Google Scholar] [CrossRef]
- Keikkala, E.; Koskinen, S.; Vuorela, P.; Laivuori, H.; Romppanen, J.; Heinonen, S.; Stenman, U.-H. First trimester serum placental growth factor and hyperglycosylated human chorionic gonadotropin are associated with pre-eclampsia: A case control study. BMC Pregnancy Childbirth 2016, 16, 378. [Google Scholar] [CrossRef] [Green Version]
- Norris, W.; Nevers, T.; Sharma, S.; Kalkunte, S. Review: hCG, preeclampsia and regulatory T cells. Placenta 2011, 32, S182–S185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murtoniemi, K.; Villa, P.M.; Matomäki, J.; Keikkala, E.; Vuorela, P.; Hämäläinen, E.; Kajantie, E.; Pesonen, A.-K.; Räikkönen, K.; Taipale, P.; et al. Prediction of pre-eclampsia and its subtypes in high-risk cohort: Hyperglycosylated human chorionic gonadotropin in multivariate models. BMC Pregnancy Childbirth 2018, 18, 279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaisbuch, E.; Whitty, J.E.; Hassan, S.S.; Romero, R.; Kusanovic, J.P.; Cotton, D.B.; Sorokin, Y.; Karumanchi, S.A. Circulating angiogenic and antiangiogenic factors in women with eclampsia. Am. J. Obstet. Gynecol. 2011, 204, 152-e1. [Google Scholar] [CrossRef] [Green Version]
- Mayhew, T.M.; Ohadike, C.; Baker, P.N.; Crocker, I.P.; Mitchell, C.; Ong, S.S. Stereological investigation of placental morphology in pregnancies complicated by pre-eclampsia with and without intrauterine growth restriction. Placenta 2003, 24, 219–226. [Google Scholar] [CrossRef]
- Jones, C.J.; Fox, H. An ultrastructural and ultrahistochemical study of the human placenta in maternal pre-eclampsia. Placenta 1980, 1, 61–76. [Google Scholar] [CrossRef]
- Riquelme, G.; Vallejos, C.; de Gregorio, N.; Morales, B.; Godoy, V.; Berrios, M.; Bastías, N.; Rodríguez, C. Lipid rafts and cytoskeletal proteins in placental microvilli membranes from preeclamptic and IUGR pregnancies. J. Membr. Biol. 2011, 241, 127–140. [Google Scholar] [CrossRef]
- Roland, C.S.; Hu, J.; Ren, C.-E.; Chen, H.; Li, J.; Varvoutis, M.S.; Leaphart, L.W.; Byck, D.B.; Zhu, X.; Jiang, S.-W. Morphological changes of placental syncytium and their implications for the pathogenesis of preeclampsia. Cell. Mol. Life Sci. 2016, 73, 365–376. [Google Scholar] [CrossRef] [Green Version]
- Huppertz, B. Placental origins of preeclampsia: challenging the current hypothesis. Hypertension 2008, 51, 970–975. [Google Scholar] [CrossRef]
- Baczyk, D.; Drewlo, S.; Proctor, L.; Dunk, C.; Lye, S.; Kingdom, J. Glial cell missing-1 transcription factor is required for the differentiation of the human trophoblast. Cell Death Differ. 2009, 16, 719–727. [Google Scholar] [CrossRef]
- Chen, C.-P.; Chen, C.-Y.; Yang, Y.-C.; Su, T.-H.; Chen, H. Decreased placental GCM1 (glial cells missing) gene expression in pre-eclampsia. Placenta 2004, 25, 413–421. [Google Scholar] [CrossRef]
- Holder, B.S.; Tower, C.L.; Abrahams, V.M.; Aplin, J.D. Syncytin 1 in the human placenta. Placenta 2012, 33, 460–466. [Google Scholar] [CrossRef] [PubMed]
- Keith, J.C.; Pijnenborg, R.; Van Assche, F.A. Placental syncytin expression in normal and preeclamptic pregnancies. Am. J. Obstet. Gynecol. 2002, 187, 1122–1123. [Google Scholar] [CrossRef]
