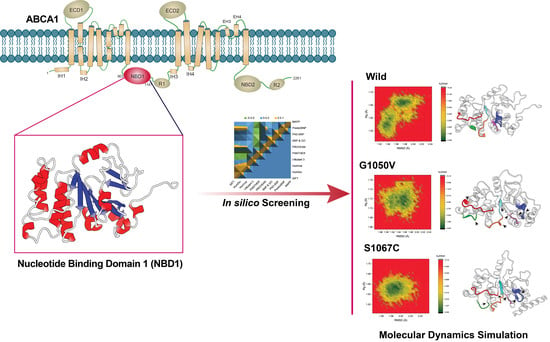
Computational SNP Analysis and Molecular Simulation Revealed the Most Deleterious Missense Variants in the NBD1 Domain of Human ABCA1 Transporter
Abstract
:1. Introduction
2. Results
2.1. Dataset Selection and Screening
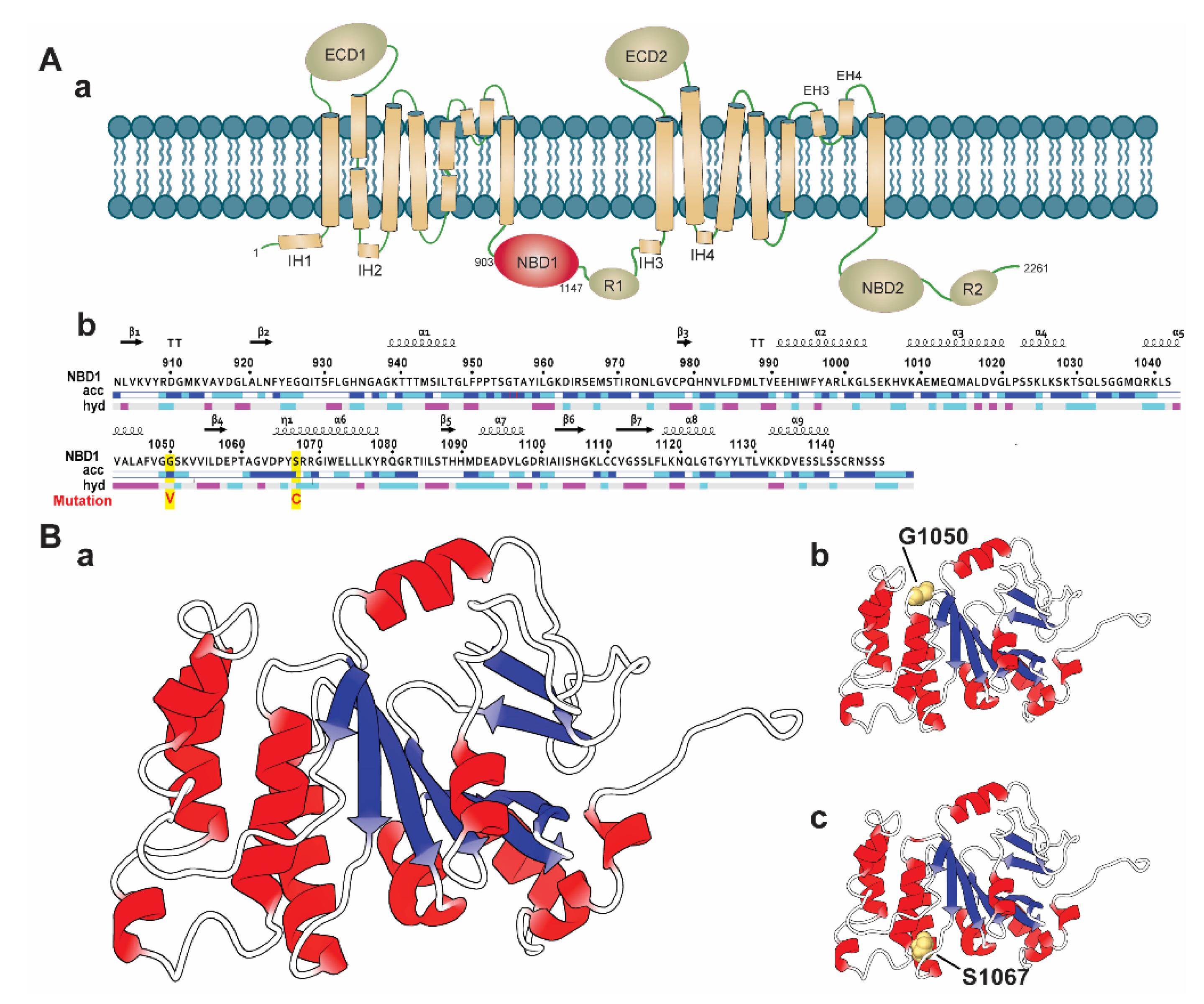
2.2. Identification of Most Deleterious nsSNPs
2.3. Evolutionary Conservancy
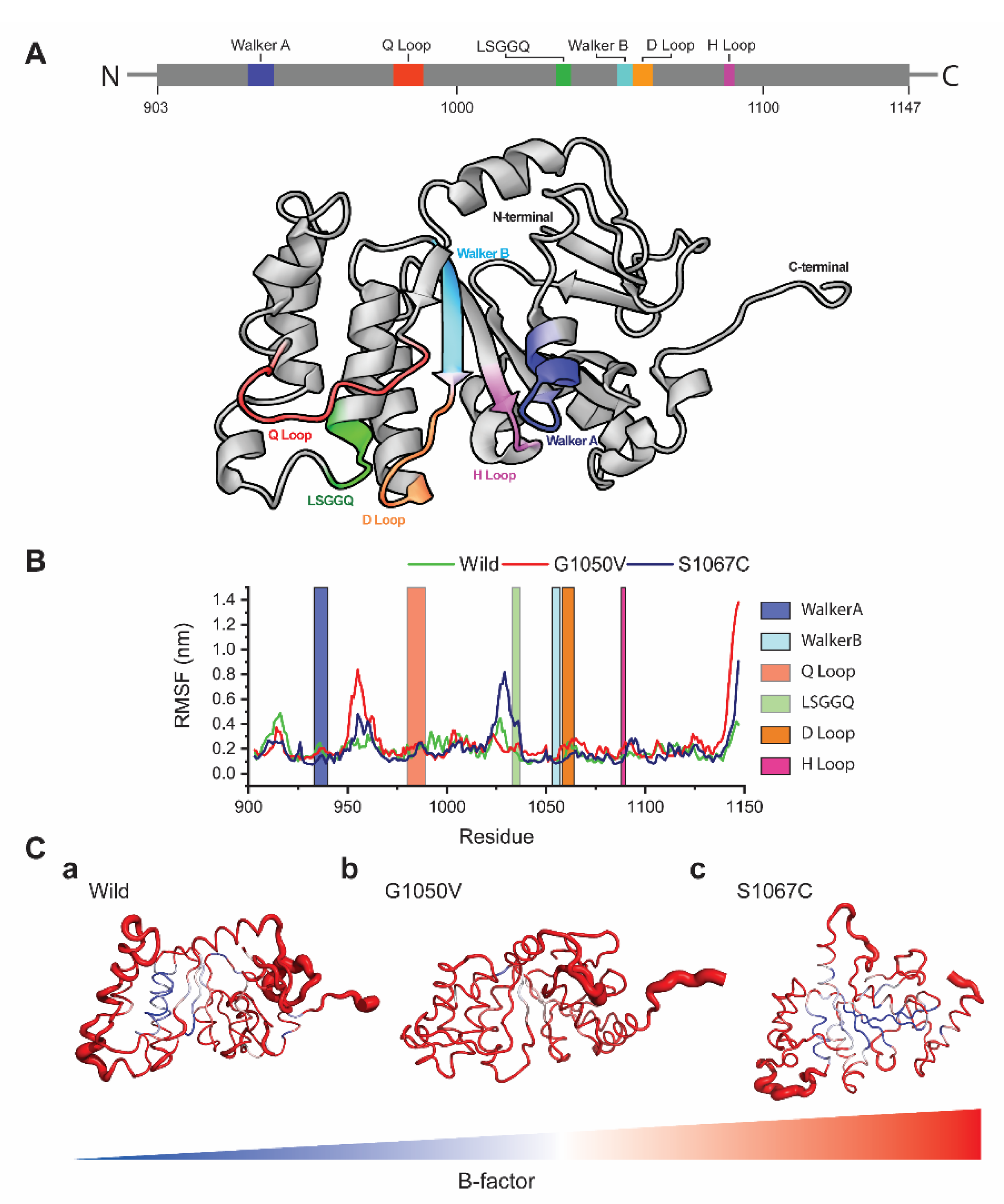
2.4. Molecular Dynamics (MD) Simulation
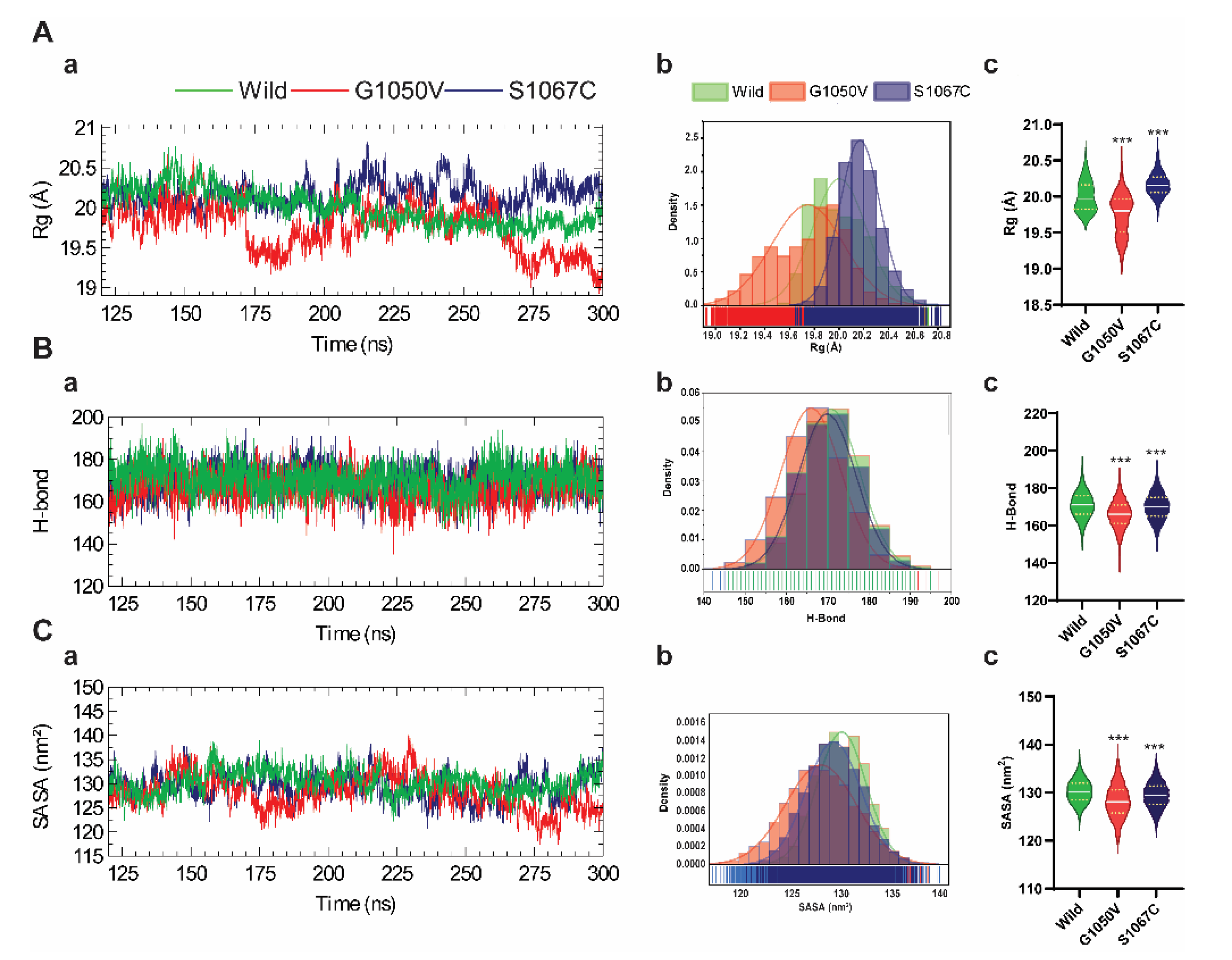
2.4.1. Effects of Variants on Conformational Dynamics
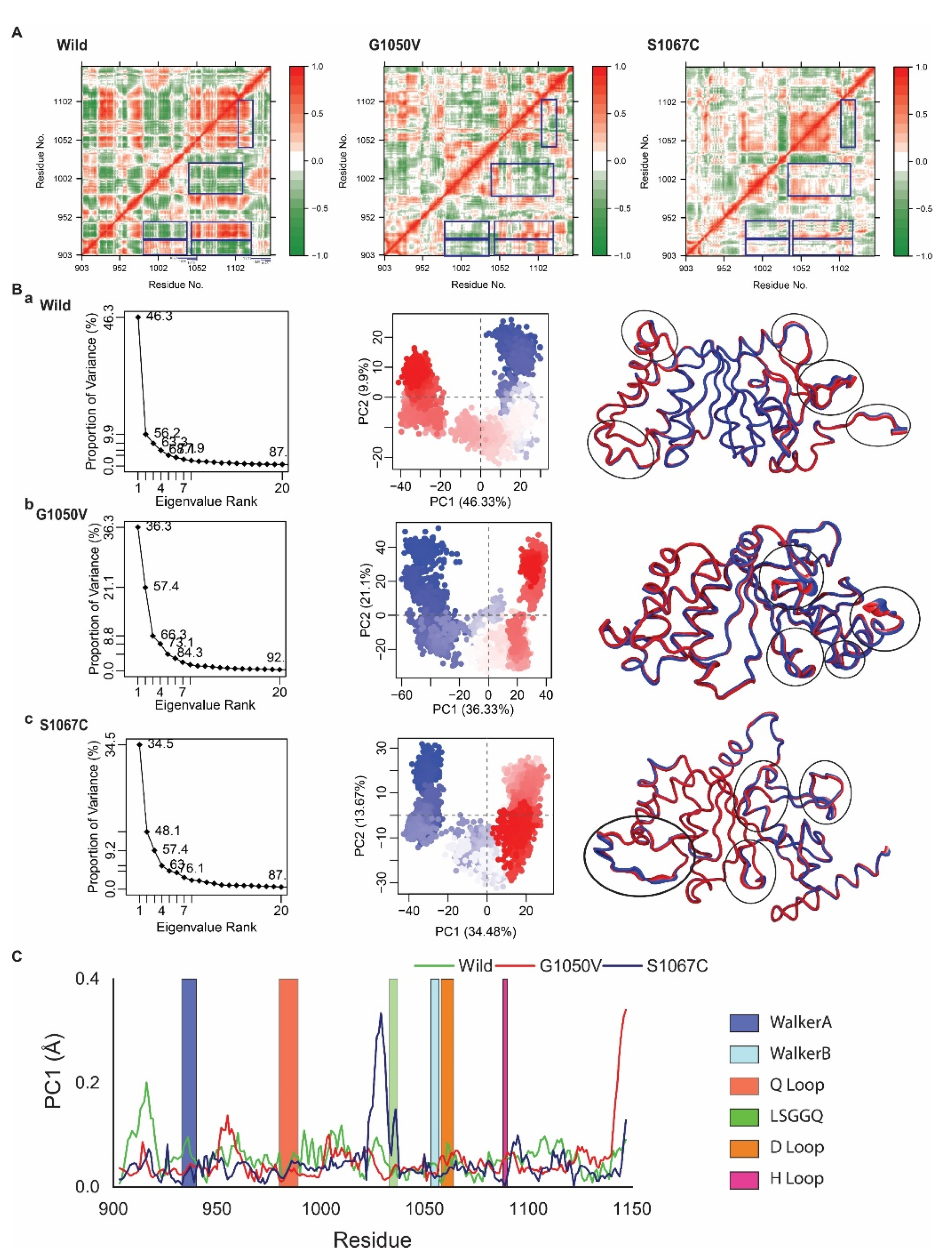
2.4.2. Effects of Variants on Protein Dynamics
2.4.3. Effect of Variants on Intra-Protein Communication
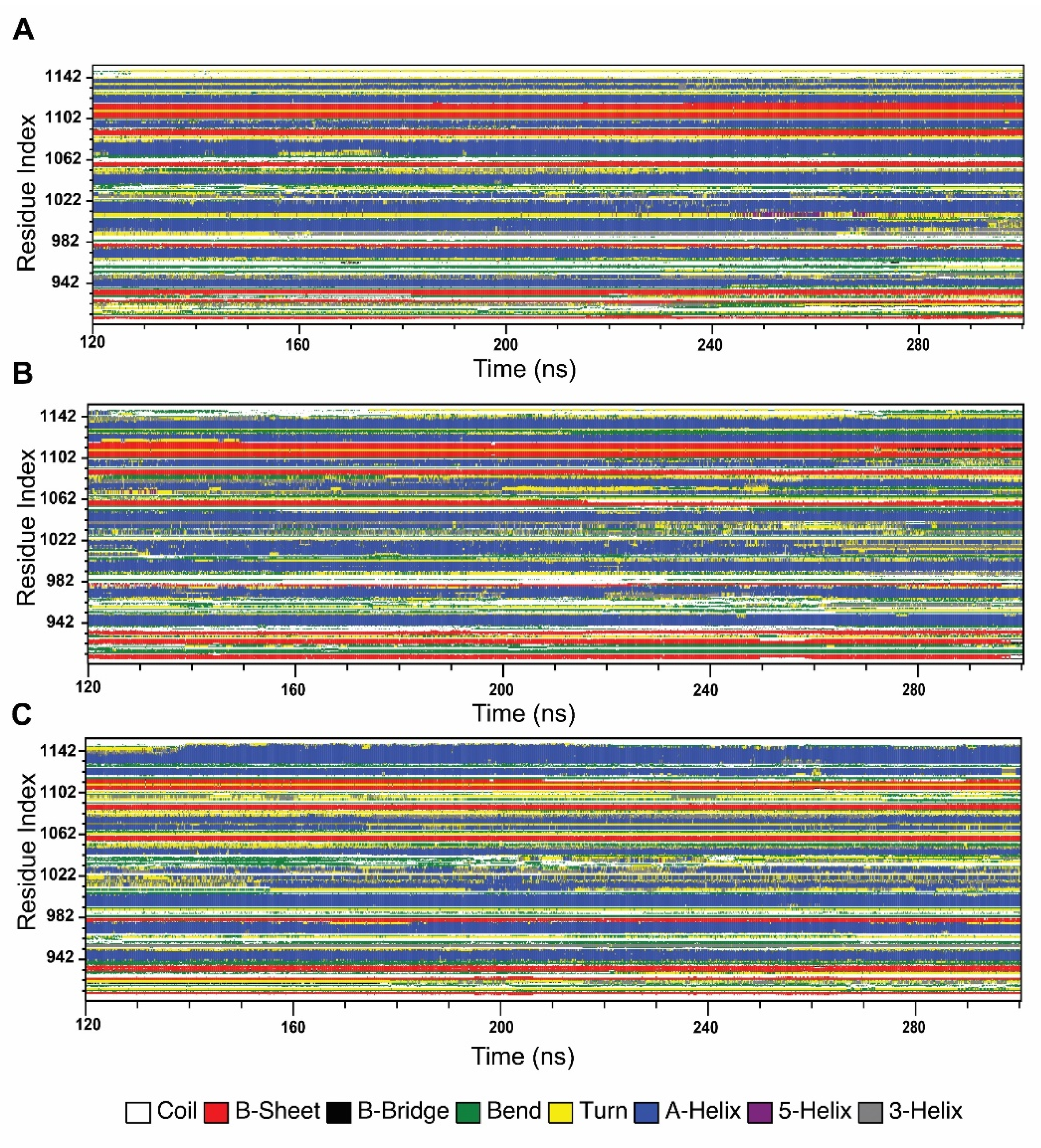
2.4.4. Effect of Variants in Protein Secondary Structure Elements
2.4.5. Free Energy Landscapes (FELs)
3. Discussion
4. Materials and Methods
4.1. Data Retrieval and In Silico Deleterious nsSNP Prediction
4.2. Conservation Analysis
4.3. Molecular Dynamic Simulation
4.3.1. Preparation of Simulation System
4.3.2. Dynamic Cross-Correlation Map (DCCM) and Principle Component Analysis (PCA)
4.3.3. Dynamic Residue Network Analysis
4.3.4. Free Energy Landscape (FEL) Analysis
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Khera, A.V.; Cuchel, M.; De La Llera-Moya, M.; Rodrigues, A.; Burke, M.F.; Jafri, K.; French, B.C.; Phillips, J.A.; Mucksavage, M.L.; Wilensky, R.L. Cholesterol efflux capacity, high-density lipoprotein function, and atherosclerosis. N. Engl. J. Med. 2011, 364, 127–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hozoji, M.; Kimura, Y.; Kioka, N.; Ueda, K. Formation of two intramolecular disulfide bonds is necessary for ApoA-I-dependent cholesterol efflux mediated by ABCA1. J. Biol. Chem. 2009, 284, 11293–11300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brunham, L.R.; Singaraja, R.R.; Duong, M.; Timmins, J.M.; Fievet, C.; Bissada, N.; Kang, M.H.; Samra, A.; Fruchart, J.C.; McManus, B.; et al. Tissue-Specific roles of ABCA1 influence susceptibility to atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2009, 29, 548–554. [Google Scholar] [CrossRef] [Green Version]
- Lawn, R.M.; Wade, D.P.; Couse, T.L.; Wilcox, J.N. Localization of human ATP-binding cassette transporter 1 (ABC1) in normal and atherosclerotic tissues. Arterioscler. Thromb. Vasc. Biol. 2001, 21, 378–385. [Google Scholar] [CrossRef]
- Kielar, D.; Dietmaier, W.; Langmann, T.; Aslanidis, C.; Probst, M.; Naruszewicz, M.; Schmitz, G. Rapid quantification of human ABCA1 mRNA in various cell types and tissues by real-time reverse transcription-PCR. Clin. Chem. 2001, 47, 2089–2097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ceccanti, M.; Cambieri, C.; Frasca, V.; Onesti, E.; Biasiotta, A.; Giordano, C.; Bruno, S.M.; Testino, G.; Lucarelli, M.; Arca, M.; et al. A novel mutation in ABCA1 gene causing tangier disease in an Italian family with uncommon neurological presentation. Front. Neurol. 2016, 7, 185. [Google Scholar] [CrossRef]
- Costet, P.; Luo, Y.; Wang, N.; Tall, A.R. Sterol-Dependent transactivation of the ABC1 promoter by the liver X receptor/retinoid X receptor. J. Biol. Chem. 2000, 275, 28240–28245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delgado-Lista, J.; Perez-Martinez, P.; Perez-Jimenez, F.; Garcia-Rios, A.; Fuentes, F.; Marin, C.; Gómez-Luna, P.; Camargo, A.; Parnell, L.D.; Ordovas, J.M.; et al. ABCA1 gene variants regulate postprandial lipid metabolism in healthy men. Arterioscler. Thromb. Vasc. Biol. 2010, 30, 1051–1057. [Google Scholar] [CrossRef]
- Anastasius, M.; Kockx, M.; Jessup, W.; Sullivan, D.; Rye, K.A.; Kritharides, L. Cholesterol efflux capacity: An introduction for clinicians. Am. Heart J. 2016, 180, 54–63. [Google Scholar] [CrossRef]
- Lewis, G.F.; Rader, D.J. New insights into the regulation of HDL metabolism and reverse cholesterol transport. Circ. Res. 2005, 96, 1221–1232. [Google Scholar] [CrossRef] [Green Version]
