Jasmonic Acid at the Crossroads of Plant Immunity and Pseudomonas syringae Virulence
Abstract
:1. Introduction
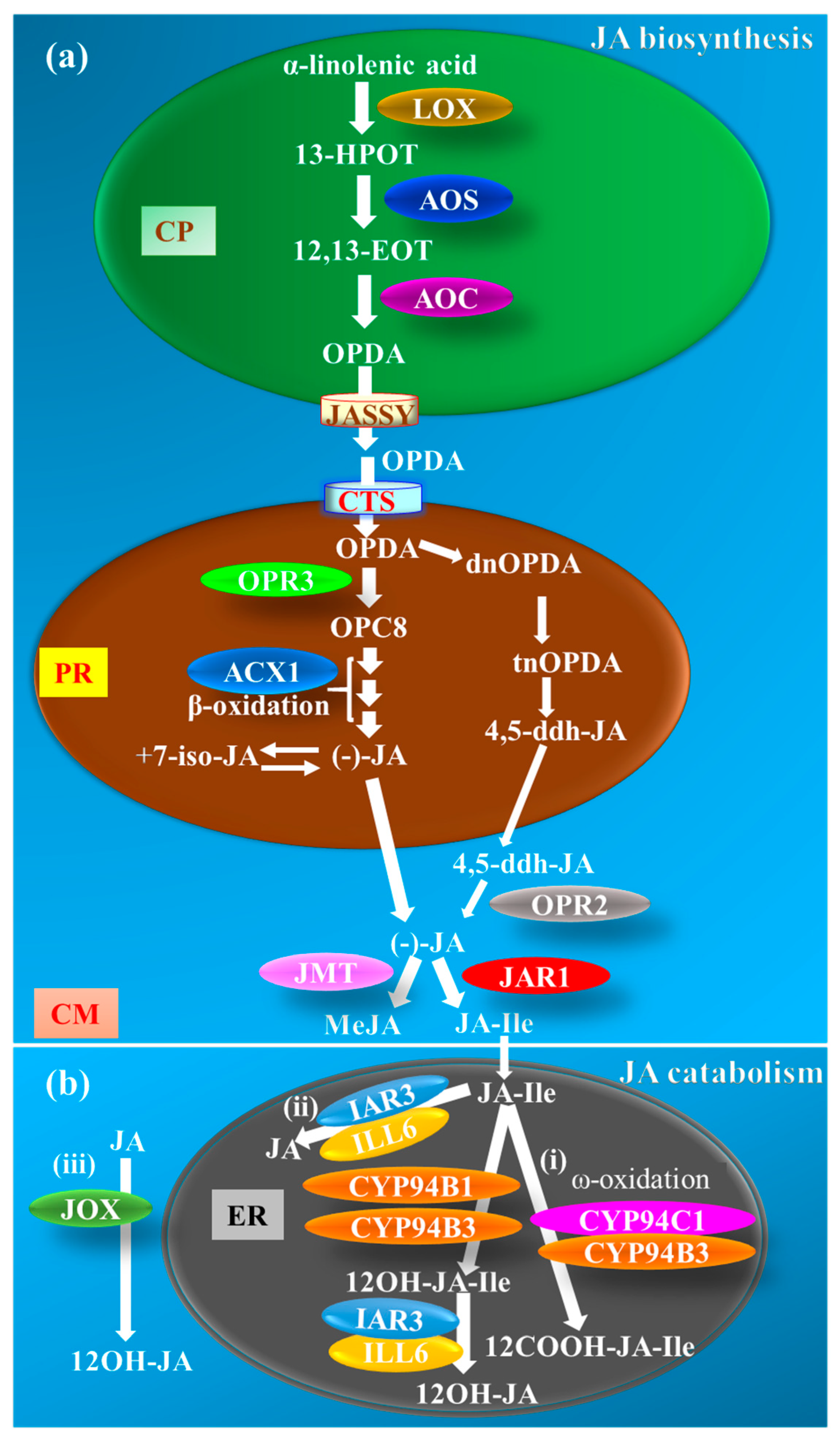
2. JA Metabolism
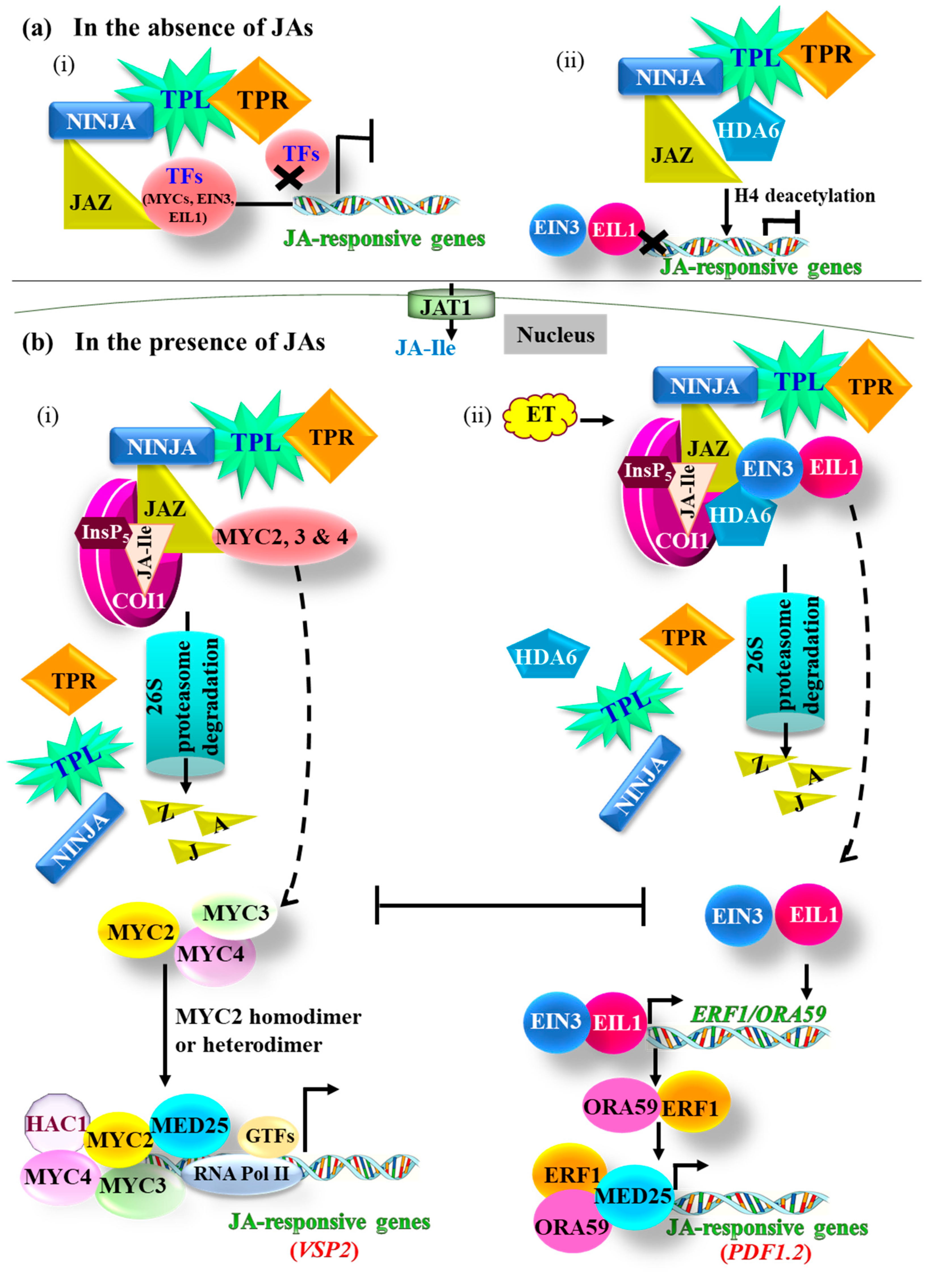
3. JA Signaling
3.1. The MYC Branch
3.2. The ERF Branch
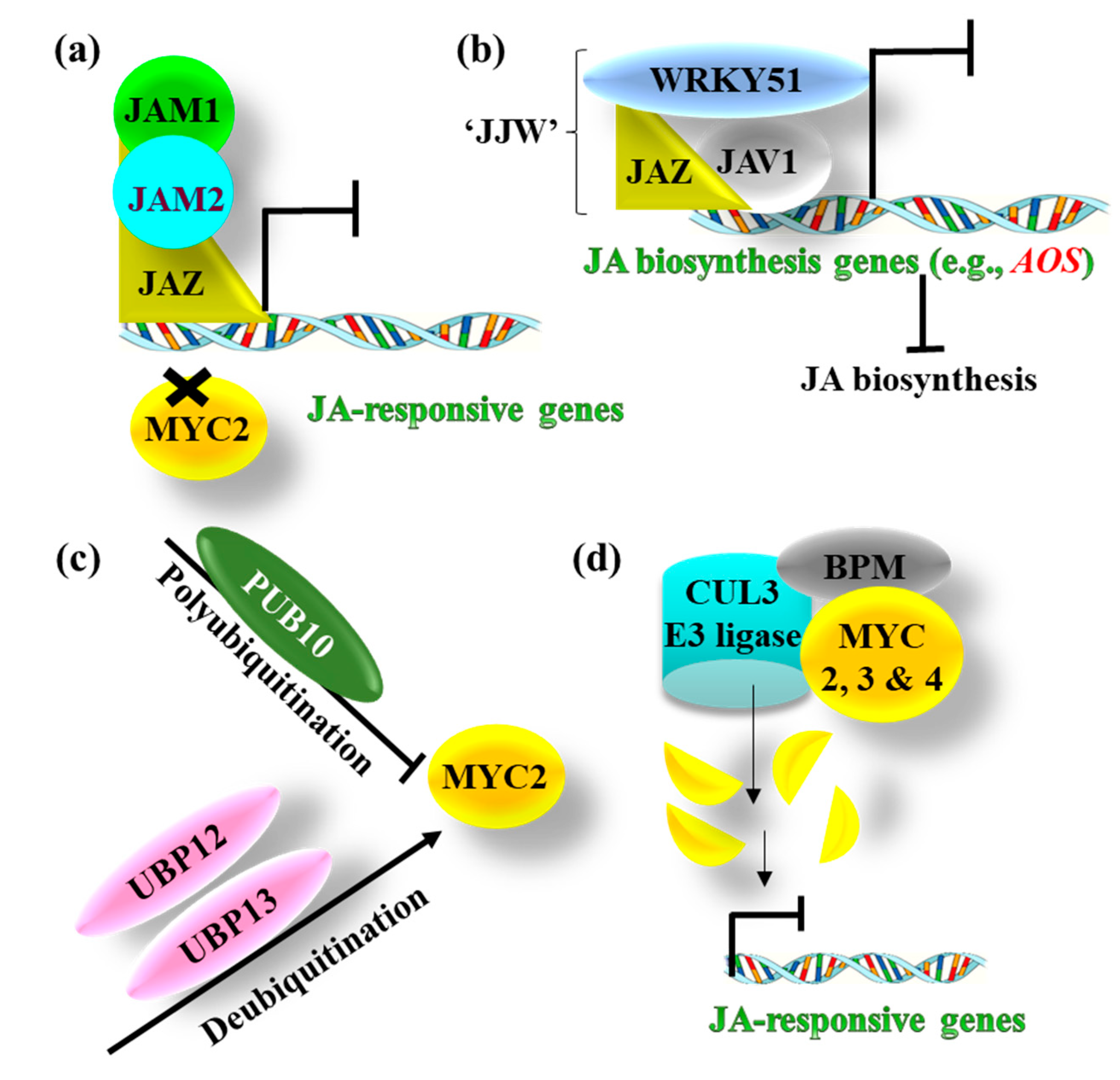
3.3. Negative Regulation of JA Signaling
4. Role of JAs in Plant Defense against P. syringae and Some Other Major Hemibiotrophs
4.1. JAs Inhibit Plant Immunity
4.2. JAs Positively Mediate Plant Immunity
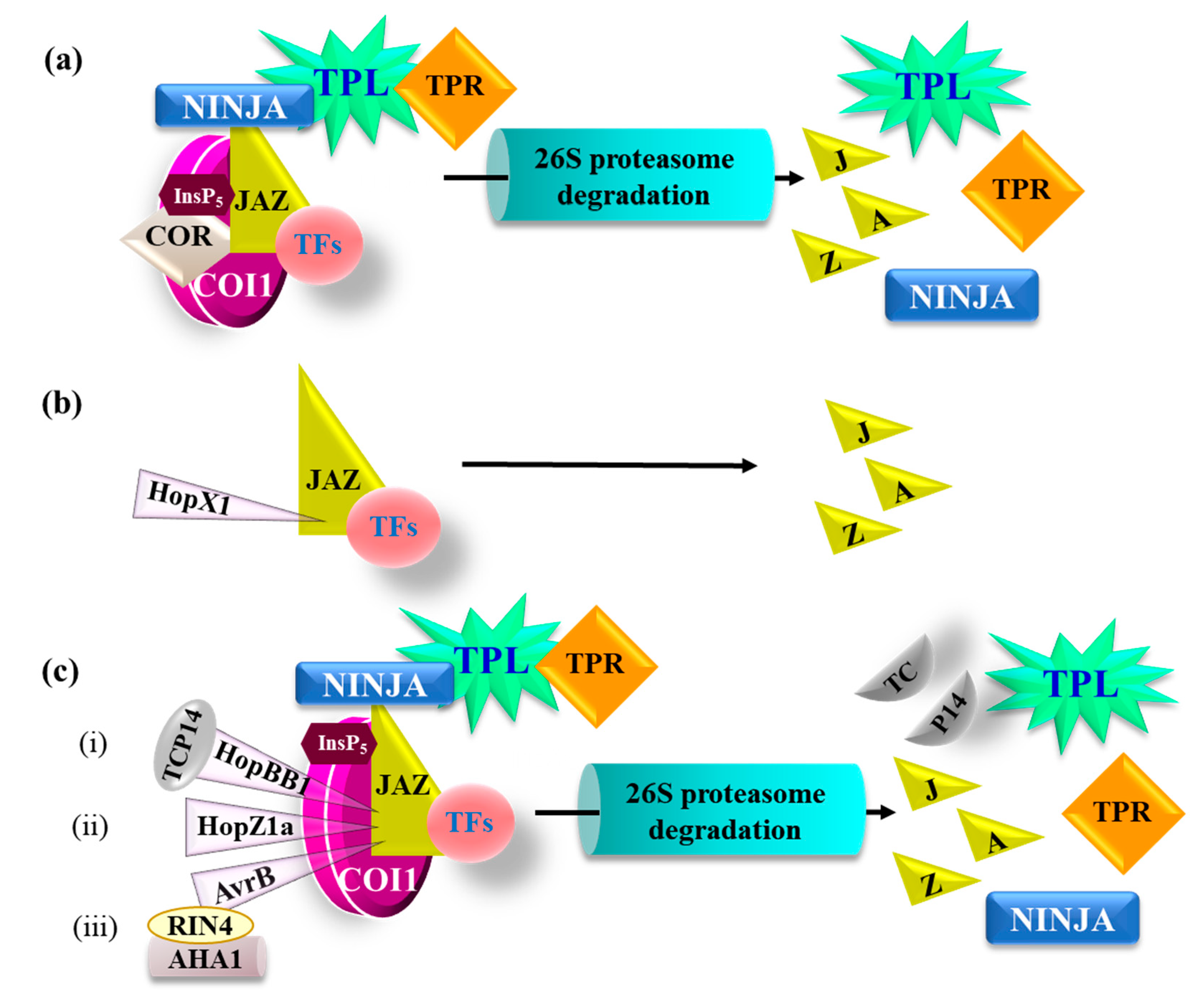
5. P. syringae Hijacks JA Signaling to Counter Plant Defense
6. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Wasternack, C.; Strnad, M. Jasmonates: News on occurrence, biosynthesis, metabolism and action of an ancient group of signaling compounds. Int. J. Mol. Sci. 2018, 19, 2539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glazebrook, J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 2005, 43, 205–227. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Qiu, J.; Zhou, Y.; Bhandari, D.D.; Zhao, C.; Bautor, J.; Parker, J.E. Antagonism of transcription factor MYC2 by EDS1/PAD4 complexes bolsters salicylic acid defense in Arabidopsis effector-triggered immunity. Mol. Plant. 2018, 11, 1053–1066. [Google Scholar] [CrossRef] [PubMed]
- Thomma, B.P.H.J.; Eggermont, K.; Penninckx, I.A.M.A.; Mauch-Mani, B.; Vogelsang, R.; Cammue, B.P.A.; Broekaert, W.F. Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl. Acad. Sci. USA 1998, 95, 15107–15111. [Google Scholar] [CrossRef] [Green Version]
- Klessig, D.F.; Choi, H.W.; Dempsey, D.M.A. Systemic acquired resistance and salicylic acid: Past, present, and future. Mol. Plant-Microbe Interact. 2018, 31, 871–888. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Duan, G.; Li, C.; Liu, L.; Han, G.; Zhang, Y.; Wang, C. The crosstalks between jasmonic acid and other plant hormone signaling highlight the involvement of jasmonic acid as a core component in plant response to biotic and abiotic stresses. Front. Plant Sci. 2019, 10, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Katsir, L.; Schilmiller, A.L.; Staswick, P.E.; He, S.Y.; Howe, G.A. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. USA 2008, 105, 7100–7105. [Google Scholar] [CrossRef] [Green Version]
- Jiang, S.; Yao, J.; Ma, K.; Zhou, H.; Song, J.; He, S.Y.; Ma, W. Bacterial effector activates jasmonate signaling by directly targeting JAZ transcriptional repressors. PLoS Pathog. 2013, 9, e1003715. [Google Scholar] [CrossRef] [Green Version]
- Sheard, L.B.; Tan, X.; Mao, H.; Withers, J.; Ben-nissan, G.; Hinds, T.R.; Kobayashi, Y.; Hsu, F.; Sharon, M.; Browse, J.; et al. Jasmonate perception by inositol phosphate-potentiated COI1- JAZ co-receptor. Nature 2010, 468, 400–405. [Google Scholar] [CrossRef]
- Liu, L.; Sonbol, F.M.; Huot, B.; Gu, Y.; Withers, J.; Mwimba, M.; Yao, J.; He, S.Y.; Dong, X. Salicylic acid receptors activate jasmonic acid signalling through a non-canonical pathway to promote effector-triggered immunity. Nat. Commun. 2016, 7, 13099. [Google Scholar] [CrossRef] [Green Version]
