Wheat Transcription Factor TaSNAC11-4B Positively Regulates Leaf Senescence through Promoting ROS Production in Transgenic Arabidopsis
Abstract
:1. Introduction
2. Results
2.1. TaSNAC11-4B Expression Is Associated with Leaf Senescence and Can Be Induced by Stress Treatments
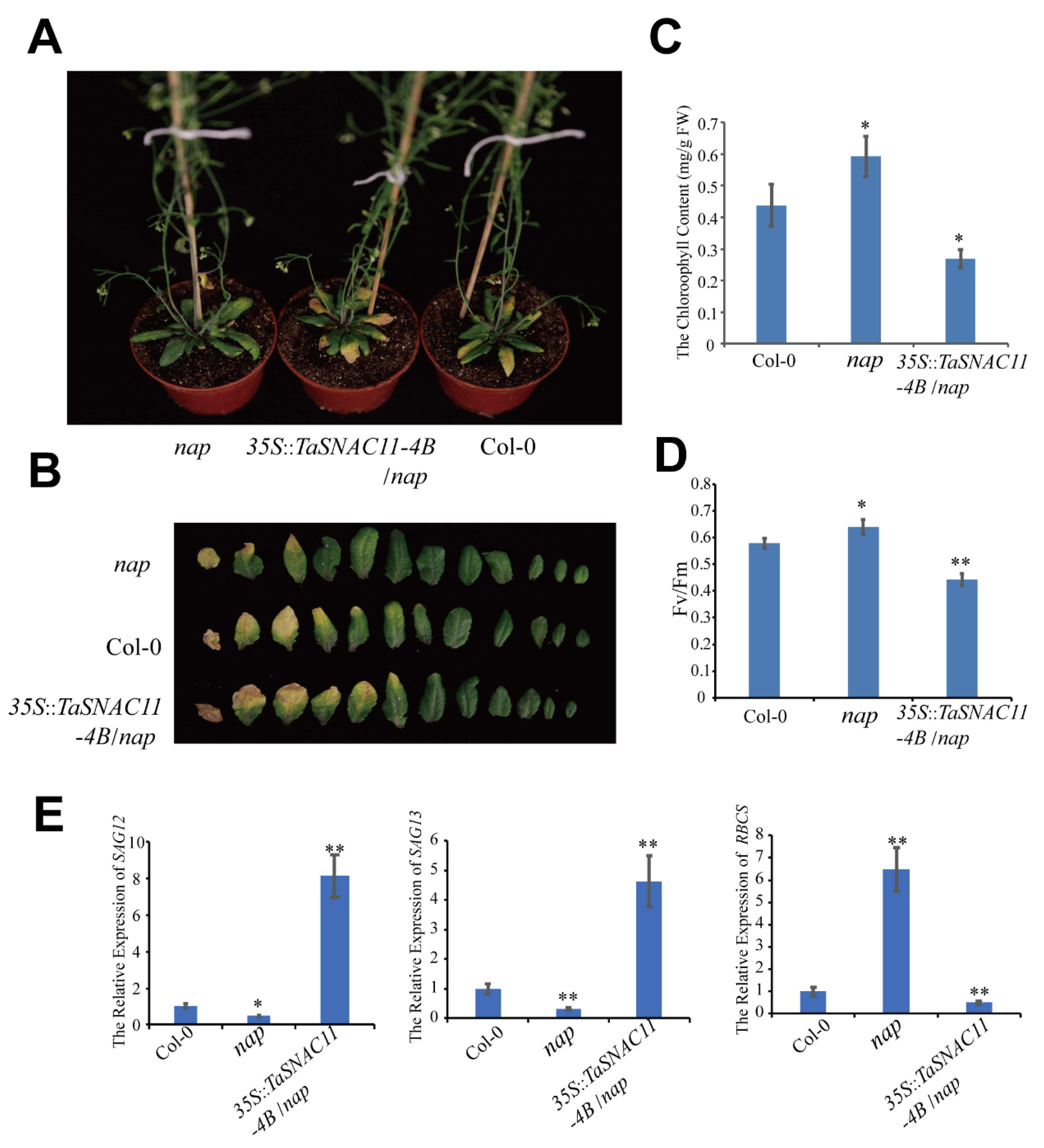
2.2. TaSNAC11-4B Is a Functional Homolog of AtNAP
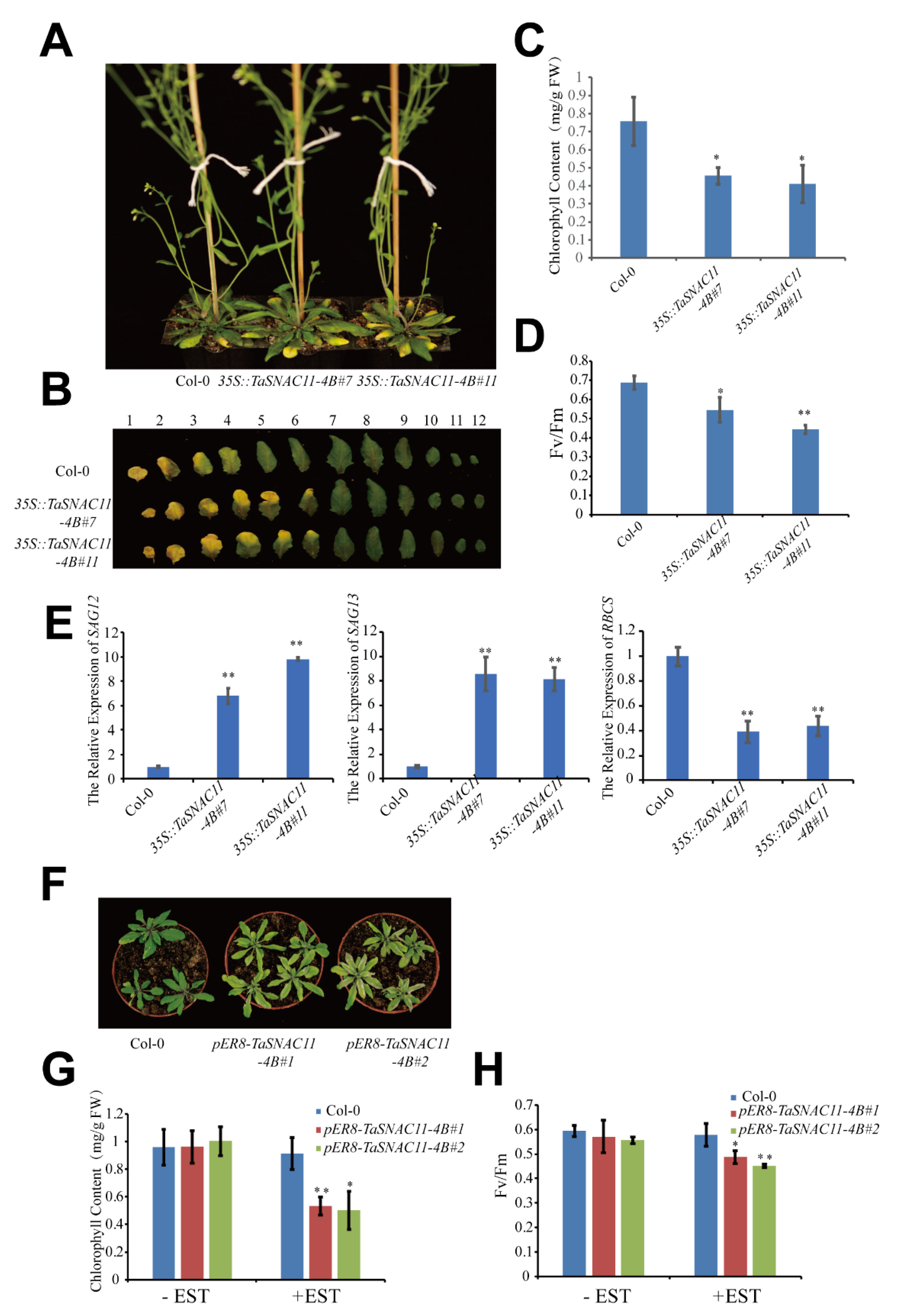
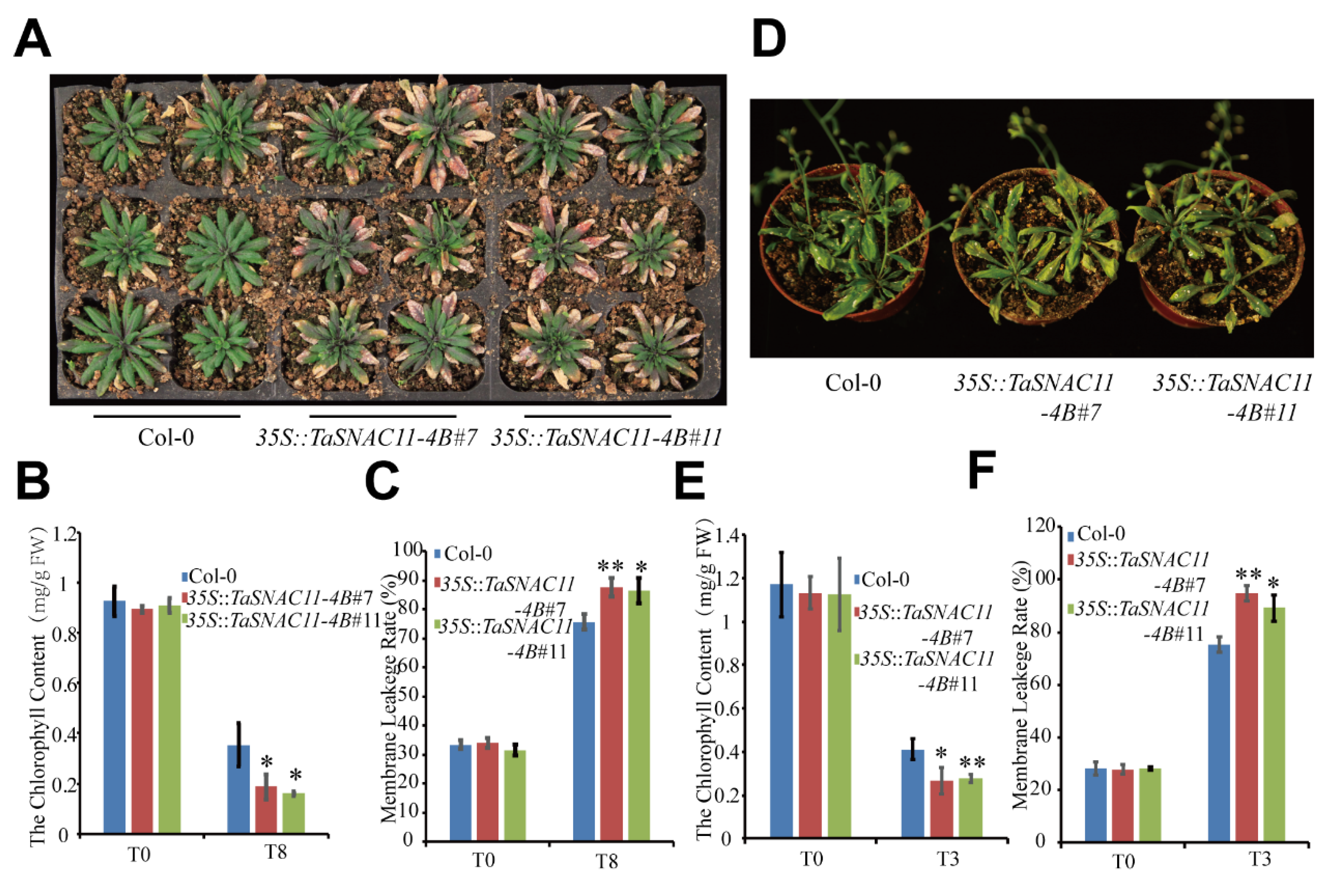
2.3. TaSNAC11-4B Plays a Positive Role in Regulating Leaf Senescence
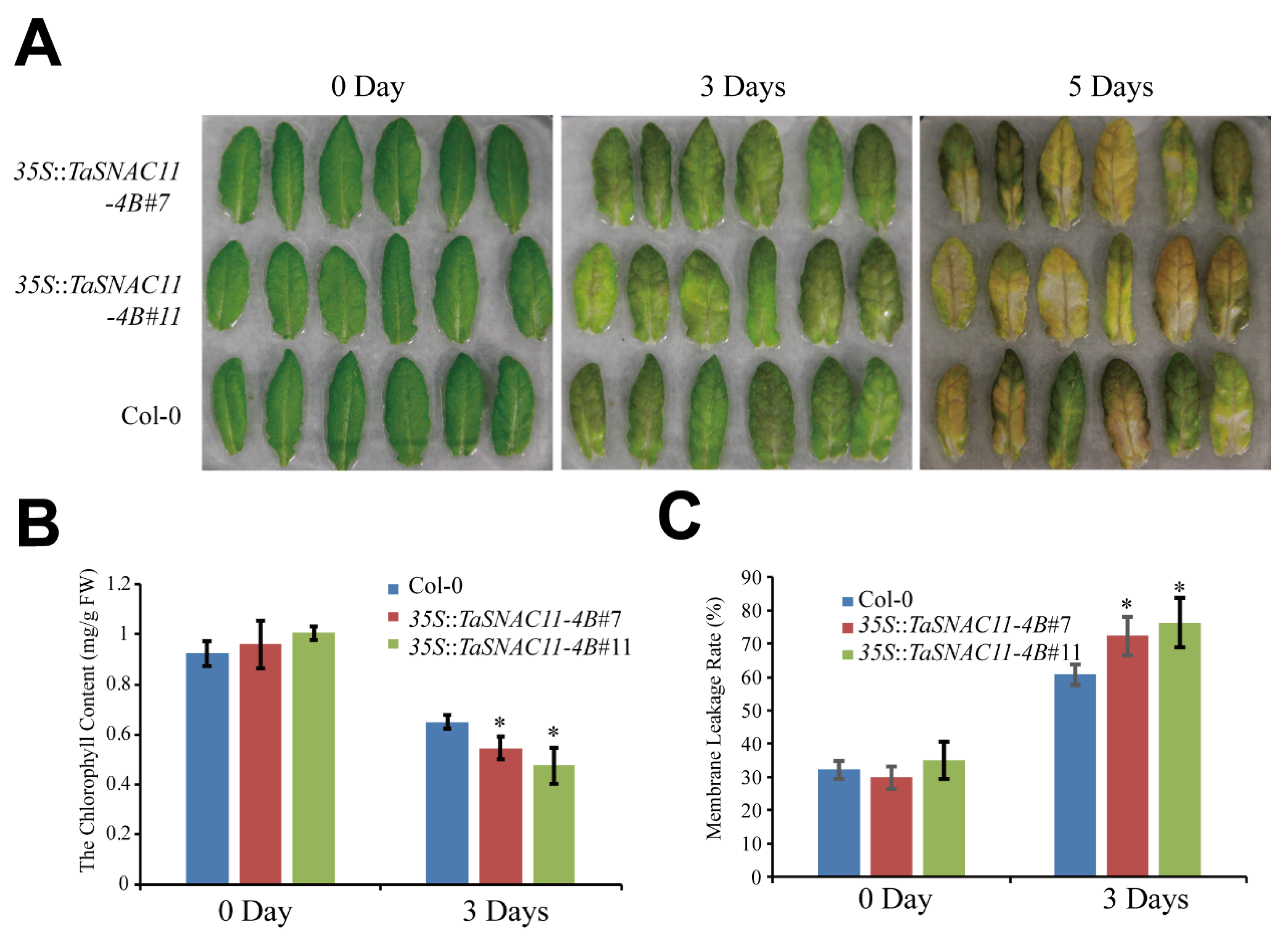
2.4. TaSNAC11-4B Functions to Promote Drought-Induced Leaf Senescence
2.5. TaSNAC11-4B Is Involved in ABA-Induced Leaf Senescence
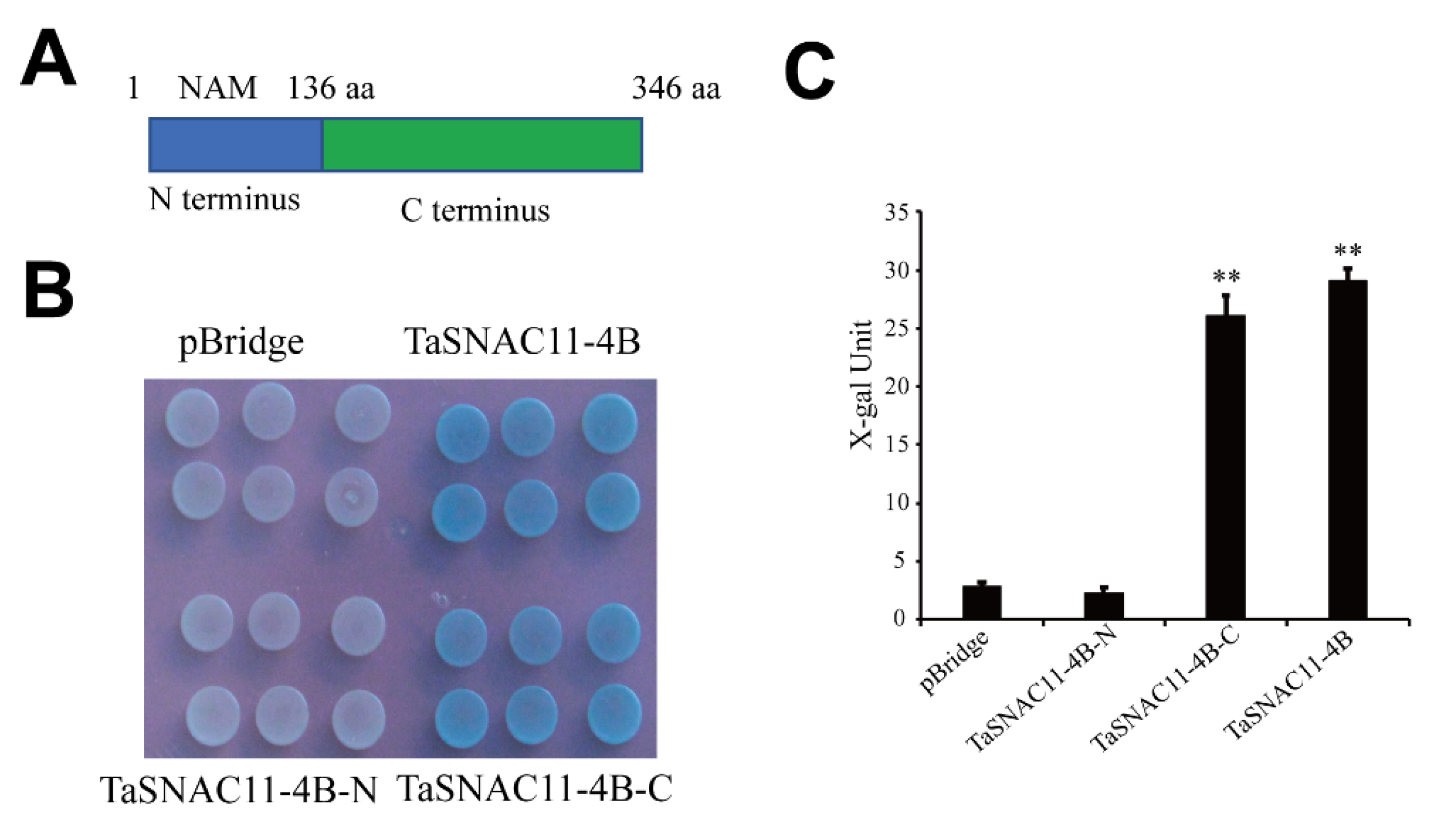
2.6. Transcriptional Activity of TaSNAC11-4B
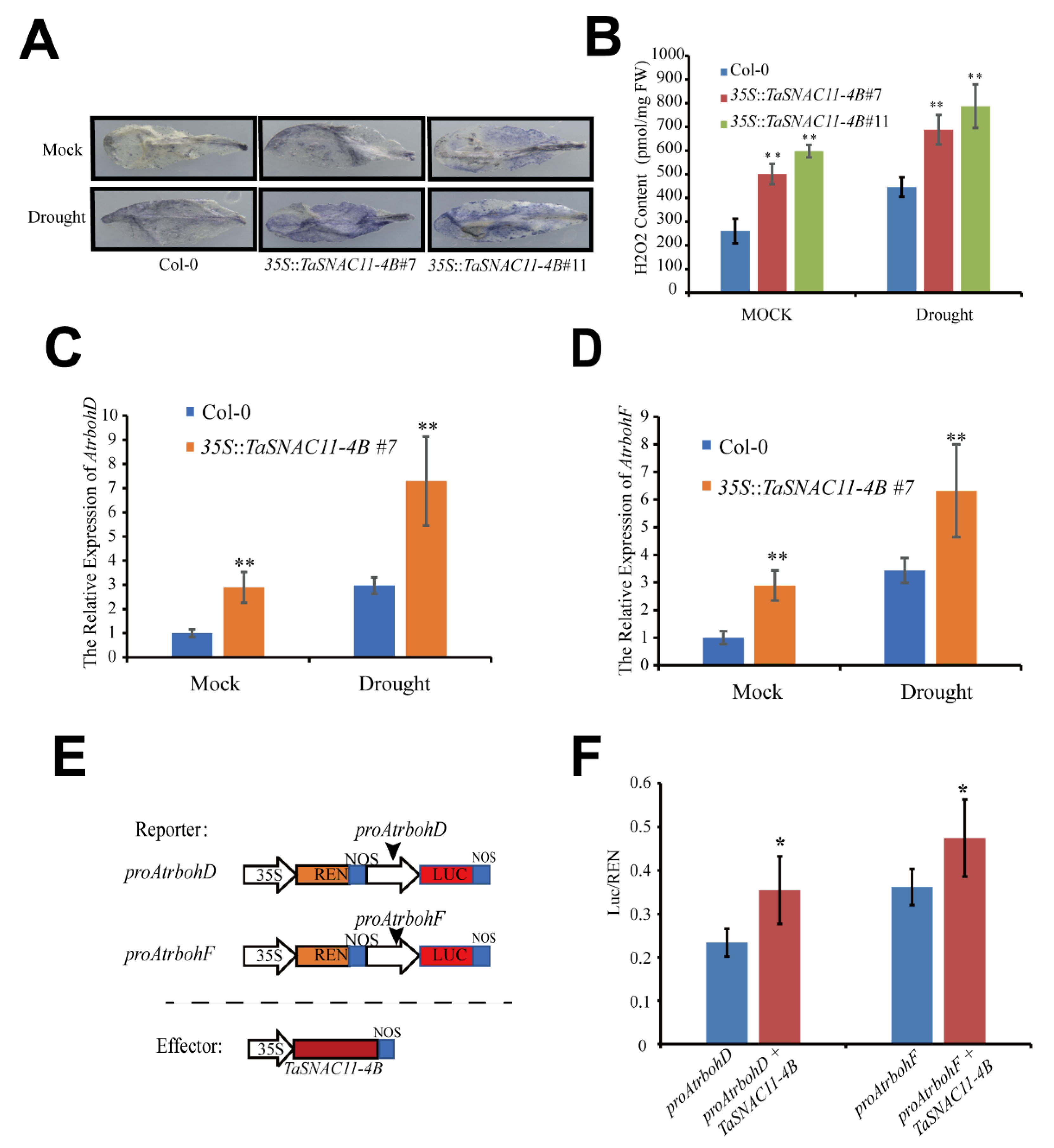
2.7. TaSNAC11-4B Enhances ROS Accumulation by Activating Atrboh Genes
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Phytohormone and Stress Treatments
4.3. Construct Generation and Arabidopsis Transformation
4.4. β-Estradiol (EST) Treatments
4.5. Chlorophyll, Fv/Fm, and Electrolyte (Ion) Leakage Rate Measurement
4.6. Quantitative RT-PCR Assay
4.7. TF Transcriptional Activity
4.8. Dual-Luciferase Assay
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ABA | Abscisic acid |
| ROS | Reactive oxygen species |
| GA | Gibberellins |
| NADPH | Nicotinamide adenine dinucleotide phosphate |
| MeJA | Methyl jasmonate |
| SA | Salicylic acid |
| IAA | Indole-3-acetic acid |
References
- Balazadeh, S.; Parlitz, S.; Mueller-Roeber, B.; Meyer, R.C. Natural developmental variations in leaf and plant senescence inArabidopsis thaliana. Plant Biol. 2008, 10, 136–147. [Google Scholar] [CrossRef]
- Gregersen, P.L.; Holm, P.B.; Krupinska, K. Leaf senescence and nutrient remobilisation in barley and wheat. Plant Biol. 2008, 10, 37–49. [Google Scholar] [CrossRef]
- Keech, O.; Pesquet, E.; Gutierrez, L.; Ahad, A.; Bellini, C.; Smith, A.M.; Gardeström, P. Leaf Senescence Is Accompanied by an Early Disruption of the Microtubule Network in Arabidopsis. Plant Physiol. 2010, 154, 1710–1720. [Google Scholar] [CrossRef] [Green Version]
- Ali, A.; Gao, X.; Guo, Y. Initiation, Progression, and Genetic Manipulation of Leaf Senescence. In Plant Senescence; Humana Press: New York, NY, USA, 2018; pp. 9–31. [Google Scholar]
- Lim, P.O.; Kim, H.J.; Gil Nam, H. Leaf Senescence. Annu. Rev. Plant Biol. 2007, 58, 115–136. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, S. Molecular regulation of leaf senescence. Curr. Opin. Plant Biol. 2003, 6, 79–84. [Google Scholar] [CrossRef]