- Lee, X.; Keith, J.C.; Stumm, N.; Moutsatsos, I.; McCoy, J.M.; Crum, C.P.; Genest, D.; Chin, D.; Ehrenfels, C.; Pijnenborg, R.; et al. Downregulation of placental syncytin expression and abnormal protein localization in pre-eclampsia. Placenta 2001, 22, 808–812. [Google Scholar] [CrossRef] [PubMed]
- Ruebner, M.; Strissel, P.L.; Ekici, A.B.; Stiegler, E.; Dammer, U.; Goecke, T.W.; Faschingbauer, F.; Fahlbusch, F.B.; Beckmann, M.W.; Strick, R. Reduced syncytin-1 expression levels in placental syndromes correlates with epigenetic hypermethylation of the ERVW-1 promoter region. PLoS ONE 2013, 8, e56145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toufaily, C.; Vargas, A.; Lemire, M.; Lafond, J.; Rassart, E.; Barbeau, B. MFSD2a, the Syncytin-2 receptor, is important for trophoblast fusion. Placenta 2013, 34, 85–88. [Google Scholar] [CrossRef]
- Várkonyi, T.; Nagy, B.; Füle, T.; Tarca, A.L.; Karászi, K.; Schönléber, J.; Hupuczi, P.; Mihalik, N.; Kovalszky, I.; Rigó, J.; et al. Microarray profiling reveals that placental transcriptomes of early-onset HELLP syndrome and preeclampsia are similar. Placenta 2011, 32, S21–S29. [Google Scholar] [CrossRef] [Green Version]
- Su, A.I.; Wiltshire, T.; Batalov, S.; Lapp, H.; Ching, K.A.; Block, D.; Zhang, J.; Soden, R.; Hayakawa, M.; Kreiman, G.; et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. USA 2004, 101, 6062–6067. [Google Scholar] [CrossRef] [Green Version]
- Lichtner, B.; Knaus, P.; Lehrach, H.; Adjaye, J. BMP10 as a potent inducer of trophoblast differentiation in human embryonic and induced pluripotent stem cells. Biomaterials 2013, 34, 9789–9802. [Google Scholar] [CrossRef]
- Maruo, T.; Matsuo, H.; Otani, T.; Mochizuki, M. Role of epidermal growth factor (EGF) and its receptor in the development of the human placenta. Reprod. Fertil. Dev. 1995, 7, 1465–1470. [Google Scholar] [CrossRef]
- Bai, Q.; Assou, S.; Haouzi, D.; Ramirez, J.-M.; Monzo, C.; Becker, F.; Gerbal-Chaloin, S.; Hamamah, S.; De Vos, J. Dissecting the first transcriptional divergence during human embryonic development. Stem Cell Rev. Rep. 2012, 8, 150–162. [Google Scholar] [CrossRef] [Green Version]
- Baczyk, D.; Satkunaratnam, A.; Nait-Oumesmar, B.; Huppertz, B.; Cross, J.C.; Kingdom, J.C.P. Complex patterns of GCM1 mRNA and protein in villous and extravillous trophoblast cells of the human placenta. Placenta 2004, 25, 553–559. [Google Scholar] [CrossRef] [PubMed]
- Apps, R.; Sharkey, A.; Gardner, L.; Male, V.; Trotter, M.; Miller, N.; North, R.; Founds, S.; Moffett, A. Genome-wide expression profile of first trimester villous and extravillous human trophoblast cells. Placenta 2011, 32, 33–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bilban, M.; Haslinger, P.; Prast, J.; Klinglmüller, F.; Woelfel, T.; Haider, S.; Sachs, A.; Otterbein, L.E.; Desoye, G.; Hiden, U.; et al. Identification of novel trophoblast invasion-related genes: Heme oxygenase-1 controls motility via peroxisome proliferator-activated receptor gamma. Endocrinology 2009, 150, 1000–1013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tilburgs, T.; Crespo, Â.C.; van der Zwan, A.; Rybalov, B.; Raj, T.; Stranger, B.; Gardner, L.; Moffett, A.; Strominger, J.L. Human HLA-G+ extravillous trophoblasts: Immune-activating cells that interact with decidual leukocytes. Proc. Natl. Acad. Sci. USA 2015, 112, 7219–7224. [Google Scholar] [CrossRef] [Green Version]