- Gupta, A.; Hisano, H.; Hojo, Y.; Matsuura, T.; Ikeda, Y.; Mori, I.C.; Senthil-Kumar, M. Global profiling of phytohormone dynamics during combined drought and pathogen stress in Arabidopsis thaliana reveals ABA and JA as major regulators. Sci. Rep. 2017, 7, 4017. [Google Scholar] [CrossRef] [PubMed]
- Vick, B.A.; Zimmerman, D.O.N.C. Biosynthesis of jasmonic acid by several plant species. Plant Physiol. 1984, 75, 458–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guan, L.; Denkert, N.; Eisa, A.; Lehmann, M.; Sjuts, I.; Weiberg, A.; Soll, J.; Meinecke, M.; Schwenkert, S. JASSY, a chloroplast outer membrane protein required for jasmonate biosynthesis. Proc. Natl. Acad. Sci. USA 2019, 116, 10568–10575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Theodoulou, F.L.; Job, K.; Slocombe, S.P.; Footitt, S.; Holdsworth, M.; Baker, A.; Larson, T.R.; Graham, I.A. Jasmonic acid levels are reduced in COMATOSE ATP-binding cassette transporter mutants. Implications for transport of jasmonate precursors into peroxisomes. Plant Physiol. 2005, 137, 835–840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seo, H.S.; Song, J.T.; Cheong, J.; Lee, Y.; Lee, Y.; Hwang, I.; Lee, J.S.; Choi, Y. Do Jasmonic acid carboxyl methyltransferase: A key enzyme for jasmonate-regulated plant responses. Proc. Natl. Acad. Sci. USA 2001, 98, 4788–4793. [Google Scholar] [CrossRef] [Green Version]
- Staswick, P.E.; Tiryaki, I. The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 2004, 16, 2117–2127. [Google Scholar] [CrossRef] [Green Version]
- Fonseca, S.; Chico, J.M.; Solano, R. The jasmonate pathway: The ligand, the receptor and the core signalling module. Curr. Opin. Plant Biol. 2009, 12, 539–547. [Google Scholar] [CrossRef]
- Chini, A.; Monte, I.; Zamarreño, A.M.; Hamberg, M.; Lassueur, S.; Reymond, P.; Weiss, S.; Stintzi, A.; Schaller, A.; Porzel, A.; et al. An OPR3-independent pathway uses 4,5-didehydrojasmonate for jasmonate synthesis. Nat. Chem. Ecol. 2018, 14, 171–178. [Google Scholar] [CrossRef] [Green Version]
- Wasternack, C.; Hause, B. A bypass in jasmonate biosynthesis—The OPR3-independent formation. Trends Plant Sci. 2018, 23, 276–279. [Google Scholar] [CrossRef]
- Aubert, Y.; Widemann, E.; Miesch, L.; Pinot, F.; Heitz, T. CYP94-mediated jasmonoyl-isoleucine hormone oxidation shapes jasmonate profiles and attenuates defence responses to Botrytis cinerea infection. J. Exp. Bot. 2015, 66, 3879–3892. [Google Scholar] [CrossRef] [Green Version]
- Koo, A.J.K.; Cooke, T.F.; Howe, G.A. Cytochrome P450 CYP94B3 mediates catabolism and inactivation of the plant hormone jasmonoyl-L-isoleucine. Proc. Natl. Acad. Sci. USA 2011, 108, 9298–9303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koo, A.J.; Thireault, C.; Zemelis, S.; Poudel, A.N.; Zhang, T.; Kitaoka, N.; Brandizzi, F.; Matsuura, H.; Howe, G.A. Endoplasmic reticulum-associated inactivation of the hormone jasmonoyl-L-isoleucine by multiple members of the cytochrome P450 94 family in Arabidopsis. J. Biol. Chem. 2014, 289, 29728–29738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heitz, T.; Widemann, E.; Lugan, R.; Miesch, L.; Ullmann, P.; Désaubry, L.; Holder, E.; Grausem, B.; Kandel, S.; Miesch, M.; et al. Cytochromes P450 CYP94C1 and CYP94B3 