- Srivalli, S.; Khanna-Chopra, R. Delayed wheat flag leaf senescence due to removal of spikelets is associated with increased activities of leaf antioxidant enzymes, reduced glutathione/oxidized glutathione ratio and oxidative damage to mitochondrial proteins. Plant Physiol. Biochem. 2009, 47, 663–670. [Google Scholar] [CrossRef]
- Schippers, J.H.; Kunkowska, A.B.; Wagstaff, C.; Jing, H.-C. Living to Die and Dying to Live: The Survival Strategy behind Leaf Senescence. Plant Physiol. 2015, 169, 914–930. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Kim, J.H.; Lyu, J.I.; Woo, H.R.; Lim, P.O. New insights into the regulation of leaf senescence in Arabidopsis. J. Exp. Bot. 2017, 69, 787–799. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Li, X.; Chao, J.; Zhang, Z.; Wang, W.; Guo, Y. NAC Family Transcription Factors in Tobacco and Their Potential Role in Regulating Leaf Senescence. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [Green Version]
- Podzimska-Sroka, D.; O’Shea, C.; Gregersen, P.L.; Skriver, K. NAC Transcription Factors in Senescence: From Molecular Structure to Function in Crops. Plants 2015, 4, 412–448. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Gan, S. AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. 2006, 46, 601–612. [Google Scholar] [CrossRef]
- Rauf, M.; Arif, M.; Dortay, H.; Matallana-Ramírez, L.P.; Waters, M.T.; Gil Nam, H.; Lim, P.-O.; Mueller-Roeber, B.; Balazadeh, S. ORE1 balances leaf senescence against maintenance by antagonizing G2-like-mediated transcription. EMBO Rep. 2013, 14, 382–388. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.-S.; Sakuraba, Y.; Han, S.; Yoo, S.-C.; Paek, N.-C. Mutation of the Arabidopsis NAC016 Transcription Factor Delays Leaf Senescence. Plant Cell Physiol. 2013, 54, 1660–1672. [Google Scholar] [CrossRef]
- Garapati, P.; Xue, G.-P.; Munné-Bosch, S.; Balazadeh, S. Transcription Factor ATAF1 in Arabidopsis Promotes Senescence by Direct Regulation of Key Chloroplast Maintenance and Senescence Transcriptional Cascades. Plant Physiol. 2015, 168, 1122–1139. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Gao, J.; Yao, L.; Ren, G.; Zhu, X.; Gao, S.; Qiu, K.; Zhou, X.; Kuai, B. The role of ANAC072 in the regulation of chlorophyll degradation during age- and dark-induced leaf senescence. Plant Cell Rep. 2016, 35, 1729–1741. [Google Scholar] [CrossRef]