- Hubert, M.A.; Sherritt, S.L.; Bachurski, C.J.; Handwerger, S. Involvement of transcription factor NR2F2 in human trophoblast differentiation. PLoS ONE 2010, 5, e9417. [Google Scholar] [CrossRef]
- Maere, S.; Heymans, K.; Kuiper, M. BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 2005, 21, 3448–3449. [Google Scholar] [CrossRef] [Green Version]
- Gauster, M.; Maninger, S.; Siwetz, M.; Deutsch, A.; El-Heliebi, A.; Kolb-Lenz, D.; Hiden, U.; Desoye, G.; Herse, F.; Prokesch, A. Downregulation of p53 drives autophagy during human trophoblast differentiation. Cell. Mol. Life Sci. 2018, 75, 1839–1855. [Google Scholar] [CrossRef] [Green Version]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinf. 2008, 9, 559. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Horvath, S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 2005, 4, 17. [Google Scholar] [CrossRef]
- Kubota, K.; Kent, L.N.; Rumi, M.A.K.; Roby, K.F.; Soares, M.J. Dynamic Regulation of AP-1 Transcriptional Complexes Directs Trophoblast Differentiation. Mol. Cell. Biol. 2015, 35, 3163–3177. [Google Scholar] [CrossRef] [Green Version]
- Kent, L.N.; Rumi, M.A.K.; Kubota, K.; Lee, D.-S.; Soares, M.J. FOSL1 is integral to establishing the maternal-fetal interface. Mol. Cell. Biol. 2011, 31, 4801–4813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Renaud, S.J.; Chakraborty, D.; Mason, C.W.; Rumi, M.A.K.; Vivian, J.L.; Soares, M.J. OVO-like 1 regulates progenitor cell fate in human trophoblast development. Proc. Natl. Acad. Sci. USA 2015, 112, E6175–E6184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karg, N.J.; Csaba, I.F.; Than, G.N.; Arany, A.A.; Szabó, D.G. The prognosis of the possible foetal and placental complications during delivery by measuring maternal serum levels of pregnancy-specific beta-1-glycoprotein (SP1). Arch. Gynecol. 1981, 231, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Fülöp, V.; Vermes, G.; Demeter, J. The relationship between inflammatory and immunological processes during pregnancy. Practical aspects. Orv. Hetil. 2019, 160, 1247–1259. [Google Scholar] [CrossRef]
- Kovács, K.; Vásárhelyi, B.; Gyarmati, B.; Karvaly, G. Estrogen metabolism during pregnancy. Orv. Hetil. 2019, 160, 1007–1014. [Google Scholar] [CrossRef]
- Guillaumond, F.; Gréchez-Cassiau, A.; Subramaniam, M.; Brangolo, S.; Peteri-Brünback, B.; Staels, B.; Fiévet, C.; Spelsberg, T.C.; Delaunay, F.; Teboul, M. Kruppel-like factor KLF10 is a link between the circadian clock and metabolism in liver. Mol. Cell. Biol. 2010, 30, 3059–3070. [Google Scholar] [CrossRef] [Green Version]
- Yang, N.; Chen, J.; Zhang, H.; Wang, X.; Yao, H.; Peng, Y.; Zhang, W. LncRNA OIP5-AS1 loss-induced microRNA-410 accumulation regulates cell proliferation and apoptosis by targeting KLF10 via activating PTEN/PI3K/AKT pathway in multiple myeloma. Cell Death Dis. 2017, 8, e2975. [Google Scholar] [CrossRef]
