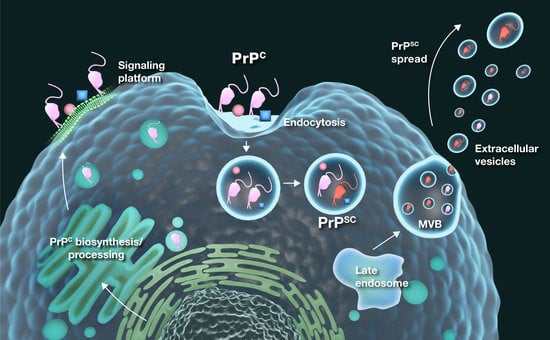
A New Take on Prion Protein Dynamics in Cellular Trafficking
Abstract
1. Introduction
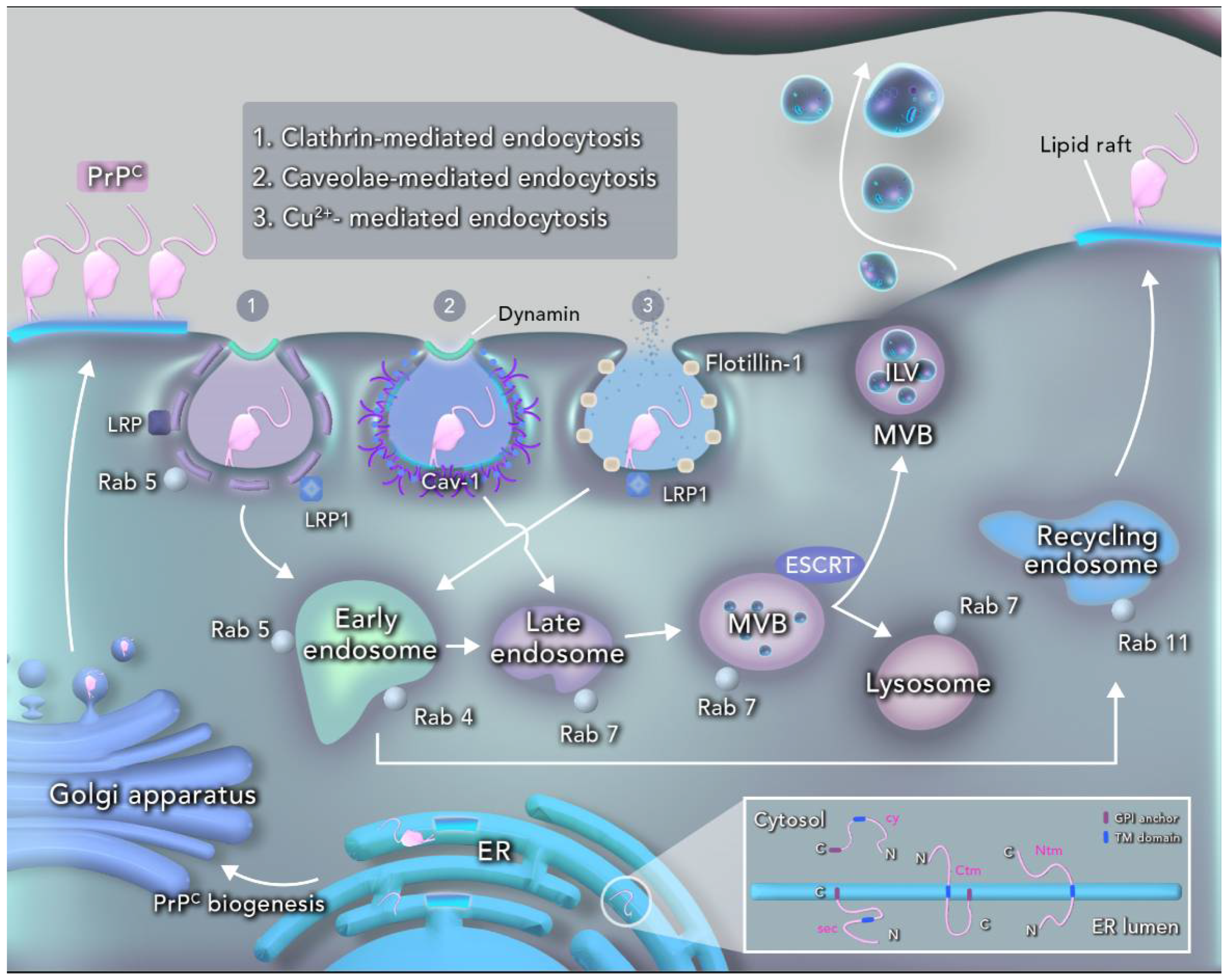
2. PrPC Structure and Cellular Processing
3. PrPC in Intracellular Trafficking
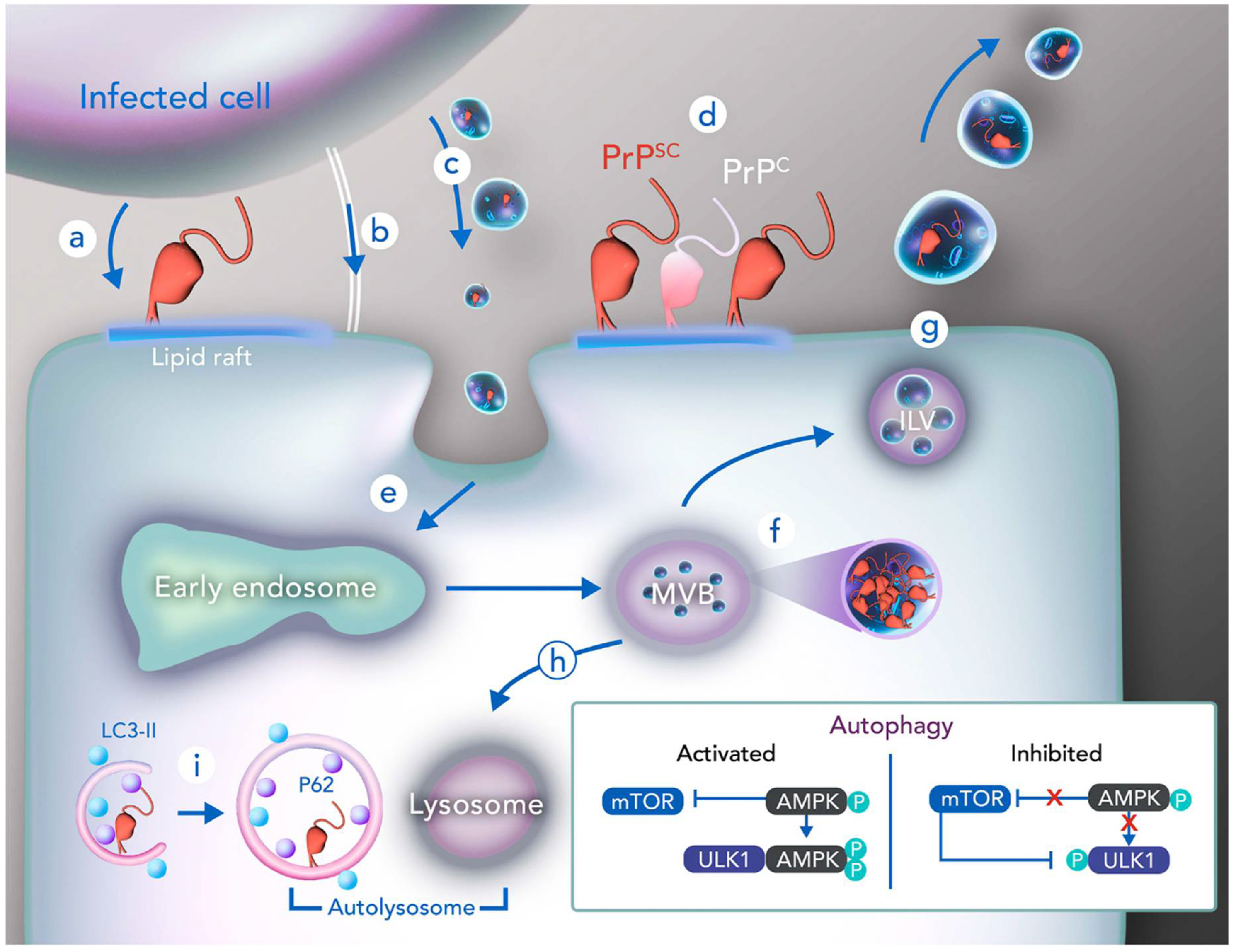
4. Cellular Trafficking in PrPSc Infection
5. Prion and Autophagy
6. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Hinze, C.; Boucrot, E. Endocytosis in proliferating, quiescent and terminally differentiated cells. J. Cell Sci. 2018, 131, jcs216804. [Google Scholar] [CrossRef]
- Taylor, D.R.; Hooper, N.M. The prion protein and lipid rafts. Mol. Membr. Biol. 2006, 23, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Martins, V.R.; Beraldo, F.H.; Hajj, G.N.; Lopes, M.H.; Lee, K.S.; Prado, M.A.; Linden, R. Prion protein: Orchestrating neurotrophic activities. Curr. Issues Mol. Biol. 2010, 12, 63–86. [Google Scholar]
- Santos, T.G.; Lopes, M.H.; Martins, V.R. Targeting prion protein interactions in cancer. Prion 2015, 9, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Harris, D.A. Trafficking, turnover and membrane topology of PrP. Br. Med. Bull. 2003, 66, 71–85. [Google Scholar] [CrossRef]
- Nikles, D.; Vana, K.; Gauczynski, S.; Knetsch, H.; Ludewigs, H.; Weiss, S. Subcellular localization of prion proteins and the 37 kDa/67 kDa laminin receptor fused to fluorescent proteins. Biochim. Biophys. Acta Mol. Basis Dis. 2008, 1782, 335–340. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Godsave, S.F.; Peters, P.J.; Wille, H. Subcellular distribution of the prion protein in sickness and in health. Virus Res. 2015, 207, 136–145. [Google Scholar] [CrossRef]
- Vorberg, I.M. All the same? The secret life of prion strains within their target cells. Viruses 2019, 11, 334. [Google Scholar] [CrossRef] [PubMed]
- Prado, M.A.M.; Alves-Silva, J.; Magalhães, A.C.; Prado, V.F.; Linden, R.; Martins, V.R.; Brentani, R.R. PrPc on the road: Trafficking of the cellular prion protein. J. Neurochem. 2004, 88, 769–781. [Google Scholar] [CrossRef]
- Marijanovic, Z.; Caputo, A.; Campana, V.; Zurzolo, C. Identification of an intracellular site of prion conversion. PLoS Pathog. 2009, 5, e1000426. [Google Scholar] [CrossRef]
- Kumari, S.; Mg, S.; Mayor, S. Endocytosis unplugged: Multiple ways to enter the cell. Cell Res. 2010, 20, 256–275. [Google Scholar] [CrossRef] [PubMed]
- Ungewickell, E.J.; Hinrichsen, L. Endocytosis: Clathrin-mediated membrane budding. Curr. Opin. Cell Biol. 2007, 19, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Jimah, J.R.; Hinshaw, J.E. Structural Insights into the Mechanism of Dynamin Superfamily Proteins. Trends Cell Biol. 2019, 29, 257–273. [Google Scholar] [CrossRef]
- Langemeyer, L.; Fröhlich, F.; Ungermann, C. Rab GTPase Function in Endosome and Lysosome Biogenesis. Trends Cell Biol. 2018, 28, 957–970. [Google Scholar] [CrossRef]
- Rodman, J.S.; Wandinger-Ness, A. Rab GTPases coordinate endocytosis. J. Cell Sci. 2000, 113, 183–192. [Google Scholar]
- Jahn, R.; Scheller, R.H. SNAREs—Engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 2006, 7, 631–643. [Google Scholar] [CrossRef]
- Le Roy, C.; Wrana, J.L. Clathrin- and non-clathrin-mediated endocytic regulation of cell signalling. Nat. Rev. Mol. Cell Biol. 2005, 6, 112–126. [Google Scholar] [CrossRef]
- Elkin, S.R.; Lakoduk, A.M.; Schmid, S.L. Endocytic pathways and endosomal trafficking: A primer. Wien. Med. Wochenschr. 2016, 166, 196–204. [Google Scholar] [CrossRef]
- Redpath, G.M.I.; Betzler, V.M.; Rossatti, P.; Rossy, J. Membrane Heterogeneity Controls Cellular Endocytic Trafficking. Front. Cell Dev. Biol. 2020, 8, 757. [Google Scholar] [CrossRef]
- McNally, K.E.; Cullen, P.J. Endosomal Retrieval of Cargo: Retromer Is Not Alone. Trends Cell Biol. 2018, 28, 807–822. [Google Scholar] [CrossRef]
- Trivedi, P.C.; Bartlett, J.J.; Pulinilkunnil, T. Lysosomal Biology and Function: Modern View of Cellular Debris Bin. Cells 2020, 9, 1131. [Google Scholar] [CrossRef] [PubMed]
- Neefjes, J.; Jongsma, M.M.L.; Berlin, I. Stop or Go? Endosome Positioning in the Establishment of Compartment Architecture, Dynamics, and Function. Trends Cell Biol. 2017, 27, 580–594. [Google Scholar] [CrossRef] [PubMed]
- Piper, R.C.; Katzmann, D.J. Biogenesis and function of multivesicular bodies. Annu. Rev. Cell Dev. Biol. 2007, 23, 519–547. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Ren, D. Lysosomal physiology. Annu. Rev. Physiol. 2015, 77, 57–80. [Google Scholar] [CrossRef]
- Meldolesi, J. Exosomes and Ectosomes in Intercellular Communication. Curr. Biol. 2018, 28, R435–R444. [Google Scholar] [CrossRef]