- Wu, A.; Allu, A.D.; Garapati, P.; Siddiqui, H.; Dortay, H.; Zanor, M.-I.; Asensi-Fabado, M.A.; Munné-Bosch, S.; Antonio, C.; Tohge, T.; et al. Jungbrunnen1, a Reactive Oxygen Species–Responsive NAC Transcription Factor, Regulates Longevity in Arabidopsis. Plant Cell 2012, 24, 482–506. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.-D.; Seo, P.J.; Yoon, H.-K.; Park, C.-M. The Arabidopsis NAC Transcription Factor VNI2 Integrates Abscisic Acid Signals into Leaf Senescence via the COR/RD Genes. Plant Cell 2011, 23, 2155–2168. [Google Scholar] [CrossRef] [Green Version]
- Qiu, K.; Li, Z.; Yang, Z.; Chen, J.; Wu, S.; Zhu, X.; Gao, S.; Gao, J.; Ren, G.; Kuai, B.; et al. EIN3 and ORE1 Accelerate Degreening during Ethylene-Mediated Leaf Senescence by Directly Activating Chlorophyll Catabolic Genes in Arabidopsis. PLoS Genet. 2015, 11, e1005399. [Google Scholar] [CrossRef]
- Kim, H.; Kim, H.J.; Vu, Q.T.; Jung, S.; McClung, C.R.; Hong, S.; Gil Nam, H. Circadian control of ORE1 by PRR9 positively regulates leaf senescence in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, 8448–8453. [Google Scholar] [CrossRef] [Green Version]
- Borrill, P.; Harrington, S.A.; Uauy, C. Genome-Wide Sequence and Expression Analysis of the NAC Transcription Factor Family in Polyploid Wheat. G3 2017, 7, 3019–3029. [Google Scholar] [CrossRef] [Green Version]
- Uauy, C.; Distelfeld, A.; Fahima, T.; Blechl, A.; Dubcovsky, J. A NAC Gene Regulating Senescence Improves Grain Protein, Zinc, and Iron Content in Wheat. Science 2006, 314, 1298–1301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, X.; Qu, B.; Li, W.; Zhao, X.; Teng, W.; Ma, W.; Ren, Y.; Li, B.; Li, Z.; Tong, Y. The nitrate inducible NAC transcription factor TaNAC2-5A controls nitrate response and increases wheat yield. Plant Physiol. 2015, 169, 1991–2005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, B.; Wei, J.; Song, N.; Wang, N.; Zhao, J.; Kang, Z. A novel wheat NAC transcription factor, Ta NAC30, negatively regulates resistance of wheat to stripe rust. J. Integr. Plant Biol. 2018, 60, 432–443. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Duan, X.; Zhang, Q.; Li, X.; Wang, B.; Huang, L.; Wang, X.; Kang, Z. The target gene of tae-miR164, a novel NAC transcription factor from the NAM subfamily, negatively regulates resistance of wheat to stripe rust. Mol. Plant Pathol. 2014, 15, 284–296. [Google Scholar] [CrossRef]
- Liu, Y.; Hou, J.; Wang, X.; Li, T.; Majeed, U.; Hao, C.; Zhang, X. The Nac Transcription Factor Nac019-A1 Is a Negative Regulator of Starch Synthesis in Wheat Developing Endosperm. J. Exp. Bot. 2020, 71, 5794–5807. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Zhang, H.; Qian, X.; Li, A.; Zhao, G.; Jing, R. TaNAC2, a NAC-type wheat transcription factor conferring enhanced multiple abiotic stress tolerances in Arabidopsis. J. Exp. Bot. 2012, 63, 2933–2946. [Google Scholar] [CrossRef]