catalyze two successive oxidation steps of plant hormone jasmonoyl-isoleucine for catabolic turnover. J. Biol. Chem. 2012, 287, 6296–6306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruckhoff, V.; Haroth, S.; Feussner, K.; König, S.; Brodhun, F.; Feussner, I. Functional characterization of CYP94-genes and identification of a novel jasmonate catabolite in flowers. PLoS ONE 2016, 11, 1–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kitaoka, N.; Matsubara, T.; Sato, M.; Takahashi, K.; Wakuta, S.; Kawaide, H.; Matsui, H.; Nabeta, K.; Matsuura, H. Arabidopsis CYP94B3 encodes jasmonyl-l-isoleucine 12-hydroxylase, a key enzyme in the oxidative catabolism of jasmonate. Plant Cell Physiol. 2011, 52, 1757–1765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marquis, V.; Smirnova, E.; Poirier, L.; Zumsteg, J.; Schweizer, F.; Reymond, P.; Heitz, T. Stress- and pathway-specific impacts of impaired jasmonoyl-isoleucine (JA-Ile) catabolism on defense signalling and biotic stress resistance. Plant Cell Environ. 2020, 1–13. [Google Scholar] [CrossRef]
- Caarls, L.; Elberse, J.; Awwanah, M.; Ludwig, N.R.; De Vries, M.; Zeilmaker, T.; Van Wees, S.C.M.; Schuurink, R.C.; Van den Ackerveken, G. Arabidopsis JASMONATE-INDUCED OXYGENASES down-regulate plant immunity by hydroxylation and inactivation of the hormone jasmonic acid. Proc. Natl. Acad. Sci. USA 2017, 114, 6388–6393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Widemann, E.; Miesch, L.; Lugan, R.; Holder, E.; Heinrich, C.; Aubert, Y.; Miesch, M.; Pinot, F.; Heitz, T. The amidohydrolases IAR3 and ILL6 contribute to jasmonoyl-isoleucine hormone turnover and generate 12-hydroxyjasmonic acid upon wounding in Arabidopsis. J. Biol. Chem. 2013, 288, 31701–31714. [Google Scholar] [CrossRef] [Green Version]
- Chini, A.; Fonseca, S.; Fernández, G.; Adie, B.; Chico, J.M.; Lorenzo, O.; García-Casado, G.; López-Vidriero, I.; Lozano, F.M.; Ponce, M.R.; et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007, 448, 666–671. [Google Scholar] [CrossRef]
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.; Liu, G.; Nomura, K.; He, S.Y.; Howe, G.A.; Browse, J. JAZ repressor proteins are targets of the SCF COI1 complex during jasmonate signalling. Nature 2007, 448, 661–665. [Google Scholar] [CrossRef]
- Wasternack, C.; Song, S. Jasmonates: Biosynthesis, metabolism, and signaling by proteins activating and repressing transcription. J. Exp. Bot. 2017, 68, 1303–1321. [Google Scholar] [CrossRef] [PubMed]
- Pauwels, L.; Fernández Barbero, G.; Geerinck, J.; Tilleman, S.; Grunewald, W.; Pérez, A.C.; Chico, J.M.; Bossche, R.V.; Sewell, J.; Gil, E.; et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 2010, 464, 788–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; An, F.; Feng, Y.; Li, P.; Xue, L.; A, M.; Jiang, Z.; Kim, J.-M.; To, T.K.; Li, W.; et al. Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 12539–12544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, K.; Zhang, L.; Zhou, C.; Yu, C.; Chaikam, V. HDA6 is required for jasmonate response, senescence and flowering in Arabidopsis. J. Exp. Bot. 2008, 59, 225–234. [Google Scholar] [CrossRef]