- Hurley, J.H. ESCRT s are everywhere. EMBO J. 2015, 34, 2398–2407. [Google Scholar] [CrossRef]
- Porto-Carreiro, I.; Février, B.; Paquet, S.; Vilette, D.; Raposo, G. Prions and exosomes: From PrPc trafficking to PrPsc propagation. Blood Cells Mol. Dis. 2005, 35, 143–148. [Google Scholar] [CrossRef]
- Hartmann, A.; Muth, C.; Dabrowski, O.; Krasemann, S.; Glatzel, M. Exosomes and the prion protein: More than one truth. Front. Neurosci. 2017, 11, 194. [Google Scholar] [CrossRef]
- Hooper, N.M.; Taylor, D.R.; Watt, N.T. Mechanism of the metal-mediated endocytosis of the prion protein. Biochem. Soc. Trans. 2008, 36, 1272–1276. [Google Scholar] [CrossRef]
- Haucke, V.; Neher, E.; Sigrist, S.J. Protein scaffolds in the coupling of synaptic exocytosis and endocytosis. Nat. Rev. Neurosci. 2011, 12, 127–138. [Google Scholar] [CrossRef]
- Campana, V.; Sarnataro, D.; Zurzolo, C. The highways and byways of prion protein trafficking. Trends Cell Biol. 2005, 15, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Baral, P.K.; Yin, J.; Aguzzi, A.; James, M.N.G. Transition of the prion protein from a structured cellular form (PrPC) to the infectious scrapie agent (PrPSc). Protein Sci. 2019, 28, 2055–2063. [Google Scholar] [CrossRef] [PubMed]
- Borchelt, D.R.; Taraboulos, A.; Prusiner, S.B. Evidence for synthesis of scrapie prion proteins in the endocytic pathway. J. Biol. Chem. 1992, 267, 16188–16199. [Google Scholar]
- Riek, R.; Hornemann, S.; Wider, G.; Billeter, M.; Glockshuber, R.; Wüthrich, K. NMR structure of the mouse prion protein domain PrP(121-231). Nature 1996, 382, 180–182. [Google Scholar] [CrossRef]
- Tatzelt, J.; Winklhofer, K.F. Folding and misfolding of the prion protein in the secretory pathway. Amyloid 2004, 11, 162–172. [Google Scholar] [CrossRef] [PubMed]
- Acevedo-Morantes, C.Y.; Wille, H. The structure of human prions: From biology to structural models—Considerations and pitfalls. Viruses 2014, 6, 3875–3892. [Google Scholar] [CrossRef]
- Heller, U.; Winklhofer, K.F.; Heske, J.; Reintjes, A.; Tatzel, J. Post-translational import of the prion protein into the endoplasmic reticulum interferes with cell viability: A critical role for the putative transmembrane domain. J. Biol. Chem. 2003, 278, 36139–36147. [Google Scholar] [CrossRef]
- Song, Z.Q.; Zhao, D.M.; Yang, L.F. Metabolism of minor isoforms of prion proteins: Cytosolic prion protein and transmembrane prion protein. Neural Regen. Res. 2013, 8, 2868–2878. [Google Scholar]
- Stewart, R.S.; Piccardo, P.; Ghetti, B.; Harris, D.A. Neurodegenerative illness in transgenic mice expressing a transmembrane form of the prion protein. J. Neurosci. 2005, 25, 3469–3477. [Google Scholar] [CrossRef]
- Hegde, R.S.; Mastrianni, J.A.; Scott, M.R.; DeFea, K.A.; Tremblay, P.; Torchia, M.; DeArmond, S.J.; Prusiner, S.B.; Lingappa, V.R. A transmembrane form of the prion protein in neurodegenerative disease. Science 1998, 279, 827–834. [Google Scholar] [CrossRef]
- Stewart, R.S.; Harris, D.A. Mutational Analysis of Topological Determinants in Prion Protein (PrP) and Measurement of Transmembrane and Cytosolic PrP during Prion Infection. J. Biol. Chem. 2003, 278, 45960–45968. [Google Scholar] [CrossRef]
- Gilch, S.; Nunziante, M.; Ertmer, A.; Wopfner, F.; Laszlo, L.; Schätzl, H.M. Recognition of lumenal prion protein aggregates by post-ER quality control mechanisms is mediated by the preoctarepeat region of PrP. Traffic 2004, 5, 300–313. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, O.; Hegde, R.S. Functional Depletion of Mahogunin by Cytosolically Exposed Prion Protein Contributes to Neurodegeneration. Cell 2009, 137, 1136–1147. [Google Scholar] [CrossRef] [PubMed]
- Dubnikov, T.; Ben-Gedalya, T.; Reiner, R.; Hoepfner, D.; Cabral, W.A.; Marini, J.C.; Cohen, E. PrP-containing aggresomes are cytosolic components of an ER quality control mechanism. J. Cell Sci. 2016, 129, 3635–3647. [Google Scholar] [CrossRef]
- Castle, A.R.; Gill, A.C. Physiological functions of the cellular prion protein. Front. Mol. Biosci. 2017, 4, 19. [Google Scholar] [CrossRef] [PubMed]
- Wulf, M.A.; Senatore, A.; Aguzzi, A. The biological function of the cellular prion protein: An update. BMC Biol. 2017, 15, 34. [Google Scholar] [CrossRef]
- Linsenmeier, L.; Altmeppen, H.C.; Wetzel, S.; Mohammadi, B.; Saftig, P.; Glatzel, M. Diverse functions of the prion protein—Does proteolytic processing hold the key? Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 2128–2137. [Google Scholar] [CrossRef] [PubMed]
- Sarnataro, D.; Pepe, A.; Zurzolo, C. Cell Biology of Prion Protein. Prog. Mol. Biol. Transl. Sci. 2017, 150, 57–82. [Google Scholar] [PubMed]