- Huang, Q.; Wang, Y.; Li, B.; Chang, J.; Chen, M.; Li, K.; Yang, G.; He, G. TaNAC29, a NAC transcription factor from wheat, enhances salt and drought tolerance in transgenic Arabidopsis. BMC Plant Biol. 2015, 15, 268. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.J.; Hong, S.H.; Kim, Y.W.; Lee, I.H.; Jun, J.H.; Phee, B.-K.; Rupak, T.; Jeong, H.; Lee, Y.; Hong, B.S.; et al. Gene regulatory cascade of senescence-associated NAC transcription factors activated by ETHYLENE-INSENSITIVE2-mediated leaf senescence signalling in Arabidopsis. J. Exp. Bot. 2014, 65, 4023–4036. [Google Scholar] [CrossRef] [Green Version]
- Kou, X.; Watkins, C.B.; Gan, S.-S. Arabidopsis AtNAP regulates fruit senescence. J. Exp. Bot. 2012, 63, 6139–6147. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Gan, S.-S. Translational researches on leaf senescence for enhancing plant productivity and quality. J. Exp. Bot. 2014, 65, 3901–3913. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.; Xia, X.; Zhang, Y.; Gan, S.-S. An ABA-regulated and Golgi-localized protein phosphatase controls water loss during leaf senescence in Arabidopsis. Plant J. 2011, 69, 667–678. [Google Scholar] [CrossRef] [PubMed]
- Lei, W.; Li, Y.; Yao, X.; Qiao, K.; Wei, L.; Liu, B.; Zhang, D.; Lin, H.-H. NAP is involved in GA-mediated chlorophyll degradation and leaf senescence by interacting with DELLAs in Arabidopsis. Plant Cell Rep. 2019, 39, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Gan, S.-S. An Abscisic Acid-AtNAP Transcription Factor-SAG113 Protein Phosphatase 2C Regulatory Chain for Controlling Dehydration in Senescing Arabidopsis Leaves. Plant Physiol. 2011, 158, 961–969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, C.; Wang, Y.; Zhu, Y.; Tang, J.; Hu, B.; Liu, L.; Ou, S.; Wu, H.; Sun, X.; Chu, J.; et al. OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc. Natl. Acad. Sci. USA 2014, 111, 10013–10018. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Wang, Y.; Lv, B.; Li, J.; Luo, L.; Lu, S.; Zhang, X.; Ma, H.; Ming, F. The NAC Family Transcription Factor OsNAP Confers Abiotic Stress Response Through the ABA Pathway. Plant Cell Physiol. 2014, 55, 604–619. [Google Scholar] [CrossRef] [Green Version]
- Levine, A.; Tenhaken, R.; Dixon, R.; Lamb, C. H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 1994, 79, 583–593. [Google Scholar] [CrossRef]
