A Screen for PKN3 Substrates Reveals an Activating Phosphorylation of ARHGAP18
Abstract
1. Introduction
2. Results
2.1. Phosphoproteomic Screen for Novel PKN3 Substrates
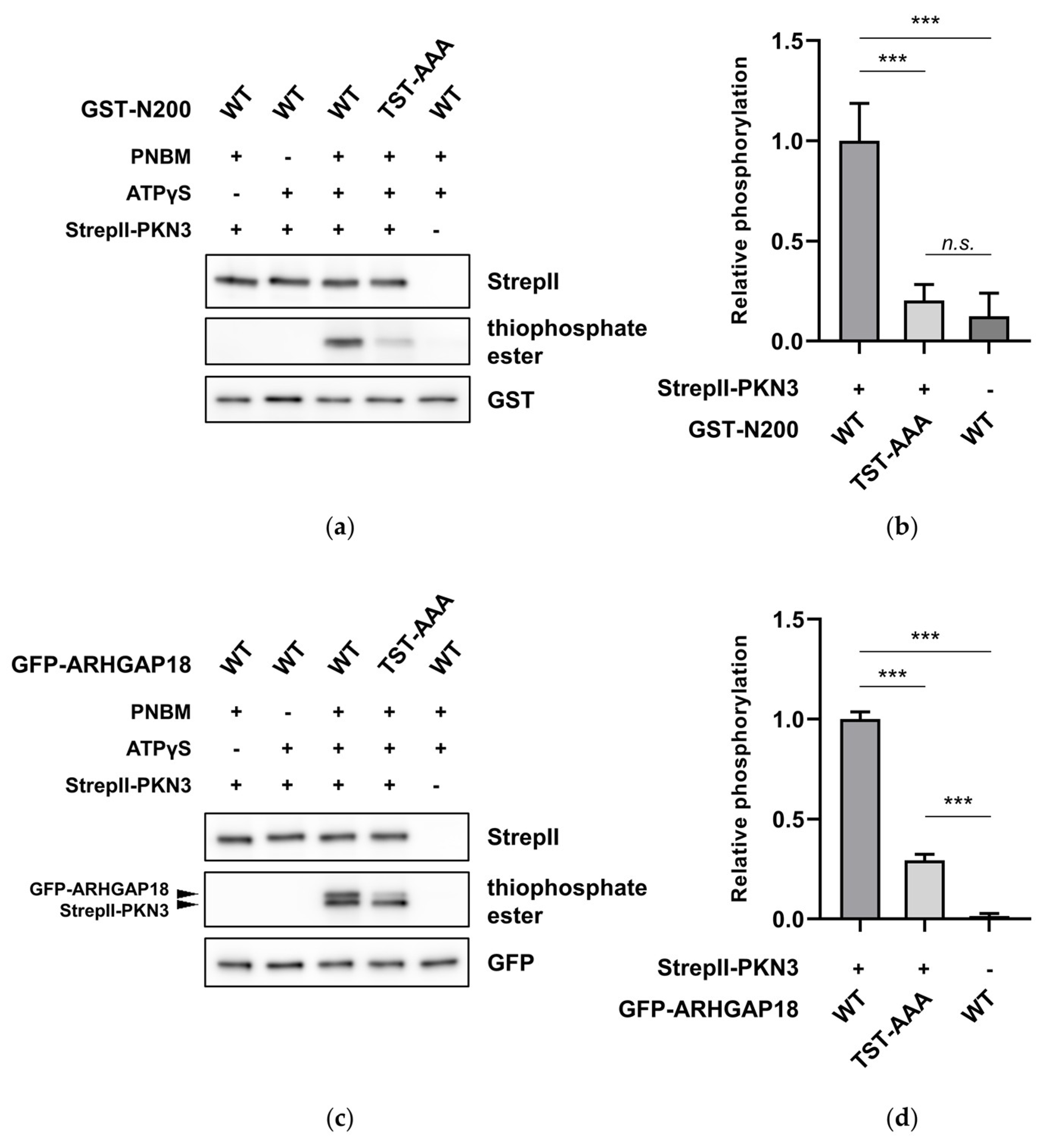
2.2. ARHGAP18 is Phosphorylated by PKN3