- Sarnataro, D.; Caputo, A.; Casanova, P.; Puri, C.; Paladino, S.; Tivodar, S.S.; Campana, V.; Tacchetti, C.; Zurzolo, C. Lipid rafts and clathrin cooperate in the internalization of PrPC in epithelial FRT cells. PLoS ONE 2009, 4, e5829. [Google Scholar] [CrossRef]
- Taraboulos, A.; Raeber, A.J.; Borchelt, D.R.; Serban, D.; Prusiner, S.B. Synthesis and trafficking of prion proteins in cultured cells. Mol. Biol. Cell 1992, 3, 851–863. [Google Scholar] [CrossRef]
- Caetano, F.A.; Lopes, M.H.; Hajj, G.N.M.; Machado, C.F.; Arantes, C.P.; Magalhães, A.C.; Vieira, M.D.P.B.; Américo, T.A.; Massensini, A.R.; Priola, S.A.; et al. Endocytosis of prion protein is required for ERK1/2 signaling induced by stress-inducible protein 1. J. Neurosci. 2008, 28, 6691–6702. [Google Scholar] [CrossRef]
- Paitel, E.; Alves da Costa, C.; Vilette, D.; Grassi, J.; Checler, F. Overexpression of PrPc triggers caspase 3 activation: Potentiation by proteasome inhibitors and blockade by anti-PrP antibodies. J. Neurochem. 2002, 83, 1208–1214. [Google Scholar] [CrossRef]
- Sunyach, C.; Checler, F. Combined pharmacological, mutational and cell biology approaches indicate that p53-dependent caspase 3 activation triggered by cellular prion is dependent on its endocytosis. J. Neurochem. 2005, 92, 1399–1407. [Google Scholar] [CrossRef]
- Lewis, V.; Hooper, N.M. The role of lipid rafts in prion protein biology. Front. Biosci. 2011, 16, 151–168. [Google Scholar] [CrossRef]
- Monnet, C.; Gavard, J.; Mège, R.M.; Sobel, A. Clustering of cellular prion protein induces ERK1/2 and stathmin phosphorylation in GT1-7 neuronal cells. FEBS Lett. 2004, 576, 114–118. [Google Scholar] [CrossRef] [PubMed]
- Hugel, B.; Martínez, M.C.; Kunzelmann, C.; Blättler, T.; Aguzzi, A.; Freyssinet, J.M. Modulation of signal transduction through the cellular prion protein is linked to its incorporation in lipid rafts. Cell. Mol. Life Sci. 2004, 61, 2998–3007. [Google Scholar] [CrossRef]
- Sarnataro, D.; Campana, V.; Paladino, S.; Stornaiuolo, M.; Nitsch, L.; Zurzolo, C. PrPC association with lipid rafts in the early secretory pathway stabilizes its cellular conformation. Mol. Biol. Cell 2004, 15, 4031–4042. [Google Scholar] [CrossRef] [PubMed]
- Lakhan, S.E.; Sabharanjak, S.; De, A. Endocytosis of glycosylphosphatidylinositol-anchored proteins. J. Biomed. Sci. 2009, 16, 93. [Google Scholar] [CrossRef] [PubMed]
- Shyng, S.L.; Heuser, J.E.; Harris, D.A. A glycolipid-anchored prion protein is endocytosed via clathrin-coated pits. J. Cell Biol. 1994, 125, 1239–1250. [Google Scholar] [CrossRef]
- Sunyach, C.; Jen, A.; Deng, J.; Fitzgerald, K.T.; Frobert, Y.; Grassi, J.; McCaffrey, M.W.; Morris, R. The mechanism of internalization of glycosylphosphatidylinositol-anchored prion protein. EMBO J. 2003, 22, 3591–3601. [Google Scholar] [CrossRef]
- Taylor, D.R.; Hooper, N.M. The low-density lipoprotein receptor-related protein 1 (LRP1) mediates the endocytosis of the cellular prion protein. Biochem. J. 2007, 402, 17–23. [Google Scholar] [CrossRef]
- Gauczynski, S.; Nikles, D.; El-Gogo, S.; Papy-Garcia, D.; Rey, C.; Alban, S.; Barritault, D.; Lasmezas, C.I.; Weiss, S. The 37-kDa/67-kDa laminin receptor acts as a receptor for infectious prions and is inhibited by polysulfated glycanes. J. Infect. Dis. 2006, 194, 702–709. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.S.; Zhao, X.; Lovaas, J.; Eisenberg, E.; Greene, L.E. Clathrin-independent internalization of normal cellular prion protein in neuroblastoma cells is associated with the Arf6 pathway. J. Cell Sci. 2009, 122, 4062–4069. [Google Scholar] [CrossRef] [PubMed]
- Magalhães, A.C.; Silva, J.A.; Lee, K.S.; Martins, V.R.; Prado, H.V.F.; Ferguson, S.S.G.; Gomez, M.V.; Brentani, R.R.; Prado, M.A.M. Endocytic intermediates involved with the intracellular trafficking of a fluorescent cellular prion protein. J. Biol. Chem. 2002, 277, 33311–33318. [Google Scholar] [CrossRef]
- Peters, P.J.; Mironov, A.; Peretz, D.; Van Donselaar, E.; Leclerc, E.; Erpel, S.; DeArmond, S.J.; Burton, D.R.; Williamson, R.A.; Vey, M.; et al. Trafficking of prion proteins through a caveolae-mediated endosomal pathway. J. Cell Biol. 2003, 162, 703–717. [Google Scholar] [CrossRef] [PubMed]
- Shi, Q.; Jing, Y.Y.; Wang, S.B.; Chen, C.; Sun, H.; Xu, Y.; Gao, C.; Zhang, J.; Tian, C.; Guo, Y.; et al. PrP octarepeats region determined the interaction with caveolin-1 and phosphorylation of caveolin-1 and Fyn. Med. Microbiol. Immunol. 2013, 202, 215–227. [Google Scholar] [CrossRef] [PubMed]