- Del Río, L.A.; López-Huertas, E. ROS Generation in Peroxisomes and its Role in Cell Signaling. Plant Cell Physiol. 2016, 57, pcw076. [Google Scholar] [CrossRef]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef]
- Lherminier, J.; Elmayan, T.; Fromentin, J.; Elaraqui, K.T.; Vesa, S.; Morel, J.; Verrier, J.-L.; Cailleteau, B.; Blein, J.-P.; Simon-Plas, F. NADPH Oxidase-Mediated Reactive Oxygen Species Production: Subcellular Localization and Reassessment of Its Role in Plant Defense. Mol. Plant-Microbe Interactions 2009, 22, 868–881. [Google Scholar] [CrossRef] [Green Version]
- Kaya, H.; Takeda, S.; Kobayashi, M.J.; Kimura, S.; Iizuka, A.; Imai, A.; Hishinuma, H.; Kawarazaki, T.; Mori, K.; Yamamoto, Y.; et al. Comparative analysis of the reactive oxygen species-producing enzymatic activity of Arabidopsis NADPH oxidases. Plant J. 2019, 98, 291–300. [Google Scholar] [CrossRef]
- Müller, K.; Carstens, A.C.; Linkies, A.; Torres, M.A.; Leubner-Metzger, G. The NADPH-oxidase AtrbohB plays a role in Arabidopsis seed after-ripening. New Phytol. 2009, 184, 885–897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeda, S.; Gapper, C.; Kaya, H.; Bell, E.; Kuchitsu, K.; Dolan, L. Local Positive Feedback Regulation Determines Cell Shape in Root Hair Cells. Science 2008, 319, 1241–1244. [Google Scholar] [CrossRef] [PubMed]
- Krieger, G.; Shkolnik, D.; Miller, G.; Fromm, H. Reactive oxygen species tune root tropic responses. Plant Physiol. 2016, 172, 1209–1220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwak, J.M.; Mori, I.C.; Pei, Z.; Leonhardt, N.; Torres, M.A.; Dangl, J.L.; Bloom, R.E.; Bodde, S.; Jones, J.D.; Schroeder, J.I. NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 2003, 22, 2623–2633. [Google Scholar] [CrossRef]
- Maruta, T.; Inoue, T.; Tamoi, M.; Yabuta, Y.; Yoshimura, K.; Ishikawa, T.; Shigeoka, S. Arabidopsis NADPH oxidases, AtrbohD and AtrbohF, are essential for jasmonic acid-induced expression of genes regulated by MYC2 transcription factor. Plant Sci. 2011, 180, 655–660. [Google Scholar] [CrossRef]
- Davletova, S.; Rizhsky, L.; Liang, H.; Shengqiang, Z.; Oliver, D.J.; Coutu, J.; Shulaev, V.; Schlauch, K.; Mittler, R. Cytosolic Ascorbate Peroxidase 1 is a Central Component of the Reactive Oxygen Gene Network of Arabidopsis. Plant Cell 2004, 17, 268–281. [Google Scholar] [CrossRef] [Green Version]