- Schmitz, G.; Grandl, M. The molecular mechanisms of HDL and associated vesicular trafficking mechanisms to mediate cellular lipid homeostasis. Arterioscler. Thromb. Vasc. Biol. 2009, 29, 1718–1722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Babashamsi, M.M.; Koukhaloo, S.Z.; Halalkhor, S.; Salimi, A.; Babashamsi, M. ABCA1 and metabolic syndrome; a review of the ABCA1 role in HDL-VLDL production, insulin-glucose homeostasis, inflammation and obesity. Diabetes Metab. Syndr. 2019, 13, 1529–1534. [Google Scholar] [CrossRef] [PubMed]
- Phillips, M.C. Molecular mechanisms of cellular cholesterol efflux. J. Biol. Chem. 2014, 289, 24020–24029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oram, J.F. HDL apolipoproteins and ABCA1: Partners in the removal of excess cellular cholesterol. Arterioscler. Thromb. Vasc. Biol. 2003, 23, 720–727. [Google Scholar] [CrossRef] [Green Version]
- Fitzgerald, M.L.; Mujawar, Z.; Tamehiro, N. ABC transporters, atherosclerosis and inflammation. Atherosclerosis 2010, 211, 361–370. [Google Scholar] [CrossRef] [Green Version]
- Hirsch-Reinshagen, V.; Zhou, S.; Burgess, B.L.; Bernier, L.; McIsaac, S.A.; Chan, J.Y.; Tansley, G.H.; Cohn, J.S.; Hayden, M.R.; Wellington, C.L. Deficiency of ABCA1 impairs apolipoprotein E metabolism in brain. J. Biol. Chem. 2004, 279, 41197–41207. [Google Scholar] [CrossRef] [Green Version]
- Chroni, A.; Liu, T.; Fitzgerald, M.L.; Freeman, M.W.; Zannis, V.I. Cross-linking and lipid efflux properties of apoA-I mutants suggest direct association between apoA-I helices and ABCA1. Biochemistry 2004, 43, 2126–2139. [Google Scholar] [CrossRef]
- Bodzioch, M.; Orsó, E.; Klucken, J.; Langmann, T.; Böttcher, A.; Diederich, W.; Drobnik, W.; Barlage, S.; Büchler, C.; Porsch-Özcürümez, M. The gene encoding ATP-binding cassette transporter 1 is mutated in Tangier disease. Nat. Genet. 1999, 22, 347–351. [Google Scholar] [CrossRef]
- Clee, S.M.; Zwinderman, A.H.; Engert, J.C.; Zwarts, K.Y.; Molhuizen, H.O.; Roomp, K.; Jukema, J.W.; van Wijland, M.; van Dam, M.; Hudson, T.J.; et al. Common genetic variation in ABCA1 is associated with altered lipoprotein levels and a modified risk for coronary artery disease. Circulation 2001, 103, 1198–1205. [Google Scholar] [CrossRef] [Green Version]
- Murano, T.; Yamaguchi, T.; Tatsuno, I.; Suzuki, M.; Noike, H.; Takanami, T.; Yoshida, T.; Suzuki, M.; Hashimoto, R.; Maeno, T.; et al. Subfraction analysis of circulating lipoproteins in a patient with Tangier disease due to a novel ABCA1 mutation. Clin. Chim. Acta Int. J. Clin. Chem. 2016, 452, 167–172. [Google Scholar] [CrossRef]
- Maranghi, M.; Truglio, G.; Gallo, A.; Grieco, E.; Verrienti, A.; Montali, A.; Gallo, P.; Alesini, F.; Arca, M.; Lucarelli, M. A novel splicing mutation in the ABCA1 gene, causing Tangier disease and familial HDL deficiency in a large family. Biochem. Biophys. Res. Commun. 2019, 508, 487–493. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Razek, O.; Sadananda, S.N.; Li, X.; Cermakova, L.; Frohlich, J.; Brunham, L.R. Increased prevalence of clinical and subclinical atherosclerosis in patients with damaging mutations in ABCA1 or APOA1. J. Clin. Lipidol. 2018, 12, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Jiang, X.; Cao, Y.; Juan, J.; Wu, T.; Hu, Y. Interaction between an ATP-Binding Cassette A1 (ABCA1) variant and egg consumption for the risk of ischemic stroke and carotid atherosclerosis: A family-based study in the Chinese population. J. Atheroscler. Thromb. 2019, 26, 835–845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacLeod, M.J.; De Lange, R.P.; Breen, G.; Meiklejohn, D.; Lemmon, H.; Clair, D.S. Lack of association between apolipoprotein E genoype and ischaemic stroke in a Scottish population. Eur. J. Clin. Investig. 2001, 31, 570–573. [Google Scholar] [CrossRef]