- Pantera, B.; Bini, C.; Cirri, P.; Paoli, P.; Camici, G.; Manao, G.; Caselli, A. PrPc activation induces neurite outgrowth and differentiation in PC12 cells: Role for caveolin-1 in the signal transduction pathway. J. Neurochem. 2009, 110, 194–207. [Google Scholar] [CrossRef] [PubMed]
- Toni, M.; Spisni, E.; Griffoni, C.; Santi, S.; Riccio, M.; Lenaz, P.; Tomasi, V. Cellular prion protein and caveolin-1 interaction in a neuronal cell line precedes Fyn/Erk 1/2 signal transduction. J. Biomed. Biotechnol. 2006, 69469. [Google Scholar] [CrossRef]
- Santuccione, A.; Sytnyk, V.; Leshchyns’ka, I.; Schachner, M. Prion protein recruits its neuronal receptor NCAM to lipid rafts to activate p59fyn and to enhance neurite outgrowth. J. Cell Biol. 2005, 169, 341–354. [Google Scholar] [CrossRef]
- Galvan, C.; Camoletto, P.G.; Dotti, C.G.; Aguzzi, A.; Ledesma, M.D. Proper axonal distribution of PrPC depends on cholesterol-sphingomyelin-enriched membrane domains and is developmentally regulated in hippocampal neurons. Mol. Cell. Neurosci. 2005, 30, 304–315. [Google Scholar] [CrossRef]
- Salzano, G.; Giachin, G.; Legname, G. Structural Consequences of Copper Binding to the Prion Protein. Cells 2019, 8, 770. [Google Scholar] [CrossRef]
- Ren, K.; Gao, C.; Zhang, J.; Wang, K.; Xu, Y.; Wang, S.B.; Wang, H.; Tian, C.; Shi, Q.; Dong, X.P. Flotillin-1 mediates PrPC endocytosis in the cultured cells during Cu2+ stimulation through molecular interaction. Mol. Neurobiol. 2013, 48, 631–646. [Google Scholar] [CrossRef] [PubMed]
- Fevrier, B.; Vilette, D.; Archer, F.; Loew, D.; Faigle, W.; Vidal, M.; Laude, H.; Raposo, G. Cells release prions in association with exosomes. Proc. Natl. Acad. Sci. USA 2004, 101, 9683–9688. [Google Scholar] [CrossRef]
- Vella, L.J.; Greenwood, D.L.V.; Cappai, R.; Scheerlinck, J.P.Y.; Hill, A.F. Enrichment of prion protein in exosomes derived from ovine cerebral spinal fluid. Vet. Immunol. Immunopathol. 2008, 124, 385–393. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, A.J.; Crawford, D.M.; Ferguson, D.J.P.; Burthem, J.; Roberts, D.J. Normal prion protein is expressed on exosomes isolated from human plasma. Br. J. Haematol. 2013, 163, 678–680. [Google Scholar] [CrossRef] [PubMed]
- Brenna, S.; Altmeppen, H.C.; Mohammadi, B.; Rissiek, B.; Schlink, F.; Ludewig, P.; Failla, A.V.; Schneider, C.; Glatzel, M.; Puig, B.; et al. Brain-Derived Extracellular Vesicles are Highly Enriched in the Prion Protein and Its C1 Fragment: Relevance for Cellular Uptake and Implications in Stroke. CSHL 2020, in press. [Google Scholar]
- Leng, B.; Sun, H.; Zhao, J.; Liu, Y.; Shen, T.; Liu, W.; Liu, X.; Tan, M.; Li, F.; Zhang, J.; et al. Plasma exosomal prion protein levels are correlated with cognitive decline in PD patients. Neurosci. Lett. 2020, 723, 134866. [Google Scholar] [CrossRef] [PubMed]
- Peggion, C.; Stella, R.; Chemello, F.; Massimino, M.L.; Arrigoni, G.; Cagnin, S.; Biancotto, G.; Franchin, C.; Sorgato, M.C.; Bertoli, A. The Prion Protein Regulates Synaptic Transmission by Controlling the Expression of Proteins Key to Synaptic Vesicle Recycling and Exocytosis. Mol. Neurobiol. 2019, 56, 3420–3436. [Google Scholar] [CrossRef]
- Horner, D.S.; Pasini, M.E.; Beltrame, M.; Mastrodonato, V.; Morelli, E.; Vaccari, T. ESCRT genes and regulation of developmental signaling. Semin. Cell Dev. Biol. 2018, 74, 29–39. [Google Scholar] [CrossRef]
- Ballmer, B.A.; Moos, R.; Liberali, X.P.; Pelkmans, L.; Hornemann, S.; Aguzzi, A. Modifiers of prion protein biogenesis and recycling identified by a highly parallel endocytosis kinetics assay. J. Biol. Chem. 2017, 292, 8356–8368. [Google Scholar] [CrossRef]
- Majumder, P.; Chakrabarti, O. Mahogunin regulates fusion between amphisomes/MVBs and lysosomes via ubiquitination of TSG101. Cell Death Dis. 2015, 6, e1970. [Google Scholar] [CrossRef]
- Guo, B.B.; Bellingham, S.A.; Hill, A.F. The neutral sphingomyelinase pathway regulates packaging of the prion protein into exosomes. J. Biol. Chem. 2015, 290, 3455–3467. [Google Scholar] [CrossRef] [PubMed]
- Dias, M.V.S.; Teixeira, B.L.; Rodrigues, B.R.; Sinigaglia-Coimbra, R.; Porto-Carreiro, I.; Roffé, M.; Hajj, G.N.M.; Martins, V.R. PRNP/prion protein regulates the secretion of exosomes modulating CAV1/caveolin-1-suppressed autophagy. Autophagy 2016, 12, 2113–2128. [Google Scholar] [CrossRef] [PubMed]
- Linden, R.; Martins, V.R.; Prado, M.a.M.; Cammarota, M.; Izquierdo, I.; Brentani, R.R. Physiology of the prion protein. Physiol. Rev. 2008, 88, 673–728. [Google Scholar] [CrossRef]
- Le, N.T.T.; Wu, B.; Harris, D.A. Prion neurotoxicity. Brain Pathol. 2019, 29, 263–277. [Google Scholar] [CrossRef] [PubMed]