- Qi, W.; Zhang, L.; Feng, W.; Xu, H.; Wang, L.; Jiao, Z. ROS and ABA Signaling Are Involved in the Growth Stimulation Induced by Low-Dose Gamma Irradiation in Arabidopsis Seedling. Appl. Biochem. Biotechnol. 2014, 175, 1490–1506. [Google Scholar] [CrossRef] [PubMed]
- Rivero, R.M.; Kojima, M.; Gepstein, A.; Sakakibara, H.; Mittler, R.; Gepstein, S.; Blumwald, E. Delayed leaf senescence induces extreme drought tolerance in a flowering plant. Proc. Natl. Acad. Sci. USA 2007, 104, 19631–19636. [Google Scholar] [CrossRef] [Green Version]
- Joshi, S.; Choukimath, A.; Isenegger, D.; Panozzo, J.; Spangenberg, G.; Kant, S. Improved Wheat Growth and Yield by Delayed Leaf Senescence Using Developmentally Regulated Expression of a Cytokinin Biosynthesis Gene. Front. Plant Sci. 2019, 10, 1285. [Google Scholar] [CrossRef] [Green Version]
- Mao, H.; Li, S.; Wang, Z.; Cheng, X.; Li, F.; Mei, F.; Chen, N.; Kang, Z. Regulatory changes in TaSNAC8-6A are associated with drought tolerance in wheat seedlings. Plant Biotechnol. J. 2019, 18, 1078–1092. [Google Scholar] [CrossRef] [Green Version]
- Nakashima, K.; Takasaki, H.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 2012, 1819, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Seok, H.-Y.; Woo, D.-H.; Nguyen, L.V.; Tran, H.T.; Tarte, V.N.; Mehdi, S.M.M.; Lee, S.-Y.; Moon, Y.-H. Arabidopsis AtNAP functions as a negative regulator via repression of AREB1 in salt stress response. Planta 2016, 245, 329–341. [Google Scholar] [CrossRef] [PubMed]
- Manaa, A.; Ben Ahmed, H.; Smiti, S.; Faurobert, M. Salt-Stress Induced Physiological and Proteomic Changes in Tomato (Solanum lycopersicum) Seedlings. OMICS 2011, 15, 801–809. [Google Scholar] [CrossRef] [PubMed]
- Eyang, L.; Ezhao, X.; Ezhu, H.; Epaul, M.; Ezu, Y.; Tang, Z. Exogenous trehalose largely alleviates ionic unbalance, ROS burst, and PCD occurrence induced by high salinity in Arabidopsis seedlings. Front. Plant Sci. 2014, 5, 570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Chan, Z.; Gao, J.; Xing, L.; Cao, M.; Yu, C.; Hu, Y.; You, J.; Shi, H.; Zhu, Y.; et al. ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc. Natl. Acad. Sci. USA 2016, 113, 1949–1954. [Google Scholar] [CrossRef] [Green Version]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 2005, 10, 79–87. [Google Scholar] [CrossRef]
- Wang, Y.; Lin, A.; Loake, G.J.; Chu, C. H2O2-induced Leaf Cell Death and the Crosstalk of Reactive Nitric/Oxygen SpeciesF. J. Integr. Plant Biol. 2013, 55, 202–208. [Google Scholar] [CrossRef]
- Cheng, F.; Wang, F.; Zhao, Q.; Liu, J.; Cheng, F. Involvement of NADPH oxidase isoforms in the production of O2− manipulated by ABA in the senescing leaves of early-senescence-leaf (esl) mutant rice (Oryza sativa). PLoS ONE 2018, 13, e0190161. [Google Scholar] [CrossRef]