- Li, Q.; Zheng, J.; Li, S.; Huang, G.; Skilling, S.J.; Wang, L.; Li, L.; Li, M.; Yuan, L.; Liu, P. Transporter-mediated nuclear entry of jasmonoyl-isoleucine is essential for jasmonate signaling. Mol. Plant 2017, 10, 695–708. [Google Scholar] [CrossRef] [Green Version]
- Mosblech, A.; Thurow, C.; Gatz, C.; Feussner, I.; Heilmann, I.; Karspu, U. Jasmonic acid perception by COI1 involves inositol polyphosphates in Arabidopsis thaliana. Plant J. 2011, 65, 949–957. [Google Scholar] [CrossRef]
- Chen, R.; Jiang, H.; Li, L.; Zhai, Q.; Qi, L.; Zhou, W.; Liu, X.; Li, H.; Zheng, W.; Sun, J.; et al. The Arabidopsis mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell 2012, 24, 2898–2916. [Google Scholar] [CrossRef] [Green Version]
- Zarei, A.; Korbes, A.P.; Younessi, P.; Montiel, G.; Champion, A.; Memelink, J. Two GCC boxes and AP2/ERF-domain transcription factor ORA59 in jasmonate/ethylene-mediated activation of the PDF1.2 promoter in Arabidopsis. Plant Mol. Biol. 2011, 75, 321–331. [Google Scholar] [CrossRef] [Green Version]
- Lorenzo, O.; Chico, J.M.; Sa, J.J. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 2004, 16, 1938–1950. [Google Scholar] [CrossRef] [Green Version]
- Lorenzo, O.; Solano, R. Molecular players regulating the jasmonate signalling network. Curr. Opin. Plant Biol. 2005, 8, 532–540. [Google Scholar] [CrossRef] [PubMed]
- Verhage, A.; Vlaardingerbroek, I.; Raaymakers, C.; Van Dam, N.M.; Dicke, M.; Van Wees, S.C.M.; Pieterse, C.M.J. Rewiring of the jasmonate signaling pathway in Arabidopsis during insect herbivory. Front. Plant Sci. 2011, 2, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, S.; Huang, H.; Gao, H.; Wang, J.; Wu, D.; Liu, X.; Yang, S.; Zhai, Q.; Li, C.; Qi, T.; et al. Interaction between MYC2 and ETHYLENE INSENSITIVE3 modulates antagonism between jasmonate and ethylene signaling in Arabidopsis. Plant Cell 2014, 26, 263–279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandez-Calvo, P.; Chini, A.; Fernandez-Barbero, G.; Chico, J.-M.; Gimenez-ibanez, S.; Geerinck, J.; Eeckhout, D.; Schweizer, F.; Godoy, M.; Franco-zorrilla, J.M.; et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 2011, 23, 701–715. [Google Scholar] [CrossRef] [Green Version]
- Kidd, B.N.; Edgar, C.I.; Kumar, K.K.; Aitken, E.A.; Schenk, P.M.; Manners, J.M.; Kazan, K. The mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in Arabidopsis. Plant Cell 2009, 21, 2237–2252. [Google Scholar] [CrossRef] [Green Version]
- An, C.; Li, L.; Zhai, Q.; You, Y.; Deng, L.; Wu, F.; Chen, R.; Jiang, H. Mediator subunit MED25 links the jasmonate receptor to transcriptionally active chromatin. Proc. Natl. Acad. Sci. USA 2017, E8930–E8939. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Yao, J.; Ke, J.; Zhang, L.; Lam, V.Q.; Xin, X.-F.; Zhou, X.E.; Chen, J.; Brunzelle, J.; Griffin, P.R.; et al. Structural basis of JAZ repression of MYC transcription factors in jasmonate signaling. Nature 2015, 344, 1173–1178. [Google Scholar] [CrossRef]
- Broekaert, W.F.; Delaur, S.L.; De Bolle, M.F.C.; Cammue, B.P.A. The role of ethylene in host-pathogen interactions. Annu. Rev. Phytopathol. 2006, 44, 393–416. [Google Scholar] [CrossRef]