- Mallucci, G.R. Prion neurodegeneration: Starts and stops at the synapse. Prion 2009, 3, 195–201. [Google Scholar] [CrossRef]
- Pan, K.M.; Baldwin, M.; Nguyen, J.; Gasset, M.; Serban, A.; Groth, D.; Mehlhorn, I.; Huang, Z.; Fletterick, R.J.; Cohen, F.E.; et al. Conversion of α-helices into β-sheets features in the formation of the scrapie prion proteins. Proc. Natl. Acad. Sci. USA 1993, 90, 10962–10966. [Google Scholar] [CrossRef]
- Cheng, L.; Zhao, W.; Hill, A.F. Exosomes and their role in the intercellular trafficking of normal and disease associated prion proteins. Mol. Aspects Med. 2018, 60, 62–68. [Google Scholar] [CrossRef]
- Wadia, J.S.; Schaller, M.; Williamson, R.A.; Dowdy, S.F. Pathologic prion protein infects cells by lipid-raft dependent macropinocytosis. PLoS ONE 2008, 3, e3314. [Google Scholar] [CrossRef]
- Yim, Y.I.; Park, B.C.; Yadavalli, R.; Zhao, X.; Eisenberg, E.; Greene, L.E. The multivesicular body is the major internal site of prion conversion. J. Cell Sci. 2015, 128, 1434–1443. [Google Scholar] [CrossRef]
- Van Der Kamp, M.W.; Daggett, V. Influence of pH on the human prion protein: Insights into the early steps of misfolding. Biophys. J. 2010, 99, 2289–2298. [Google Scholar] [CrossRef] [PubMed]
- Kanu, N.; Imokawa, Y.; Drechsel, D.N.; Williamson, R.A.; Birkett, C.R.; Bostock, C.J.; Brockes, J.P. Transfer of scrapie prion infectivity by cell contact in culture. Curr. Biol. 2002, 12, 523–530. [Google Scholar] [CrossRef]
- Gousset, K.; Schiff, E.; Langevin, C.; Marijanovic, Z.; Caputo, A.; Browman, D.T.; Chenouard, N.; de Chaumont, F.; Martino, A.; Enninga, J.; et al. Prions hijack tunnelling nanotubes for intercellular spread. Nat. Cell Biol. 2009, 11, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Vella, L.J.; Sharples, R.A.; Lawson, V.A.; Masters, C.L.; Cappai, R.; Hill, A.F. Packaging of prions into exosomes is associated with a novel pathway of PrP processing. J. Pathol. 2007, 211, 582–890. [Google Scholar] [CrossRef]
- Glatzel, M.; Aguzzi, A. PrP(C) expression in the peripheral nervous system is a determinant of prion neuroinvasion. J. Gen. Virol. 2000, 81, 2813–2821. [Google Scholar] [CrossRef] [PubMed]
- Marshall, K.E.; Hughson, A.; Vascellari, S.; Priola, S.A.; Sakudo, A.; Onodera, T.; Baron, G.S. PrP Knockout Cells Expressing Transmembrane PrP Resist Prion Infection. J. Virol. 2017, 91, e01616–e01686. [Google Scholar] [CrossRef]
- Taraboulos, A.; Scott, M.; Semenov, A.; Avraham, D.; Laszlo, L.; Prusiner, S.B. Cholesterol depletion and modification of COOH-terminal targeting sequence of the prion protein inhibit formation of the scrapie isoform. J. Cell Biol. 1995, 129, 121–132. [Google Scholar] [CrossRef]
- Rangel, A.; Race, B.; Klingeborn, M.; Striebel, J.; Chesebro, B. Unusual cerebral vascular prion protein amyloid distribution in scrapie-infected transgenic mice expressing anchorless prion protein. Acta Neuropathol. Commun. 2014, 1, 25. [Google Scholar] [CrossRef]
- Chesebro, B.; Race, B.; Meade-White, K.; LaCasse, R.; Race, R.; Klingeborn, M.; Striebel, J.; Dorward, D.; McGovern, G.; Jeffrey, M. Fatal transmissible amyloid encephalopathy: A new type of prion disease associated with lack of prion protein membrane anchoring. PLoS Pathog. 2010, 6, e1000800. [Google Scholar] [CrossRef]
- Jansen, C.; Parchi, P.; Capellari, S.; Vermeij, A.J.; Corrado, P.; Baas, F.; Strammiello, R.; Van Gool, W.A.; Van Swieten, J.C.; Rozemuller, A.J.M. Prion protein amyloidosis with divergent phenotype associated with two novel nonsense mutations in PRNP. Acta Neuropathol. 2010, 119, 189–197. [Google Scholar] [CrossRef]
- Vilette, D.; Courte, J.; Peyrin, J.M.; Coudert, L.; Schaeffer, L.; Andréoletti, O.; Leblanc, P. Cellular mechanisms responsible for cell-to-cell spreading of prions. Cell. Mol. Life Sci. 2018, 75, 2557–2574. [Google Scholar] [CrossRef]
- Zhu, S.; Victoria, G.S.; Marzo, L.; Ghosh, R.; Zurzolo, C. Prion aggregates transfer through tunneling nanotubes in endocytic vesicles. Prion 2015, 9, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Vilette, D.; Laulagnier, K.; Huor, A.; Alais, S.; Simoes, S.; Maryse, R.; Provansal, M.; Lehmann, S.; Andreoletti, O.; Schaeffer, L.; et al. Efficient inhibition of infectious prions multiplication and release by targeting the exosomal pathway. Cell. Mol. Life Sci. 2015, 72, 4409–4427. [Google Scholar] [CrossRef] [PubMed]
- Kaul, Z.; Chakrabarti, O. Tumor susceptibility gene 101 regulates predisposition to apoptosis via ESCRT machinery accessory proteins. Mol. Biol. Cell 2017, 28, 2106–2122. [Google Scholar] [CrossRef]