- Ben Rejeb, K.; Benzarti, M.; Debez, A.; Bailly, C.; Savouré, A.; Abdelly, C. NADPH oxidase-dependent H2O2 production is required for salt-induced antioxidant defense in Arabidopsis thaliana. J. Plant Physiol. 2015, 174, 5–15. [Google Scholar] [CrossRef]
- Wu, Y.-S.; Yang, C.-Y. Physiological Responses and Expression Profile of NADPH Oxidase in Rice (Oryza Sativa) Seedlings under Different Levels of Submergence. Rice 2016, 9, 2. [Google Scholar] [CrossRef] [Green Version]
- Fu, M.; Yuan, C.; Song, A.; Lu, J.; Wang, X.; Sun, S. AtWDS1 negatively regulates age-dependent and dark-induced leaf senescence in Arabidopsis. Plant Sci. 2019, 285, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Luoni, S.B.; Astigueta, F.H.; Nicosia, S.; Moschen, S.; Fernandez, P.; Heinz, R. Transcription Factors Associated with Leaf Senescence in Crops. Plants 2019, 8, 411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balazadeh, S.; Kwasniewski, M.; Caldana, C.; Mehrnia, M.; Zanor, M.I.; Xue, G.-P.; Mueller-Roeber, B. ORS1, an H2O2-Responsive NAC Transcription Factor, Controls Senescence in Arabidopsis thaliana. Mol. Plant 2011, 4, 346–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balazadeh, S.; Siddiqui, H.; Allu, A.D.; Matallana-Ramirez, L.P.; Caldana, C.; Mehrnia, M.; Zanor, M.I.; Kohler, B.; Mueller-Roeber, B. A Gene Regulatory Network Controlled by the Nac Transcription Factor Anac092/Atnac2/Ore1 during Salt-Promoted Senescence. Plant J. 2010, 62, 250–264. [Google Scholar] [CrossRef] [PubMed]
- Oda-Yamamizo, C.; Mitsuda, N.; Sakamoto, S.; Ogawa, D.; Ohme-Takagi, M.; Ohmiya, A. Corrigendum: The Nac Transcription Factor Anac046 Is a Positive Regulator of Chlorophyll Degradation and Senescence in Arabidopsis Leaves. Sci. Rep. 2016, 6, 35125. [Google Scholar] [CrossRef]
- Xue, G.-P.; Way, H.M.; Richardson, T.; Drenth, J.; Joyce, P.A.; McIntyre, C.L. Overexpression of TaNAC69 Leads to Enhanced Transcript Levels of Stress Up-Regulated Genes and Dehydration Tolerance in Bread Wheat. Mol. Plant 2011, 4, 697–712. [Google Scholar] [CrossRef]
- Mao, X.; Chen, S.; Li, A.; Zhai, C.; Jing, R. Novel NAC Transcription Factor TaNAC67 Confers Enhanced Multi-Abiotic Stress Tolerances in Arabidopsis. PLoS ONE 2014, 9, e84359. [Google Scholar] [CrossRef] [Green Version]
- Bulgakov, V.P.; Wu, H.-C.; Jinn, T.-L. Coordination of ABA and Chaperone Signaling in Plant Stress Responses. Trends Plant Sci. 2019, 24, 636–651. [Google Scholar] [CrossRef]