- Pereira de Sousa, F.L.; Chaiwangyen, W.; Morales-Prieto, D.M.; Ospina-Prieto, S.; Weber, M.; Photini, S.M.; Sass, N.; Daher, S.; Schleussner, E.; Markert, U.R. Involvement of STAT1 in proliferation and invasiveness of trophoblastic cells. Reprod. Biol. 2017, 17, 218–224. [Google Scholar] [CrossRef]
- Hemberger, M.; Udayashankar, R.; Tesar, P.; Moore, H.; Burton, G.J. ELF5-enforced transcriptional networks define an epigenetically regulated trophoblast stem cell compartment in the human placenta. Hum. Mol. Genet. 2010, 19, 2456–2467. [Google Scholar] [CrossRef] [Green Version]
- Roncalli, M.; Bulfamante, G.; Viale, G.; Springall, D.R.; Alfano, R.; Comi, A.; Maggioni, M.; Polak, J.M.; Coggi, G. C-myc and tumour suppressor gene product expression in developing and term human trophoblast. Placenta 1994, 15, 399–409. [Google Scholar] [CrossRef]
- Haider, S.; Meinhardt, G.; Saleh, L.; Fiala, C.; Pollheimer, J.; Knöfler, M. Notch1 controls development of the extravillous trophoblast lineage in the human placenta. Proc. Natl. Acad. Sci. USA 2016, 113, E7710–E7719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, F.; Tian, F.; Zeng, W.; Liu, X.; Fan, J.; Lin, Y.; Zhang, Y. Role of peroxiredoxin2 downregulation in recurrent miscarriage through regulation of trophoblast proliferation and apoptosis. Cell Death Dis. 2017, 8, e2908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michita, R.T.; de Lima Kaminski, V.; Chies, J.A.B. Genetic Variants in Preeclampsia: Lessons from Studies in Latin-American Populations. Front. Physiol. 2018, 9, 1771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giannakou, K.; Evangelou, E.; Papatheodorou, S.I. Genetic and non-genetic risk factors for pre-eclampsia: Umbrella review of systematic reviews and meta-analyses of observational studies. Ultrasound Obstet. Gynecol. 2018, 51, 720–730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alexander, G.R.; Himes, J.H.; Kaufman, R.B.; Mor, J.; Kogan, M. A United States national reference for fetal growth. Obstet. Gynecol. 1996, 87, 163–168. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Casagrande, J.T.; Thomas, P.D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 2013, 8, 1551–1566. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [Green Version]
- Ahsan, S.; Drăghici, S. Identifying Significantly Impacted Pathways and Putative Mechanisms with iPathwayGuide. Curr. Protoc. Bioinform. 2017, 57, 7.15.1–7.15.30. [Google Scholar] [CrossRef] [PubMed]
- Draghici, S.; Khatri, P.; Tarca, A.L.; Amin, K.; Done, A.; Voichita, C.; Georgescu, C.; Romero, R. A systems biology approach for pathway level analysis. Genome Res. 2007, 17, 1537–1545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donato, M.; Xu, Z.; Tomoiaga, A.; Granneman, J.G.; Mackenzie, R.G.; Bao, R.; Than, N.G.; Westfall, P.H.; Romero, R.; Draghici, S. Analysis and correction of crosstalk effects in pathway analysis. Genome Res. 2013, 23, 1885–1893. [Google Scholar] [CrossRef] [PubMed]