- Walker, W.P.; Oehler, A.; Edinger, A.L.; Wagner, K.U.; Gunn, T.M. Oligodendroglial deletion of ESCRT-I component TSG101 causes spongiform encephalopathy. Biol. Cell 2016, 108, 324–337. [Google Scholar] [CrossRef] [PubMed]
- Silvius, D.; Pitstick, R.; Ahn, M.; Meishery, D.; Oehler, A.; Barsh, G.S.; DeArmond, S.J.; Carlson, G.A.; Gunn, T.M. Levels of the Mahogunin Ring Finger 1 E3 Ubiquitin Ligase Do Not Influence Prion Disease. PLoS ONE 2013, 8, e55575. [Google Scholar] [CrossRef]
- Ashok, A.; Hegde, R.S. Selective processing and metabolism of disease-causing mutant prion proteins. PLoS Pathog. 2009, 5, e1000479. [Google Scholar] [CrossRef] [PubMed]
- Satpute-Krishnan, P.; Ajinkya, M.; Bhat, S.; Itakura, E.; Hegde, R.S.; Lippincott-Schwartz, J. ER stress-induced clearance of misfolded GPI-anchored proteins via the secretory pathway. Cell 2014, 158, 522–533. [Google Scholar] [CrossRef] [PubMed]
- Goold, R.; McKinnon, C.; Rabbanian, S.; Collinge, J.; Schiavo, G.; Tabrizi, S. Alternative fates of newly formed PrPSc upon prion conversion on the plasma membrane. J. Cell Sci. 2013, 126, 3552–3562. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.B.; Bellingham, S.A.; Hill, A.F. Stimulating the release of exosomes increases the intercellular transfer of prions. J. Biol. Chem. 2016, 291, 5128–5137. [Google Scholar] [CrossRef]
- Klöhn, P.C.; Castro-Seoane, R.; Collinge, J. Exosome release from infected dendritic cells: A clue for a fast spread of prions in the periphery? J. Infect. 2013, 67, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Cervenakova, L.; Saá, P.; Yakovleva, O.; Vasilyeva, I.; de Castro, J.; Brown, P.; Dodd, R. Are prions transported by plasma exosomes? Transfus. Apher. Sci. 2016, 55, 70–83. [Google Scholar] [CrossRef]
- Coleman, B.M.; Hanssen, E.; Lawson, V.A.; Hill, A.F. Prion-infected cells regulate the release of exosomes with distinct ultrastructural features. FASEB J. 2012, 26, 4160–4173. [Google Scholar] [CrossRef] [PubMed]
- Gabizon, R.; McKinley, M.P.; Prusiner, S.B. Purified prion proteins and scrapie infectivity copartition into liposomes. Proc. Natl. Acad. Sci. USA 1987, 84, 4017–4021. [Google Scholar] [CrossRef]
- Sun, Y.; Hung, W.C.; Lee, M.T.; Huang, H.W. Membrane-mediated amyloid formation of PrP 106-126: A kinetic study. Biochim. Biophys. Acta Biomembr. 2015, 1848, 2422–2429. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Xiao, X.; Shen, P.; Wang, Z.; Dang, J.; Adornato, A.; Zou, L.S.; Dong, Z.; Yuan, J.; Feng, J.; Cui, L.; et al. Characterization of physiochemical properties of caveolin-1 from normal and prion-infected human brains. Oncotarget 2017, 8, 53888–53898. [Google Scholar] [CrossRef]
- Fehlinger, A.; Wolf, H.; Hossinger, A.; Duernberger, Y.; Pleschka, C.; Riemschoss, K.; Liu, S.; Bester, R.; Paulsen, L.; Priola, S.A.; et al. Prion strains depend on different endocytic routes for productive infection. Sci. Rep. 2017, 7, 6923. [Google Scholar] [CrossRef]
- Bagyinszky, E.; Van Giau, V.; Youn, Y.C.; An, S.S.A.; Kim, S. Characterization of mutations in prnp (PRION) gene and their possible roles in neurodegenerative diseases. Neuropsychiatr. Dis. Treat. 2018, 14, 2067–2085. [Google Scholar] [CrossRef]
- Magalhães, A.C.; Baron, G.S.; Lee, K.S.; Steele-Mortimer, O.; Dorward, D.; Prado, M.A.M.; Caughey, B. Uptake and neuritic transport of scrapie prion protein coincident with infection of neuronal cells. J. Neurosci. 2005, 25, 5207–5216. [Google Scholar] [CrossRef]
- Fivaz, M.; Vilbois, F.; Thurnheer, S.; Pasquali, C.; Abrami, L.; Bickel, P.E.; Parton, R.G.; Van der Goot, F.G. Differential sorting and fate of endocytosed GPI-anchored proteins. EMBO J. 2002, 21, 3989–4000. [Google Scholar] [CrossRef]
- Pimpinelli, F.; Lehmann, S.; Maridonneau-Parini, I. The scrapie prion protein is present in flotillin-1-positive vesicles in central- but not peripheral-derived neuronal cell lines. Eur. J. Neurosci. 2005, 21, 2063–2072. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, K.; Tomita, M.; Yano, M.; Chida, J.; Hara, H.; Das, N.R.; Nykjaer, A.; Sakaguchi, S. Prions amplify through degradation of the VPS10P sorting receptor sortilin. PLoS Pathog. 2017, 13, e1006470. [Google Scholar] [CrossRef] [PubMed]
- Shim, S.Y.; Karri, S.; Law, S.; Schatzl, H.M.; Gilch, S. Prion infection impairs lysosomal degradation capacity by interfering with rab7 membrane attachment in neuronal cells. Sci. Rep. 2016, 6, 21658. [Google Scholar] [CrossRef] [PubMed]