- Ghaznavi, H.; Aali, E.; Soltanpour, M.S. Association Study of the ATP—Binding cassette transporter A1 (ABCA1) Rs2230806 genetic variation with lipid profile and coronary artery disease risk in an Iranian population. Open Access Maced. J. Med. Sci. 2018, 6, 274–279. [Google Scholar] [CrossRef] [Green Version]
- Haerian, B.S.; Haerian, M.S.; Roohi, A.; Mehrad-Majd, H. ABCA1 genetic polymorphisms and type 2 diabetes mellitus and its complications. Meta Gene 2017, 13, 104–114. [Google Scholar] [CrossRef]
- Patel, D.C.; Albrecht, C.; Pavitt, D.; Paul, V.; Pourreyron, C.; Newman, S.P.; Godsland, I.F.; Valabhji, J.; Johnston, D.G. Type 2 diabetes is associated with reduced ATP-binding cassette transporter A1 gene expression, protein and function. PLoS ONE 2011, 6, e22142. [Google Scholar] [CrossRef] [Green Version]
- Hasan, M.M.; Hosen, M.B.; Rahman, M.M.; Howlader, M.Z.H.; Kabir, Y. Association of ATP binding cassette transporter 1 (ABCA 1) gene polymorphism with type 2 diabetes mellitus (T2DM) in Bangladeshi population. Gene 2019, 688, 151–154. [Google Scholar] [CrossRef]
- Cenarro, A.; Artieda, M.; Castillo, S.; Mozas, P.; Reyes, G.; Tejedor, D.; Alonso, R.; Mata, P.; Pocoví, M.; Civeira, F. A common variant in the ABCA1 gene is associated with a lower risk for premature coronary heart disease in familial hypercholesterolaemia. J. Med. Genet. 2003, 40, 163–168. [Google Scholar] [CrossRef] [Green Version]
- Xiong, T.; Xu, G.; Huang, X.L.; Lu, K.Q.; Xie, W.Q.; Yin, K.; Tu, J. ATP-Binding cassette transporter A1: A promising therapy target for prostate cancer. Mol. Clin. Oncol. 2018, 8, 9–14. [Google Scholar] [CrossRef]
- Bi, D.P.; Yin, C.H.; Zhang, X.Y.; Yang, N.N.; Xu, J.Y. MiR-183 functions as an oncogene by targeting ABCA1 in colon cancer. Oncol. Rep. 2016, 35, 2873–2879. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaneko, T.; Kanno, C.; Ichikawa-Tomikawa, N.; Kashiwagi, K.; Yaginuma, N.; Ohkoshi, C.; Tanaka, M.; Sugino, T.; Imura, T.; Hasegawa, H.; et al. Liver X receptor reduces proliferation of human oral cancer cells by promoting cholesterol efflux via up-regulation of ABCA1 expression. Oncotarget 2015, 6, 33345–33357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wollmer, M.A.; Streffer, J.R.; Lütjohann, D.; Tsolaki, M.; Iakovidou, V.; Hegi, T.; Pasch, T.; Jung, H.H.; Bergmann, K.; Nitsch, R.M.; et al. ABCA1 modulates CSF cholesterol levels and influences the age at onset of Alzheimer’s disease. Neurobiol. Aging 2003, 24, 421–426. [Google Scholar] [CrossRef]
- Allikmets, R.; Shroyer, N.F.; Singh, N.; Seddon, J.M.; Lewis, R.A.; Bernstein, P.S.; Peiffer, A.; Zabriskie, N.A.; Li, Y.; Hutchinson, A.; et al. Mutation of the Stargardt disease gene (ABCR) in age-related macular degeneration. Science 1997, 277, 1805–1807. [Google Scholar] [CrossRef] [Green Version]
- Bochem, A.E.; van der Valk, F.M.; Tolani, S.; Stroes, E.S.; Westerterp, M.; Tall, A.R. Increased systemic and plaque inflammation in ABCA1 mutation carriers with attenuation by statins. Arterioscler. Thromb. Vasc. Biol. 2015, 35, 1663–1669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gillet, J.P.; Efferth, T.; Remacle, J. Chemotherapy-Induced resistance by ATP-binding cassette transporter genes. Biochim. Biophys. Acta 2007, 1775, 237–262. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.L.; Liu, Y.H.; Meng, L.L.; Li, C.G.; Zhou, S.F. Phenotype prediction of non-synonymous single-nucleotide polymorphisms in human ATP-binding cassette transporter genes. Basic Clin. Pharmacol. Toxicol. 2011, 108, 94–114. [Google Scholar] [CrossRef]
- Frikke-Schmidt, R.; Nordestgaard, B.G.; Jensen, G.B.; Tybjaerg-Hansen, A. Genetic variation in ABC transporter A1 contributes to HDL cholesterol in the general population. J. Clin. Investig. 2004, 114, 1343–1353. [Google Scholar] [CrossRef] [Green Version]
- Marín-Martín, F.R.; Soler-Rivas, C.; Martín-Hernández, R.; Rodriguez-Casado, A. A comprehensive in silico analysis of the functional and structural impact of nonsynonymous SNPs in the ABCA1 transporter gene. Cholesterol 2014, 2014. [Google Scholar] [CrossRef] [Green Version]