- Zhou, C.; Zhang, L.; Duan, J.; Miki, B.; Wu, K. HISTONE DEACETYLASE19 is involved in jasmonic acid and ethylene signaling of pathogen response in Arabidopsis. Plant Cell 2005, 17, 1196–1204. [Google Scholar] [CrossRef] [Green Version]
- Çevik, V.; Kidd, B.N.; Zhang, P.; Hill, C.; Kiddle, S.; Denby, K.J.; Holub, E.B.; Cahill, D.M.; Manners, J.M.; Schenk, P.M.; et al. MEDIATOR25 acts as an integrative hub for the regulation of jasmonate-responsive gene expression in Arabidopsis. Plant Physiol. 2012, 160, 541–555. [Google Scholar] [CrossRef] [Green Version]
- Sasaki-Sekimoto, Y.; Jikumaru, Y.; Obayashi, T.; Saito, H.; Masuda, S.; Kamiya, Y.; Ohta, H.; Shirasu, K. Basic helix-loop-helix transcription factors JASMONATE-ASSOCIATED MYC2-LIKE1 (JAM1), JAM2, and JAM3 are negative regulators of jasmonate responses in Arabidopsis. Plant Physiol. 2013, 163, 291–304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sasaki-Sekimoto, Y.; Saito, H.; Masuda, S.; Shirasu, K.; Ohta, H. Comprehensive analysis of protein interactions between JAZ proteins and bHLH transcription factors that negatively regulate jasmonate signaling. Plant Signal. Behav. 2014, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, C.; Fan, M.; Yang, M.; Zhao, J.; Zhang, W.; Su, Y.; Xiao, L.; Deng, H.; Xie, D. Injury activates Ca2+/Calmodulin-dependent phosphorylation of JAV1-JAZ8-WRKY51 complex for jasmonate biosynthesis. Mol. Cell 2018, 70, 136–149.e7. [Google Scholar] [CrossRef] [Green Version]
- Jung, C.; Zhao, P.; Seo, J.S.; Mitsuda, N.; Deng, S.; Chua, N.H. PLANT U-BOX PROTEIN10 regulates MYC2 stability in Arabidopsis. Plant Cell 2015, 27, 2016–2031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeong, J.S.; Jung, C.; Seo, J.S.; Kim, J.K.; Chua, N.H. The deubiquitinating enzymes UBP12 and UBP13 positively regulate MYC2 levels in jasmonate responses. Plant Cell 2017, 29, 1406–1424. [Google Scholar] [CrossRef] [Green Version]
- Chico, J.M.; Lechner, E.; Fernandez-Barbero, G.; Canibano, E.; García-Casado, G.; Franco-Zorrilla, J.M.; Hammann, P.; Zamarreño, A.M.; García-Mina, J.M.; Rubio, V.; et al. CUL3BPM E3 ubiquitin ligases regulate MYC2, MYC3, and MYC4 stability and JA responses. Proc. Natl. Acad. Sci. USA 2020, 117, 6205–6215. [Google Scholar] [CrossRef]
- Liu, Y.; Du, M.; Deng, L.; Shen, J.; Fang, M.; Chen, Q.; Lu, Y.; Wang, Q.; Li, C.; Zhai, Q. MYC2 regulates the termination of jasmonate signaling via an autoregulatory negative feedback loop. Plant Cell 2019, 31, 106–127. [Google Scholar] [CrossRef] [Green Version]
- Penninckx, I.A.M.A.; Thomma, B.P.H.J.; Buchala, A.; Métraux, J.P.; Broekaert, W.F. Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 1998, 10, 2103–2113. [Google Scholar] [CrossRef] [Green Version]
- Gimenez-Ibanez, S.; Chini, A.; Solano, R. How microbes twist jasmonate signaling around their little fingers. Plants 2016, 5, 9. [Google Scholar] [CrossRef] [Green Version]
- Feys, B.J.F.; Benedetti, C.E.; Penfold, C.N.; Turner, J.G. Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 1994, 6, 751–759. [Google Scholar] [CrossRef] [Green Version]