- Béranger, F.; Mangé, A.; Goud, B.; Lehmann, S. Stimulation of PrPC retrograde transport toward the endoplasmic reticulum increases accumulation of PrPSc in prion-infected cells. J. Biol. Chem. 2002, 277, 38972–38977. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, K.; Muramatsu, N.; Yano, M.; Usui, T.; Miyata, H.; Sakaguchi, S. Prions disturb post-Golgi trafficking of membrane proteins. Nat. Commun. 2013, 4, 1846. [Google Scholar] [CrossRef] [PubMed]
- Boese, A.S.; Saba, R.; Campbell, K.; Majer, A.; Medina, S.; Burton, L.; Booth, T.F.; Chong, P.; Westmacott, G.; Dutta, S.M.; et al. MicroRNA abundance is altered in synaptoneurosomes during prion disease. Mol. Cell. Neurosci. 2016, 71, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.Z.A.; Zhao, D.; Hussain, T.; Sabir, N.; Yang, L. Regulation of MicroRNAs-mediated autophagic flux: A new regulatory avenue for neurodegenerative diseases with focus on prion diseases. Front. Aging Neurosci. 2018, 10, 139. [Google Scholar] [CrossRef]
- Liu, W.; Bai, X.; Zhang, A.; Huang, J.; Xu, S.; Zhang, J. Role of Exosomes in Central Nervous System Diseases. Front. Mol. Neurosci. 2019, 12, 240. [Google Scholar] [CrossRef]
- Heiseke, A.; Aguib, Y.; Schatzl, H.M. Autophagy, prion infection and their mutual interactions. Curr. Issues Mol. Biol. 2009, 12, 87–97. [Google Scholar] [CrossRef]
- Hara, T.; Nakamura, K.; Matsui, M.; Yamamoto, A.; Nakahara, Y.; Suzuki-Migishima, R.; Yokoyama, M.; Mishima, K.; Saito, I.; Okano, H.; et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 2006, 441, 885–889. [Google Scholar] [CrossRef]
- Abdelaziz, D.H.; Abdulrahman, B.A.; Gilch, S.; Schatzl, H.M. Autophagy pathways in the treatment of prion diseases. Curr. Opin. Pharmacol. 2019, 44, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Heiseke, A.; Aguib, Y.; Riemer, C.; Baier, M.; Schätzl, H.M. Lithium induces clearance of protease resistant prion protein in prion-infected cells by induction of autophagy. J. Neurochem. 2009, 109, 25–34. [Google Scholar] [CrossRef]
- Aguib, Y.; Heiseke, A.; Gilch, S.; Riemer, C.; Baier, M.; Schätzl, H.M.; Ertmer, A. Autophagy induction by trehalose counteracts cellular prion infection. Autophagy 2009, 5, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Jeong, J.K.; Park, S.Y. Sulforaphane-induced autophagy flux prevents prion protein-mediated neurotoxicity through AMPK pathway. Neuroscience 2014, 278, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Abdelaziz, D.H.; Thapa, S.; Abdulrahman, B.; Vankuppeveld, L.; Schatzl, H.M. Metformin reduces prion infection in neuronal cells by enhancing autophagy. Biochem. Biophys. Res. Commun. 2020, 523, 423–428. [Google Scholar] [CrossRef]
- Kim, J.; Kundu, M.; Viollet, B.; Guan, K.L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011, 13, 132–141. [Google Scholar] [CrossRef]
- Zhu, T.; Zhao, D.; Song, Z.; Yuan, Z.; Li, C.; Wang, Y.; Zhou, X.; Yin, X.; Hassan, M.F.; Yang, L. HDAC6 alleviates prion peptide-mediated neuronal death via modulating PI3K-Akt-mTOR pathway. Neurobiol. Aging 2016, 37, 91–102. [Google Scholar] [CrossRef]
- Homma, T.; Ishibashi, D.; Nakagaki, T.; Satoh, K.; Sano, K.; Atarashi, R.; Nishida, N. Increased expression of p62/SQSTM1 in prion diseases and its association with pathogenic prion protein. Sci. Rep. 2014, 4, 4504. [Google Scholar] [CrossRef]
- Jeong, J.-K.; Park, S.-Y. Neuroprotective effect of cellular prion protein (PrPC) is related with activation of alpha7 nicotinic acetylcholine receptor (α7nAchR)-mediated autophagy flux. Oncotarget 2015, 6, 24660–24674. [Google Scholar] [CrossRef]
- López-Pérez, Ó.; Otero, A.; Filali, H.; Sanz-Rubio, D.; Toivonen, J.M.; Zaragoza, P.; Badiola, J.J.; Bolea, R.; Martín-Burriel, I. Dysregulation of autophagy in the central nervous system of sheep naturally infected with classical scrapie. Sci. Rep. 2019, 9, 1911. [Google Scholar] [CrossRef] [PubMed]
- Abdulrahman, B.A.; Abdelaziz, D.H.; Schatzl, H.M. Autophagy regulates exosomal release of prions in neuronal cells. J. Biol. Chem. 2018, 293, 8956–89568. [Google Scholar] [CrossRef]
- Heisler, F.F.; Pechmann, Y.; Wieser, I.; Altmeppen, H.C.; Veenendaal, L.; Muhia, M.; Schweizer, M.; Glatzel, M.; Krasemann, S.; Kneussel, M. Muskelin Coordinates PrPC Lysosome versus Exosome Targeting and Impacts Prion Disease Progression. Neuron 2018, 99, 1155–1169. [Google Scholar] [CrossRef]
- Oh, J.M.; Shin, H.Y.; Park, S.J.; Kim, B.H.; Choi, J.K.; Choi, E.K.; Carp, R.I.; Kim, Y.S. The involvement of cellular prion protein in the autophagy pathway in neuronal cells. Mol. Cell. Neurosci. 2008, 39, 238–247. [Google Scholar] [CrossRef]
- Ertmer, A.; Huber, V.; Gilch, S.; Yoshimori, T.; Erfle, V.; Duyster, J.; Elsässer, H.P.; Schäzl, H.M. The anticancer drug imatinib induces cellular autophagy. Leukemia 2007, 21, 936–942. [Google Scholar] [CrossRef] [PubMed]