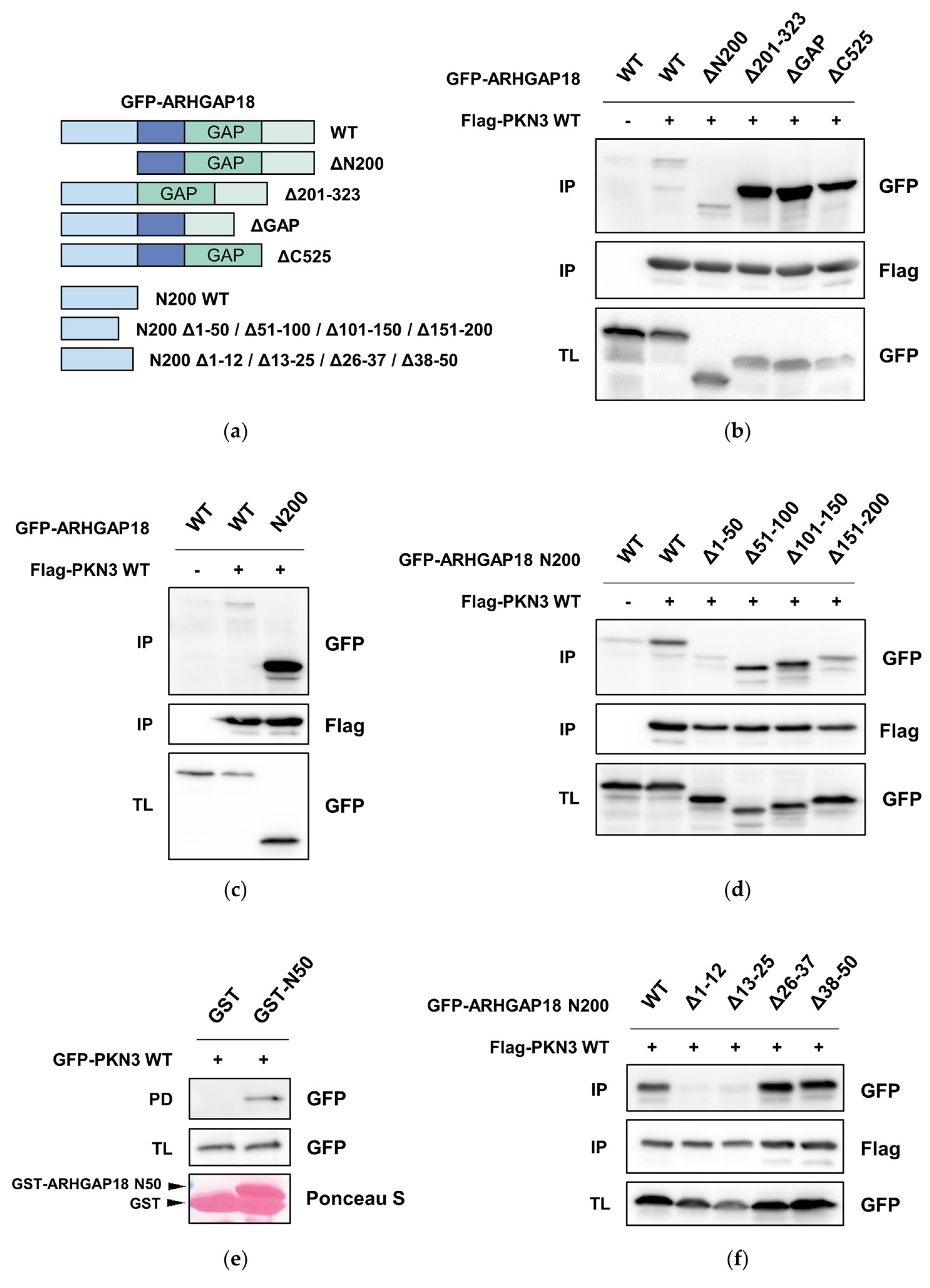
2.3. PKN3 Interacts with ARHGAP18 via its N-Terminal Region
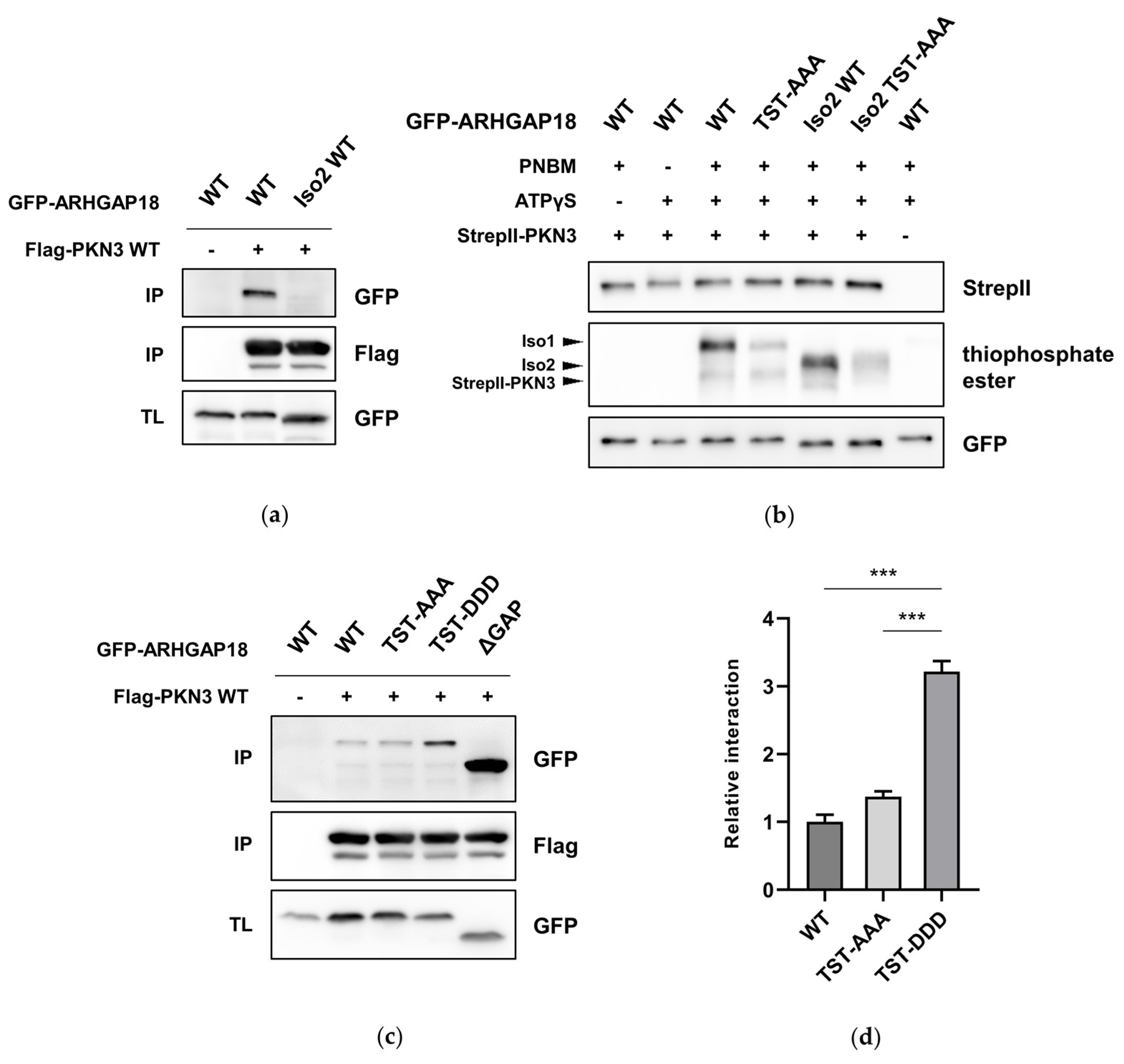