- Aldridge, J.E.; Horibe, T.; Hoogenraad, N.J. Discovery of genes activated by the mitochondrial unfolded protein response (mtUPR) and cognate promoter elements. PLoS ONE 2007, 2, e874. [Google Scholar] [CrossRef]
- Remali, J.; Aizat, W.M.; Ng, C.L.; Lim, Y.C.; Mohamed-Hussein, Z.-A.; Fazry, S. In silico analysis on the functional and structural impact of Rad50 mutations involved in DNA strand break repair. PeerJ 2020, 8, e9197. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Luo, Z.; Jia, A.; Yu, L.; Muhammad, I.; Zeng, W.; Song, Y. Associations of the ABCA1 gene polymorphisms with plasma lipid levels: A meta-analysis. Medicine 2018, 97, e13521. [Google Scholar] [CrossRef] [PubMed]
- Arifuzzaman, M.; Mitra, S.; Das, R.; Hamza, A.; Absar, N.; Dash, R. In silico analysis of nonsynonymous single-nucleotide polymorphisms (nsSNPs) of the SMPX gene. Ann. Hum. Genet. 2020, 84, 54–71. [Google Scholar] [CrossRef] [PubMed]
- Sherry, S.T.; Ward, M.-H.; Kholodov, M.; Baker, J.; Phan, L.; Smigielski, E.M.; Sirotkin, K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001, 29, 308–311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hosen, S.M.Z.; Dash, R.; Junaid, M.; Mitra, S.; Absar, N. Identification and structural characterization of deleterious non-synonymous single nucleotide polymorphisms in the human SKP2 gene. Comput. Biol. Chem. 2019, 79, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Repa, J.; Turley, S.; Lobaccaro, J.-M.; Medina, J.; Li, L.; Lustig, K.; Shan, B.; Heyman, R.; Dietschy, J.; Mangelsdorf, D. Regulation of absorption and ABC1-mediated efflux of cholesterol by RXR heterodimers. Science 2000, 289, 1524–1529. [Google Scholar] [CrossRef]
- Fitzgerald, M.L.; Morris, A.L.; Rhee, J.S.; Andersson, L.P.; Mendez, A.J.; Freeman, M.W. Naturally occurring mutations in the largest extracellular loops of ABCA1 can disrupt its direct interaction with apolipoprotein AI. J. Biol. Chem. 2002, 277, 33178–33187. [Google Scholar] [CrossRef] [Green Version]
- Biemans-Oldehinkel, E.; Doeven, M.K.; Poolman, B. ABC transporter architecture and regulatory roles of accessory domains. FEBS Lett. 2006, 580, 1023–1035. [Google Scholar] [CrossRef] [Green Version]
- Peelman, F.; Labeur, C.; Vanloo, B.; Roosbeek, S.; Devaud, C.; Duverger, N.; Denèfle, P.; Rosier, M.; Vandekerckhove, J.; Rosseneu, M. Characterization of the ABCA transporter subfamily: Identification of prokaryotic and eukaryotic members, phylogeny and topology. J. Mol. Biol. 2003, 325, 259–274. [Google Scholar] [CrossRef]
- Qian, H.; Zhao, X.; Cao, P.; Lei, J.; Yan, N.; Gong, X. Structure of the human lipid exporter ABCA1. Cell 2017, 169, 1228–1239. [Google Scholar] [CrossRef] [Green Version]
- Mitra, S.; Dash, R. Structural dynamics and quantum mechanical aspects of shikonin derivatives as CREBBP bromodomain inhibitors. J. Mol. Graph. Model. 2018, 83, 42–52. [Google Scholar] [CrossRef] [PubMed]
- Dash, R.; Choi, H.J.; Moon, I.S. Mechanistic insights into the deleterious roles of Nasu-Hakola disease associated TREM2 variants. Sci. Rep. 2020, 10, 3663. [Google Scholar] [CrossRef] [PubMed]
- Dash, R.; Junaid, M.; Mitra, S.; Arifuzzaman, M.; Hosen, S.M.Z. Structure-Based identification of potent VEGFR-2 inhibitors from in vivo metabolites of a herbal ingredient. J. Mol. Model. 2019, 25, 98. [Google Scholar] [CrossRef] [PubMed]
- Pandey, B.; Grover, S.; Goyal, S.; Kumari, A.; Singh, A.; Jamal, S.; Kaur, J.; Grover, A. Alanine mutation of the catalytic sites of Pantothenate Synthetase causes distinct conformational changes in the ATP binding region. Sci. Rep. 2018, 8, 903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, D.K.; Penkler, D.L.; Sheik Amamuddy, O.; Ross, C.; Atilgan, A.R.; Atilgan, C.; Tastan Bishop, Ö. MD-TASK: A software suite for analyzing molecular dynamics trajectories. Bioinformatics 2017, 33, 2768–2771. [Google Scholar] [CrossRef] [Green Version]