- Yun, S.W.; Ertmer, A.; Flechsig, E.; Gilch, S.; Riederer, P.; Gerlach, M.; Schätzl, H.; Klein, M. The tyrosine kinase inhibitor imatinib mesylate delays prion neuroinvasion by inhibiting prion propagation in the periphery. J. Neurovirol. 2007, 13, 328–337. [Google Scholar] [CrossRef]
- Cortes, C.J.; Qin, K.; Cook, J.; Solanki, A.; Mastrianni, J.A. Rapamycin delays disease onset and prevents PrP plaque deposition in a mouse model of Gerstmann-Sträussler-Scheinker disease. J. Neurosci. 2012, 32, 12396–12405. [Google Scholar] [CrossRef]
- Nakagaki, T.; Satoh, K.; Ishibashi, D.; Fuse, T.; Sano, K.; Kamatari, Y.O.; Kuwata, K.; Shigematsu, K.; Iwamaru, Y.; Takenouchi, T.; et al. FK506 reduces abnormal prion protein through the activation of autolysosomal degradation and prolongs survival in prion-infected mice. Autophagy 2013, 9, 1386–1394. [Google Scholar] [CrossRef] [PubMed]
- Harris, D.A. Cellular biology of prion diseases. Clin. Microbiol. Rev. 1999, 12, 429–444. [Google Scholar] [CrossRef]
- Pankiewicz, J.E.; Sanchez, S.; Kirshenbaum, K.; Kascsak, R.B.; Kascsak, R.J.; Sadowski, M.J. Anti-prion Protein Antibody 6D11 Restores Cellular Proteostasis of Prion Protein Through Disrupting Recycling Propagation of PrP Sc and Targeting PrP Sc for Lysosomal Degradation. Mol. Neurobiol. 2019, 56, 2073–2091. [Google Scholar] [CrossRef] [PubMed]
- Goold, R.; McKinnon, C.; Tabrizi, S.J. Prion degradation pathways: Potential for therapeutic intervention. Mol. Cell. Neurosci. 2015, 66, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Stefanis, L. α-Synuclein in Parkinson’s disease. Cold Spring Harb. Perspect. Med. 2012, 2, a009399. [Google Scholar] [CrossRef]
- Aulić, S.; Masperone, L.; Narkiewicz, J.; Isopi, E.; Bistaffa, E.; Ambrosetti, E.; Pastore, B.; De Cecco, E.; Scaini, D.; Zago, P.; et al. α-Synuclein Amyloids Hijack Prion Protein to Gain Cell Entry, Facilitate Cell-to-Cell Spreading and Block Prion Replication. Sci. Rep. 2017, 7, 10050. [Google Scholar] [CrossRef] [PubMed]
- Laurén, J.; Gimbel, D.A.; Nygaard, H.B.; Gilbert, J.W.; Strittmatter, S.M. Cellular prion protein mediates impairment of synaptic plasticity by amyloid-Β oligomers. Nature 2009, 457, 1128–1132. [Google Scholar] [CrossRef] [PubMed]
- Filesi, I.; Cardinale, A.; Mattei, S.; Biocca, S. Selective re-routing of prion protein to proteasomes and alteration of its vesicular secretion prevent PrPSc formation. J. Neurochem. 2007, 101, 1516–1526. [Google Scholar] [CrossRef] [PubMed]
- Morani, M.; Mai, T.D.; Krupova, Z.; Defrenaix, P.; Multia, E.; Riekkola, M.L.; Taverna, M. Electrokinetic characterization of extracellular vesicles with capillary electrophoresis: A new tool for their identification and quantification. Anal. Chim. Acta 2020, 1128, 42–51. [Google Scholar] [CrossRef]
- Vivek, A.; Bolognesi, G.; Elani, Y. Fusing artificial cell compartments and lipid domains using optical traps: A tool to modulate membrane composition and phase behaviour. Micromachines 2020, 11, 388. [Google Scholar] [CrossRef]
- Cardinale, A.; Filesi, I.; Vetrugno, V.; Pocchiari, M.; Sy, M.S.; Biocca, S. Trapping prion protein in the endoplasmic reticulum impairs PrPC maturation and prevents PrPSc accumulation. J. Biol. Chem. 2005, 280, 685–694. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alves, R.N.; Iglesia, R.P.; Prado, M.B.; Melo Escobar, M.I.; Boccacino, J.M.; Fernandes, C.F.d.L.; Coelho, B.P.; Fortes, A.C.; Lopes, M.H. A New Take on Prion Protein Dynamics in Cellular Trafficking. Int. J. Mol. Sci. 2020, 21, 7763. https://doi.org/10.3390/ijms21207763
Alves RN, Iglesia RP, Prado MB, Melo Escobar MI, Boccacino JM, Fernandes CFdL, Coelho BP, Fortes AC, Lopes MH. A New Take on Prion Protein Dynamics in Cellular Trafficking. International Journal of Molecular Sciences. 2020; 21(20):7763. https://doi.org/10.3390/ijms21207763
Chicago/Turabian StyleAlves, Rodrigo Nunes, Rebeca Piatniczka Iglesia, Mariana Brandão Prado, Maria Isabel Melo Escobar, Jacqueline Marcia Boccacino, Camila Felix de Lima Fernandes, Bárbara Paranhos Coelho, Ailine Cibele Fortes, and Marilene Hohmuth Lopes. 2020. "A New Take on Prion Protein Dynamics in Cellular Trafficking" International Journal of Molecular Sciences 21, no. 20: 7763. https://doi.org/10.3390/ijms21207763
APA StyleAlves, R. N., Iglesia, R. P., Prado, M. B., Melo Escobar, M. I., Boccacino, J. M., Fernandes, C. F. d. L., Coelho, B. P., Fortes, A. C., & Lopes, M. H. (2020). A New Take on Prion Protein Dynamics in Cellular Trafficking. International Journal of Molecular Sciences, 21(20), 7763. https://doi.org/10.3390/ijms21207763