- Du, M.; Zhai, Q.; Deng, L.; Li, S.; Li, H.; Yan, L.; Huang, Z.; Wang, B.; Jiang, H.; Huang, T.; et al. Closely related NAC transcription factors of tomato differentially regulate stomatal closure and reopening during pathogen attack. Plant Cell 2014, 26, 3167–3184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kloek, A.P.; Verbsky, M.L.; Sharma, S.B.; Schoelz, J.E.; Vogel, J.; Klessig, D.F.; Kunkel, B.N. Resistance to Pseudomonas syringae conferred by an Arabidopsis thaliana coronatine-insensitive (coi1) mutation occurs through two distinct mechanisms. Plant J. 2001, 26, 509–522. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Zhao, J.; Tzeng, D.T.W.W.; Liu, Y.; Deng, L.; Yang, T.; Zhai, Q.; Wu, F.; Huang, Z.; Zhou, M.; et al. MYC2 orchestrates a hierarchical transcriptional cascade that regulates jasmonate-mediated plant immunity in tomato. Plant Cell 2017, 29, 1883–1906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gimenez-ibanez, S.; Boter, M.; Ortigosa, A.; Garcia-Casado, G.; Chini, A.; Lewsey, M.G.; Ecker, J.R.; Ntoukakis, V.; Solano, R. JAZ2 controls stomata dynamics during bacterial invasion. New Phytol. 2017, 213, 1378–1392. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Xue, L.; Chintamanani, S.; Germain, H.; Lin, H.; Cui, H.; Cai, R.; Zuo, J.; Tang, X.; Li, X.; et al. ETHYLENE INSENSITIVE3 and ETHYLENE INSENSITIVE3-LIKE1 repress SALICYLIC ACID INDUCTION DEFICIENT2 expression to negatively regulate plant innate immunity in Arabidopsis. Plant Cell 2009, 21, 2527–2540. [Google Scholar] [CrossRef] [Green Version]
- Spoel, S.H.; Johnson, J.S.; Dong, X. Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc. Natl. Acad. Sci. USA 2007, 104, 18842–18847. [Google Scholar] [CrossRef] [Green Version]
- Laurie-berry, N.; Joardar, V.; Street, I.H.; Kunkel, B.N.; Box, C.; Drive, B.; Louis, S. The Arabidopsis thaliana JASMONATE INSENSITIVE 1 gene is required for suppression of salicylic acid-dependent defenses during infection by Pseudomonas syringae. Mol. Plant-Microbe Interact. 2006, 19, 789–800. [Google Scholar] [CrossRef] [Green Version]
- Toum, L.; Torres, P.S.; Gallego, S.M.; Benavídes, M.P.; Vojnov, A.A.; Gudesblat, G.E. Coronatine inhibits stomatal closure through guard cell-specific inhibition of NADPH oxidase-dependent ROS production. Front. Plant Sci. 2016, 7, 1851. [Google Scholar] [CrossRef] [Green Version]
- Zheng, X.; Spivey, N.W.; Zeng, W.; Liu, P.-P.; Fu, Z.Q.; Klessig, D.F.; He, S.Y.; Dong, X. Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe 2012, 11, 587–596. [Google Scholar] [CrossRef] [Green Version]
- Leon-Reyes, A.; Du, Y.; Koornneef, A.; Proietti, S.; Körbes, A.P.; Memelink, J.; Pieterse, C.M.J.; Ritsema, T. Ethylene signaling renders the jasmonate response of Arabidopsis insensitive to future suppression by salicylic acid. Mol. Plant-Microbe Interact. 2010, 23, 187–197. [Google Scholar] [CrossRef] [Green Version]
- Petersen, M.; Brodersen, P.; Naested, H.; Andreasson, E.; Lindhart, U.; Johansen, B.; Nielsen, H.B.; Lacy, M.; Austin, M.J.; Parker, J.E.; et al. Arabidopsis MAP kinase 4 negatively regulates systemic acquired resistance. Cell 2000, 103, 1111–1120. [Google Scholar] [CrossRef] [Green Version]