- Tarca, A.L.; Draghici, S.; Khatri, P.; Hassan, S.S.; Mittal, P.; Kim, J.-S.; Kim, C.J.; Kusanovic, J.P.; Romero, R. A novel signaling pathway impact analysis. Bioinformatics 2009, 25, 75–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khatri, P.; Draghici, S.; Tarca, A.L.; Hassan, S.S.; Romero, R. A System Biology Approach for the Steady-State Analysis of Gene Signaling Networks. In Progress in Pattern Recognition, Image Analysis and Applications; Rueda, L., Mery, D., Kittler, J., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; Volume 4756, pp. 32–41. ISBN 978-3-540-76724-4. [Google Scholar]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, D277–D280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neph, S.; Stergachis, A.B.; Reynolds, A.; Sandstrom, R.; Borenstein, E.; Stamatoyannopoulos, J.A. Circuitry and dynamics of human transcription factor regulatory networks. Cell 2012, 150, 1274–1286. [Google Scholar] [CrossRef] [Green Version]









| Module | Number of Genes | Odds Ratio | Up-Regulated% | Transcription Regulatory Genes | |||||
|---|---|---|---|---|---|---|---|---|---|
| All Genes | PPE Genes | PE Genes | PPE Genes | PE Genes | Number | OR | Up-Regulated% | ||
| M1: Brown | 256 | 17 | 37 | 1.54 | 1.45 | 69% | 27 | 0.91 | 44% |
| M2: Pink | 45 | 10 | 5 | 6.39 | 1.02 | 73% | 3 | 0.55 | 33% |
| M3: Magenta | 42 | 3 | 4 | 1.58 | 0.85 | 79% | 3 | 0.59 | 67% |
| M4: Turquoise | 726 | 26 | 79 | 0.65 | 0.99 | 33% | 108 | 1.71 | 17% |
| M5: Blue | 378 | 11 | 43 | 0.55 | 1.06 | 44% | 37 | 0.82 | 46% |
| M6: Green | 176 | 7 | 12 | 0.83 | 0.57 | 61% | 21 | 1.06 | 62% |
| M7: Red | 72 | 9 | 7 | 3.11 | 0.87 | 60% | 8 | 0.97 | 29% |
| M8: Yellow | 182 | 7 | 19 | 0.80 | 0.94 | 36% | 12 | 0.53 | 42% |
| M9: Black | 60 | 1 | 6 | 0.34 | 0.90 | 73% | 1 | 0.13 | 100% |
| Σ | 1937 | 91 | 212 | 47% | 220 | ||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szilagyi, A.; Gelencser, Z.; Romero, R.; Xu, Y.; Kiraly, P.; Demeter, A.; Palhalmi, J.; Gyorffy, B.A.; Juhasz, K.; Hupuczi, P.; et al. Placenta-Specific Genes, Their Regulation During Villous Trophoblast Differentiation and Dysregulation in Preterm Preeclampsia. Int. J. Mol. Sci. 2020, 21, 628. https://doi.org/10.3390/ijms21020628
Szilagyi A, Gelencser Z, Romero R, Xu Y, Kiraly P, Demeter A, Palhalmi J, Gyorffy BA, Juhasz K, Hupuczi P, et al. Placenta-Specific Genes, Their Regulation During Villous Trophoblast Differentiation and Dysregulation in Preterm Preeclampsia. International Journal of Molecular Sciences. 2020; 21(2):628. https://doi.org/10.3390/ijms21020628
Chicago/Turabian StyleSzilagyi, Andras, Zsolt Gelencser, Roberto Romero, Yi Xu, Peter Kiraly, Amanda Demeter, Janos Palhalmi, Balazs A. Gyorffy, Kata Juhasz, Petronella Hupuczi, and et al. 2020. "Placenta-Specific Genes, Their Regulation During Villous Trophoblast Differentiation and Dysregulation in Preterm Preeclampsia" International Journal of Molecular Sciences 21, no. 2: 628. https://doi.org/10.3390/ijms21020628
APA StyleSzilagyi, A., Gelencser, Z., Romero, R., Xu, Y., Kiraly, P., Demeter, A., Palhalmi, J., Gyorffy, B. A., Juhasz, K., Hupuczi, P., Kekesi, K. A., Meinhardt, G., Papp, Z., Draghici, S., Erez, O., Tarca, A. L., Knöfler, M., & Than, N. G. (2020). Placenta-Specific Genes, Their Regulation During Villous Trophoblast Differentiation and Dysregulation in Preterm Preeclampsia. International Journal of Molecular Sciences, 21(2), 628. https://doi.org/10.3390/ijms21020628