- Amusengeri, A.; Tata, R.B.; Tastan Bishop, Ö. Understanding the pyrimethamine drug resistance mechanism via combined molecular dynamics and dynamic residue network analysis. Molecules 2020, 25, 904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanyanga, T.A.; Tastan Bishop, Ö. Structural characterization of carbonic anhydrase VIII and effects of missense single nucleotide variations to protein structure and function. Int. J. Mol. Sci. 2020, 21, 2764. [Google Scholar] [CrossRef] [PubMed]
- Sheik Amamuddy, O.; Musyoka, T.M.; Boateng, R.A.; Zabo, S.; Tastan Bishop, Ö. Determining the unbinding events and conserved motions associated with the pyrazinamide release due to resistance mutations of Mycobacterium tuberculosis pyrazinamidase. Comput. Struct. Biotechnol. J. 2020, 18, 1103–1120. [Google Scholar] [CrossRef] [PubMed]
- Dash, R.; Arifuzzaman, M.; Mitra, S.; Abdul Hannan, M.; Absar, N.; Hosen, S.M.Z. Unveiling the structural insights into the selective inhibition of protein kinase D1. Curr. Pharm. Des. 2019, 25, 1059–1074. [Google Scholar] [CrossRef] [PubMed]
- Wall, S.M. The renal physiology of pendrin (SLC26A4) and its role in hypertension. Novartis Found. Symp. 2006, 273, 231–239, discussion 239–243, 234–261. [Google Scholar]
- Ferrer-Costa, C.; Orozco, M.; de la Cruz, X. Characterization of disease-associated single amino acid polymorphisms in terms of sequence and structure properties. J. Mol. Biol. 2002, 315, 771–786. [Google Scholar] [CrossRef] [PubMed]
- Chitrala, K.N.; Yeguvapalli, S. Computational screening and molecular dynamic simulation of breast cancer associated deleterious non-synonymous single nucleotide polymorphisms in TP53 gene. PLoS ONE 2014, 9, e104242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masoodi, T.A.; Al Shammari, S.A.; Al-Muammar, M.N.; Alhamdan, A.A.; Talluri, V.R. Exploration of deleterious single nucleotide polymorphisms in late-onset Alzheimer disease susceptibility genes. Gene 2013, 512, 429–437. [Google Scholar] [CrossRef] [PubMed]
- Abdul Samad, F.; Suliman, B.A.; Basha, S.H.; Manivasagam, T.; Essa, M.M. A comprehensive in silico analysis on the structural and functional impact of SNPs in the congenital heart defects associated with NKX2-5 Gene-A molecular dynamic simulation approach. PLoS ONE 2016, 11, e0153999. [Google Scholar] [CrossRef] [Green Version]
- Tanwar, H.; Kumar, D.T.; Doss, C.G.P.; Zayed, H. Bioinformatics classification of mutations in patients with Mucopolysaccharidosis IIIA. Metab. Brain Dis. 2019, 34, 1577–1594. [Google Scholar] [CrossRef] [Green Version]
- Hasnain, M.J.U.; Shoaib, M.; Qadri, S.; Afzal, B.; Anwar, T.; Abbas, S.H.; Sarwar, A.; Talha Malik, H.M.; Tariq Pervez, M. Computational analysis of functional single nucleotide polymorphisms associated with SLC26A4 gene. PLoS ONE 2020, 15, e0225368. [Google Scholar] [CrossRef]
- Chun, S.; Fay, J.C. Identification of deleterious mutations within three human genomes. Genome Res. 2009, 19, 1553–1561. [Google Scholar] [CrossRef] [Green Version]
- Singaraja, R.R.; Brunham, L.R.; Visscher, H.; Kastelein, J.J.; Hayden, M.R. Efflux and atherosclerosis: The clinical and biochemical impact of variations in the ABCA1 gene. Arterioscler. Thromb. Vasc. Biol. 2003, 23, 1322–1332. [Google Scholar] [CrossRef] [Green Version]
- Szöllősi, D.; Rose-Sperling, D.; Hellmich, U.A.; Stockner, T. Comparison of mechanistic transport cycle models of ABC exporters. Biochim. Biophys. Acta BBA Biomembr. 2018, 1860, 818–832. [Google Scholar] [CrossRef]
- Smith, P.C.; Karpowich, N.; Millen, L.; Moody, J.E.; Rosen, J.; Thomas, P.J.; Hunt, J.F. ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer. Mol. Cell 2002, 10, 139–149. [Google Scholar] [CrossRef] [Green Version]
- Higgins, C.F. ABC transporters: From microorganisms to man. Annu. Rev. Cell Biol. 1992, 8, 67–113. [Google Scholar] [CrossRef] [PubMed]