- Brodersen, P.; Petersen, M.; Nielsen, H.B.; Zhu, S.; Newman, M.A.; Shokat, K.M.; Rietz, S.; Parker, J.; Mundy, J. Arabidopsis MAP kinase 4 regulates salicylic acid- and jasmonic acid/ethylene-dependent responses via EDS1 and PAD4. Plant J. 2006, 47, 532–546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thaler, J.S.; Owen, B.; Higgins, V.J. The role of the jasmonate response in plant susceptibility to diverse pathogens with a range of lifestyles. Plant Physiol. 2004, 135, 530–538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, H.; Tsuda, K.; Parker, J.E. Effector-triggered immunity: From pathogen perception to robust defense. Annu. Rev. Plant Biol. 2015, 66, 487–511. [Google Scholar] [CrossRef]
- Kenton, P.; Mur, L.A.J.; Atzorn, R.; Wasternack, C.; Draper, J. (−)-Jasmonic acid accumulation in tobacco hypersensitive response lesions. Mol. Plant-Microbe Interact. 1999, 12, 74–78. [Google Scholar] [CrossRef] [Green Version]
- Mur, L.A.J.; Kenton, P.; Atzorn, R.; Miersch, O.; Wasternack, C. The outcomes of concentration-specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiol. 2006, 140, 249–262. [Google Scholar] [CrossRef] [Green Version]
- Martin-Rivilla, H.; Garcia-Villaraco, A.; Ramos-Solano, B.; Gutierrez-Mañero, F.J.; Lucas, J.A. Extracts from cultures of Pseudomonas fluorescens induce defensive patterns of gene expression and enzyme activity while depressing visible injury and reactive oxygen species in Arabidopsis thaliana challenged with pathogenic Pseudomonas. AoB Plants 2019, 11, 1–26. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, C.; Zhang, Y.; Sun, Y.; Mou, Z. The Arabidopsis mediator complex subunit16 positively regulates salicylate-mediated systemic acquired resistance and jasmonate/ethylene-induced defense pathways. Plant Cell 2012, 24, 4294–4309. [Google Scholar] [CrossRef] [Green Version]
- Nawrath, C.; Heck, S.; Parinthawong, N.; Métraux, J.P. EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell 2002, 14, 275–286. [Google Scholar] [CrossRef] [Green Version]
- Serrano, M.; Wang, B.; Aryal, B.; Garcion, C.; Abou-Mansour, E.; Heck, S.; Geisler, M.; Mauch, F.; Nawrath, C.; Métraux, J.P. Export of salicylic acid from the chloroplast requires the multidrug and toxin extrusion-like transporter EDS5. Plant Physiol. 2013, 162, 1815–1821. [Google Scholar] [CrossRef] [Green Version]
- Mine, A.; Nobori, T.; Salazar-rondon, M.C.; Winkelmüller, T.M.; Anver, S.; Becker, D.; Tsuda, K. An incoherent feed-forward loop mediates robustness and tunability in a plant immune network. EMBO Rep. 2017, 18, 464–476. [Google Scholar] [CrossRef] [PubMed]
- Ellis, C.; Karafyllidis, I.; Turner, J.G. Constitutive activation of jasmonate signaling in an Arabidopsis mutant correlates with enhanced resistance to Erysiphe cichoracearum, Pseudomonas syringae, and Myzus persicae. Mol. Plant-Microbe Interact. 2002, 15, 1025–1030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leon-Reyes, A.; Spoel, S.H.; De Lange, E.S.; Abe, H.; Kobayashi, M.; Tsuda, S.; Millenaar, F.F.; Welschen, R.A.M.; Ritsema, T.; Pieterse, C.M.J. Ethylene modulates the role of NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 in cross talk between salicylate and jasmonate signaling. Plant Physiol. 