- Zhao, Y.; Gao, J.; Kim, J.I.; Chen, K.; Bressan, R.A.; Zhu, J.-K. Control of Plant Water Use by ABA Induction of Senescence and Dormancy: An Overlooked Lesson from Evolution. Plant Cell Physiol. 2017, 58, 1319–1327. [Google Scholar] [CrossRef]
- Yang, J.; Worley, E.; Udvardi, M.K. A NAP-AAO3 regulatory module promotes chlorophyll degradation via ABA biosynthesis in Arabidopsis leaves. Plant Cell 2014, 26, 4862–4874. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Seo, P.J.; Lee, H.-J.; Park, C.-M. A NAC transcription factor NTL4 promotes reactive oxygen species production during drought-induced leaf senescence in Arabidopsis. Plant J. 2012, 70, 831–844. [Google Scholar] [CrossRef] [PubMed]
- Jajic, I.; Sarna, T.; Strzałka, K. Senescence, Stress, and Reactive Oxygen Species. Plants 2015, 4, 393–411. [Google Scholar] [CrossRef] [Green Version]
- Torres, M.A.; Dangl, J.L.; Jones, J.D.G. Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. USA 2001, 99, 517–522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; He, C. Regulation of plant reactive oxygen species (ROS) in stress responses: Learning from Atrbohd. Plant Cell Rep. 2016, 35, 995–1007. [Google Scholar] [CrossRef]
- Zhang, Z.; Guo, Y. Hormone Treatments in Studying Leaf Senescence. Methods Mol. Biol. 2018, 1744, 125–132. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative Pcr and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Liu, C.; Guo, Y. Wheat Transcription Factor TaSNAC11-4B Positively Regulates Leaf Senescence through Promoting ROS Production in Transgenic Arabidopsis. Int. J. Mol. Sci. 2020, 21, 7672. https://doi.org/10.3390/ijms21207672
Zhang Z, Liu C, Guo Y. Wheat Transcription Factor TaSNAC11-4B Positively Regulates Leaf Senescence through Promoting ROS Production in Transgenic Arabidopsis. International Journal of Molecular Sciences. 2020; 21(20):7672. https://doi.org/10.3390/ijms21207672
Chicago/Turabian StyleZhang, Zenglin, Chen Liu, and Yongfeng Guo. 2020. "Wheat Transcription Factor TaSNAC11-4B Positively Regulates Leaf Senescence through Promoting ROS Production in Transgenic Arabidopsis" International Journal of Molecular Sciences 21, no. 20: 7672. https://doi.org/10.3390/ijms21207672
APA StyleZhang, Z., Liu, C., & Guo, Y. (2020). Wheat Transcription Factor TaSNAC11-4B Positively Regulates Leaf Senescence through Promoting ROS Production in Transgenic Arabidopsis. International Journal of Molecular Sciences, 21(20), 7672. https://doi.org/10.3390/ijms21207672