- Melin, P.; Thoreau, V.; Norez, C.; Bilan, F.; Kitzis, A.; Becq, F. The cystic fibrosis mutation G1349D within the signature motif LSHGH of NBD2 abolishes the activation of CFTR chloride channels by genistein. Biochem. Pharmacol. 2004, 67, 2187–2196. [Google Scholar] [CrossRef] [PubMed]
- Szakács, G.; Ozvegy, C.; Bakos, E.; Sarkadi, B.; Váradi, A. Role of glycine-534 and glycine-1179 of human multidrug resistance protein (MDR1) in drug-mediated control of ATP hydrolysis. Biochem. J. 2001, 356, 71–75. [Google Scholar] [CrossRef] [PubMed]
- Loo, T.W.; Bartlett, M.C.; Clarke, D.M. The “LSGGQ” motif in each nucleotide-binding domain of human P-glycoprotein is adjacent to the opposing walker A sequence. J. Biol. Chem. 2002, 277, 41303–41306. [Google Scholar] [CrossRef] [Green Version]
- Perkins, K.J.; Byers, S.; Yogalingam, G.; Weber, B.; Hopwood, J.J. Expression and characterization of wild type and mutant recombinant human sulfamidase. Implications for Sanfilippo (Mucopolysaccharidosis IIIA) syndrome. J. Biol. Chem. 1999, 274, 37193–37199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iatan, I.; Alrasadi, K.; Ruel, I.; Alwaili, K.; Genest, J. Effect of ABCA1 mutations on risk for myocardial infarction. Curr. Atheroscler. Rep. 2008, 10, 413–426. [Google Scholar] [CrossRef] [PubMed]
- Brunham, L.R.; Singaraja, R.R.; Hayden, M.R. Variations on a gene: Rare and common variants in ABCA1 and their impact on HDL cholesterol levels and atherosclerosis. Annu. Rev. Nutr. 2006, 26, 105–129. [Google Scholar] [CrossRef]
- Frikke-Schmidt, R.; Nordestgaard, B.G.; Schnohr, P.; Steffensen, R.; Tybjaerg-Hansen, A. Mutation in ABCA1 predicted risk of ischemic heart disease in the Copenhagen City Heart Study Population. J. Am. Coll. Cardiol. 2005, 46, 1516–1520. [Google Scholar] [CrossRef] [Green Version]
- Frikke-Schmidt, R.; Nordestgaard, B.G.; Jensen, G.B.; Steffensen, R.; Tybjaerg-Hansen, A. Genetic variation in ABCA1 predicts ischemic heart disease in the general population. Arterioscler. Thromb. Vasc. Biol. 2008, 28, 180–186. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Ji, Y.; Chen, X.; Song, Y.; Huang, S.; Zhou, C.; Huang, C.; Chen, Z.; Zhang, L.; Ge, J. ABCA1 variants rs2230806 (R219K), rs4149313 (M8831I), and rs9282541 (R230C) are associated with susceptibility to coronary heart disease. J. Clin. Lab. Anal. 2019, 33, e22896. [Google Scholar] [CrossRef]
- Mokuno, J.; Hishida, A.; Morita, E.; Sasakabe, T.; Hattori, Y.; Suma, S.; Okada, R.; Kawai, S.; Naito, M.; Wakai, K. ATP-binding cassette transporter A1 (ABCA1) R219K (G1051A, rs2230806) polymorphism and serum high-density lipoprotein cholesterol levels in a large Japanese population: Cross-Sectional data from the Daiko Study. Endocr. J. 2015, 62, 543–549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, Z.; Wang, J.; Chen, W.; Wang, P.; Zeng, H.; Chen, W. Association studies of several cholesterol-related genes (ABCA1, CETP and LIPC) with serum lipids and risk of Alzheimer’s disease. Lipids Health Dis. 2012, 11, 163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puntoni, M.; Sbrana, F.; Bigazzi, F.; Sampietro, T. Tangier disease: Epidemiology, pathophysiology, and management. Am. J. Cardiovasc. Drugs Drugs Devices Other Interv. 2012, 12, 303–311. [Google Scholar] [CrossRef]
- Bhagwat, M. Searching NCBI’s dbSNP database. Curr. Protoc. Bioinform. 2010. [Google Scholar] [CrossRef]
- Ng, P.C.; Henikoff, S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003, 31, 3812–3814. [Google Scholar] [CrossRef] [Green Version]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef] [Green Version]
- Choi, Y.; Sims, G.E.; Murphy, S.; Miller, J.R.; Chan, A.P. Predicting the functional effect of amino acid substitutions and indels. PLoS ONE 2012, 7, e46688. [Google Scholar] [CrossRef] [Green Version]
- Capriotti, E.; Fariselli, P.; Casadio, R. I-Mutant2.0: Predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005, 33, W306–W310. [Google Scholar] [CrossRef] [Green Version]