2009, 149, 1797–1809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uppalapati, S.R.; Ishiga, Y.; Wangdi, T.; Kunkel, B.N.; Anand, A.; Mysore, K.S.; Bender, C.L. The phytotoxin coronatine contributes to pathogen fitness and is required for suppression of salicylic acid accumulation in tomato inoculated with Pseudomonas syringae pv. tomato DC3000. Mol. Plant-Microbe Interact. 2007, 20, 955–965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, L.; Teixeira, P.J.; Biswas, S.; Finkel, O.M.; He, Y.; Salas-Gonzalez, I.; English, M.E.; Epple, P.; Mieczkowski, P.; Dangl, J.L. Pseudomonas syringae type III effector HopBB1 promotes host transcriptional repressor degradation to regulate phytohormone responses and virulence. Cell Host Microbe 2017, 21, 156–168. [Google Scholar] [CrossRef] [Green Version]
- Wasternack, C. The Trojan horse coronatine: The COI1–JAZ2–MYC2,3,4–ANAC019,055,072 module in stomata dynamics upon bacterial infection. New Phytol. 2017, 213, 972–975. [Google Scholar] [CrossRef] [Green Version]
- Gimenez-Ibanez, S.; Boter, M.; Ferna´ndez-Barbero, G.; Chini, A.; Rathjen, J.P.; Solano, R. The bacterial effector HopX1 targets JAZ transcriptional repressors to activate jasmonate signaling and promote infection in Arabidopsis. PLoS Biol. 2014, 12, e1001792. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.; Wu, Y.; Yang, Y.; Du, M.; Zhang, X.; Guo, Y.; Li, C.; Zhou, J. An Arabidopsis plasma membrane proton ATPase modulates JA signaling and is exploited by the Pseudomonas syringae effector protein AvrB for stomatal invasion. Plant Cell 2015, 27, 2032–2041. [Google Scholar] [CrossRef] [Green Version]
- Shang, Y.; Li, X.; Cui, H.; He, P.; Thilmony, R.; Chintamanani, S.; Zwiesler-Vollick, J.; Gopalan, S.; Tang, X.; Zhou, J.M. RAR1, a central player in plant immunity, is targeted by Pseudomonas syringae effector AvrB. Proc. Natl. Acad. Sci. USA 2006, 103, 19200–19205. [Google Scholar] [CrossRef] [Green Version]
- Cui, H.; Wang, Y.; Xue, L.; Chu, J.; Yan, C.; Fu, J.; Chen, M.; Innes, R.W. Pseudomonas syringae effector protein AvrB perturbs Arabidopsis hormone signaling by activating MAP Kinase 4. Cell Host Microbe 2010, 7, 164–175. [Google Scholar] [CrossRef] [Green Version]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gupta, A.; Bhardwaj, M.; Tran, L.-S.P. Jasmonic Acid at the Crossroads of Plant Immunity and Pseudomonas syringae Virulence. Int. J. Mol. Sci. 2020, 21, 7482. https://doi.org/10.3390/ijms21207482
Gupta A, Bhardwaj M, Tran L-SP. Jasmonic Acid at the Crossroads of Plant Immunity and Pseudomonas syringae Virulence. International Journal of Molecular Sciences. 2020; 21(20):7482. https://doi.org/10.3390/ijms21207482
Chicago/Turabian StyleGupta, Aarti, Mamta Bhardwaj, and Lam-Son Phan Tran. 2020. "Jasmonic Acid at the Crossroads of Plant Immunity and Pseudomonas syringae Virulence" International Journal of Molecular Sciences 21, no. 20: 7482. https://doi.org/10.3390/ijms21207482
APA StyleGupta, A., Bhardwaj, M., & Tran, L.-S. P. (2020). Jasmonic Acid at the Crossroads of Plant Immunity and Pseudomonas syringae Virulence. International Journal of Molecular Sciences, 21(20), 7482. https://doi.org/10.3390/ijms21207482