- Bendl, J.; Stourac, J.; Salanda, O.; Pavelka, A.; Wieben, E.D.; Zendulka, J.; Brezovsky, J.; Damborsky, J. PredictSNP: Robust and accurate consensus classifier for prediction of disease-related mutations. PLoS Comput. Biol. 2014, 10, e1003440. [Google Scholar] [CrossRef]
- Thomas, P.D.; Campbell, M.J.; Kejariwal, A.; Mi, H.; Karlak, B.; Daverman, R.; Diemer, K.; Muruganujan, A.; Narechania, A. PANTHER: A library of protein families and subfamilies indexed by function. Genome Res. 2003, 13, 2129–2141. [Google Scholar] [CrossRef] [Green Version]
- Capriotti, E.; Calabrese, R.; Fariselli, P.; Martelli, P.L.; Altman, R.B.; Casadio, R. WS-SNPs&GO: A web server for predicting the deleterious effect of human protein variants using functional annotation. BMC Genom. 2013, 14, S6. [Google Scholar]
- Capriotti, E.; Calabrese, R.; Casadio, R. Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information. Bioinformatics 2006, 22, 2729–2734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stone, E.A.; Sidow, A. Physicochemical constraint violation by missense substitutions mediates impairment of protein function and disease severity. Genome Res. 2005, 15, 978–986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sunyaev, S.R.; Eisenhaber, F.; Rodchenkov, I.V.; Eisenhaber, B.; Tumanyan, V.G.; Kuznetsov, E.N. PSIC: Profile extraction from sequence alignments with position-specific counts of independent observations. Protein Eng. 1999, 12, 387–394. [Google Scholar] [CrossRef] [Green Version]
- Choi, Y.; Chan, A.P. PROVEAN web server: A tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 2015, 31, 2745–2747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzek, B.E.; Huang, H.; McGarvey, P.; Mazumder, R.; Wu, C.H. UniRef: Comprehensive and non-redundant UniProt reference clusters. Bioinformatics 2007, 23, 1282–1288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Celniker, G.; Nimrod, G.; Ashkenazy, H.; Glaser, F.; Martz, E.; Mayrose, I.; Pupko, T.; Ben-Tal, N. ConSurf: Using evolutionary data to raise testable hypotheses about protein function. Isr. J. Chem. 2013, 53, 199–206. [Google Scholar] [CrossRef]
- Ashkenazy, H.; Erez, E.; Martz, E.; Pupko, T.; Ben-Tal, N. ConSurf 2010: Calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010, 38, W529–W533. [Google Scholar] [CrossRef] [Green Version]
- Mayrose, I.; Graur, D.; Ben-Tal, N.; Pupko, T. Comparison of site-specific rate-inference methods for protein sequences: Empirical Bayesian methods are superior. Mol. Biol. Evol. 2004, 21, 1781–1791. [Google Scholar] [CrossRef] [Green Version]
- Schrödinger, LLC. Tools: Schrödinger Suite 2017-1; Schrödinger, LLC: New York, NY, USA, 2017. [Google Scholar]
- Dash, R.; Ali, M.C.; Dash, N.; Azad, M.A.K.; Hosen, S.M.Z.; Hannan, M.A.; Moon, I.S. Structural and dynamic characterizations highlight the deleterious role of SULT1A1 R213H polymorphism in substrate binding. Int. J. Mol. Sci. 2019, 20, 6256. [Google Scholar] [CrossRef] [Green Version]
- Krieger, E.; Darden, T.; Nabuurs, S.B.; Finkelstein, A.; Vriend, G. Making optimal use of empirical energy functions: Force-Field parameterization in crystal space. Proteins Struct. Funct. Bioinform. 2004, 57, 678–683. [Google Scholar] [CrossRef] [PubMed]
- Land, H.; Humble, M.S. YASARA: A tool to obtain structural guidance in biocatalytic investigations. Methods Mol. Biol. 2018, 1685, 43–67. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Harrach, M.F.; Drossel, B. Structure and dynamics of TIP3P, TIP4P, and TIP5P water near smooth and atomistic walls of different hydroaffinity. J. Chem. Phys. 2014, 140, 174501. [Google Scholar] [CrossRef] [PubMed]
- Krieger, E.; Dunbrack, R.L., Jr.; Hooft, R.W.; Krieger, B. Assignment of protonation states in proteins and ligands: Combining pKa prediction with hydrogen bonding network optimization. Methods Mol. Biol. 2012, 819, 405–421. [Google Scholar] [CrossRef]
- Krieger, E.; Nielsen, J.E.; Spronk, C.A.; Vriend, G. Fast empirical pKa prediction by Ewald summation. J. Mol. Graph. Model. 2006, 25, 481–486. [Google Scholar] [CrossRef]
- Krieger, E.; Vriend, G. New ways to boost molecular dynamics simulations. J. Comput. Chem. 2015, 36, 996–1007. [Google Scholar] [CrossRef]
- Harvey, M.J.; De Fabritiis, G. An implementation of the Smooth Particle Mesh Ewald Method on GPU hardware. J. Chem. Theory Comput. 2009, 5, 2371–2377. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef] [Green Version]
- Krieger, E.; Koraimann, G.; Vriend, G. Increasing the precision of comparative models with YASARA NOVA—A self-parameterizing force field. Proteins 2002, 47, 393–402. [Google Scholar] [CrossRef]
- Stukowski, A. Visualization and analysis of atomistic simulation data with OVITO—The Open Visualization Tool. Model. Simul. Mater. Sci. Eng. 2009, 18, 015012. [Google Scholar] [CrossRef]
- Frishman, D.; Argos, P. Knowledge-Based protein secondary structure assignment. Proteins 1995, 23, 566–579. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W.; Sander, C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 1983, 22, 2577–2637. [Google Scholar] [CrossRef] [PubMed]
- Grant, B.J.; Rodrigues, A.P.; ElSawy, K.M.; McCammon, J.A.; Caves, L.S. Bio3d: An R package for the comparative analysis of protein structures. Bioinformatics 2006, 22, 2695–2696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ichiye, T.; Karplus, M. Collective motions in proteins: A covariance analysis of atomic fluctuations in molecular dynamics and normal mode simulations. Proteins 1991, 11, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Atilgan, A.R.; Akan, P.; Baysal, C. Small-World communication of residues and significance for protein dynamics. Biophys. J. 2004, 86, 85–91. [Google Scholar] [CrossRef] [Green Version]
- Sanyanga, T.A.; Nizami, B.; Bishop, Ö.T. Mechanism of action of Non-synonymous single nucleotide variations associated with α-carbonic Anhydrase II deficiency. Molecules 2019, 24, 3987. [Google Scholar] [CrossRef] [Green Version]
- Frauenfelder, H.; Sligar, S.G.; Wolynes, P.G. The energy landscapes and motions of proteins. Science 1991, 254, 1598–1603. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
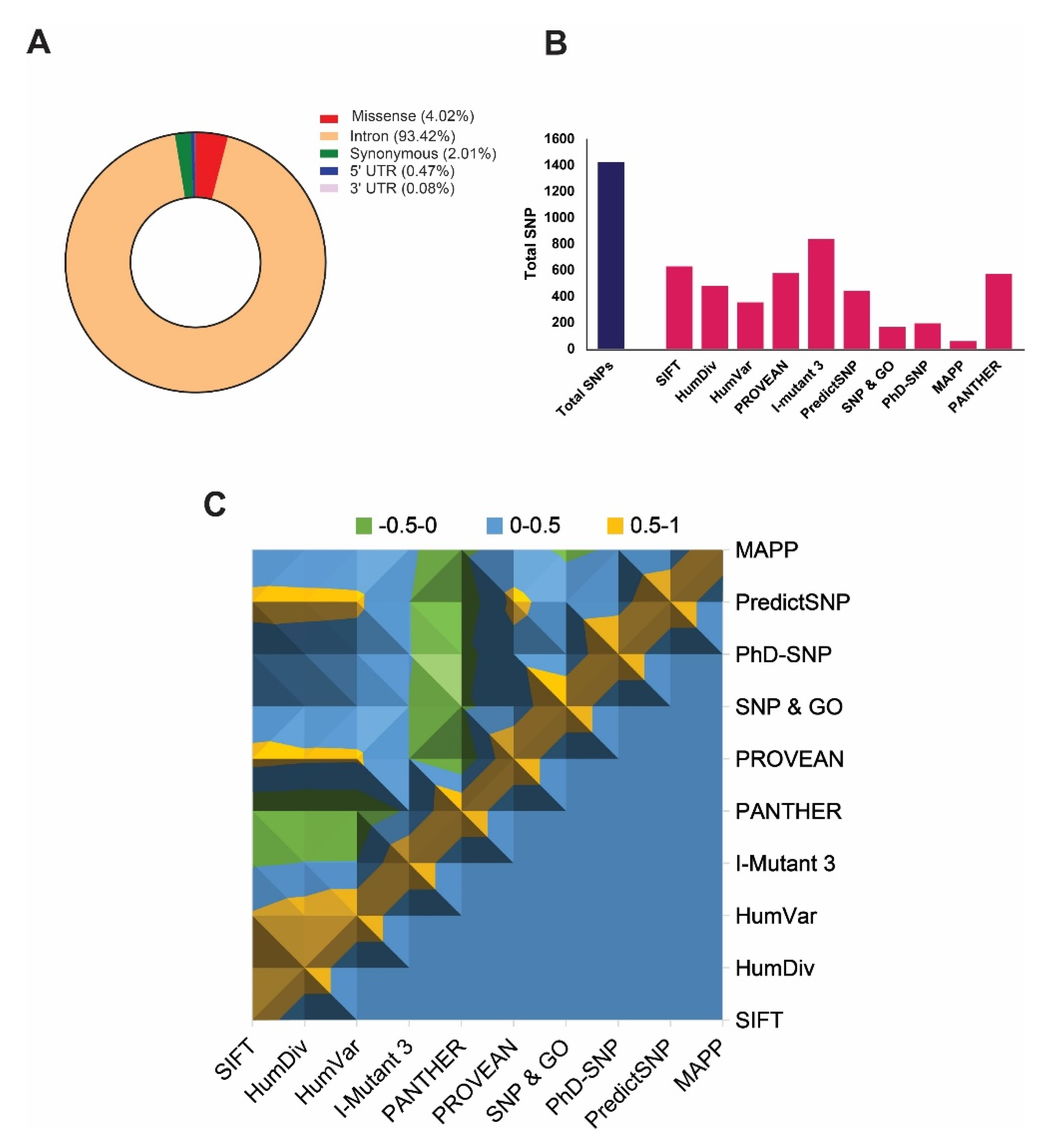


| rs ID | Substitution | SIFT | PolyPhen 2 | MAPP | PANTHER | SNP & Go | PhD-SNP | PredictSNP | PROVEAN | I-Mutant 3 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| HumDiv | HumVar | ||||||||||
| rs762112742 | S741C | 0.05 * | 1 * | 0.996 * | - | 0.663 * | 0.761 * | 0.803 * | D * | −4.265 * | −1.01 * |
| rs1461682152 | Y793C | 0.05 * | 0.994 * | 0.964 * | - | 0.654 * | - | 0.607 * | D * | −6.46 * | −0.86 * |
| rs985622413 | G1050V | 0 * | 1 * | 0.999 * | - | 0.568 * | - | 0.733 * | D * | −6.192 * | −0.58 * |
| rs1394779021 | S1067C | 0 * | 0.999 * | 0.954 * | - | 0.671 * | - | 0.858 * | D * | −3.494 * | −0.58 * |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dash, R.; Ali, M.C.; Rana, M.L.; Munni, Y.A.; Barua, L.; Jahan, I.; Haque, M.F.; Hannan, M.A.; Moon, I.S. Computational SNP Analysis and Molecular Simulation Revealed the Most Deleterious Missense Variants in the NBD1 Domain of Human ABCA1 Transporter. Int. J. Mol. Sci. 2020, 21, 7606. https://doi.org/10.3390/ijms21207606
Dash R, Ali MC, Rana ML, Munni YA, Barua L, Jahan I, Haque MF, Hannan MA, Moon IS. Computational SNP Analysis and Molecular Simulation Revealed the Most Deleterious Missense Variants in the NBD1 Domain of Human ABCA1 Transporter. International Journal of Molecular Sciences. 2020; 21(20):7606. https://doi.org/10.3390/ijms21207606
Chicago/Turabian StyleDash, Raju, Md. Chayan Ali, Md. Liton Rana, Yeasmin Akter Munni, Largess Barua, Israt Jahan, Mst. Fatema Haque, Md. Abdul Hannan, and Il Soo Moon. 2020. "Computational SNP Analysis and Molecular Simulation Revealed the Most Deleterious Missense Variants in the NBD1 Domain of Human ABCA1 Transporter" International Journal of Molecular Sciences 21, no. 20: 7606. https://doi.org/10.3390/ijms21207606