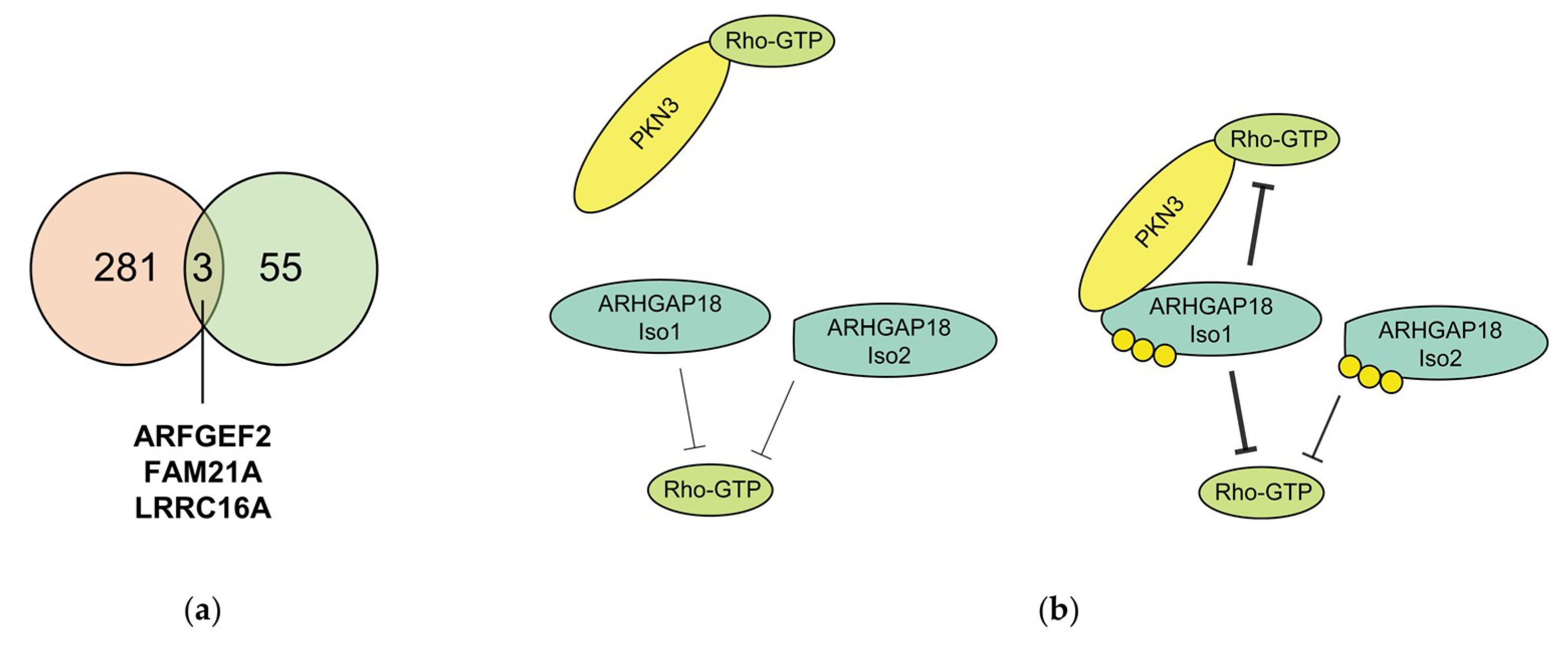
2.4. Phosphorylation of ARHGAP18 Isoform1 but not Isoform2, Promotes Interaction with PKN3
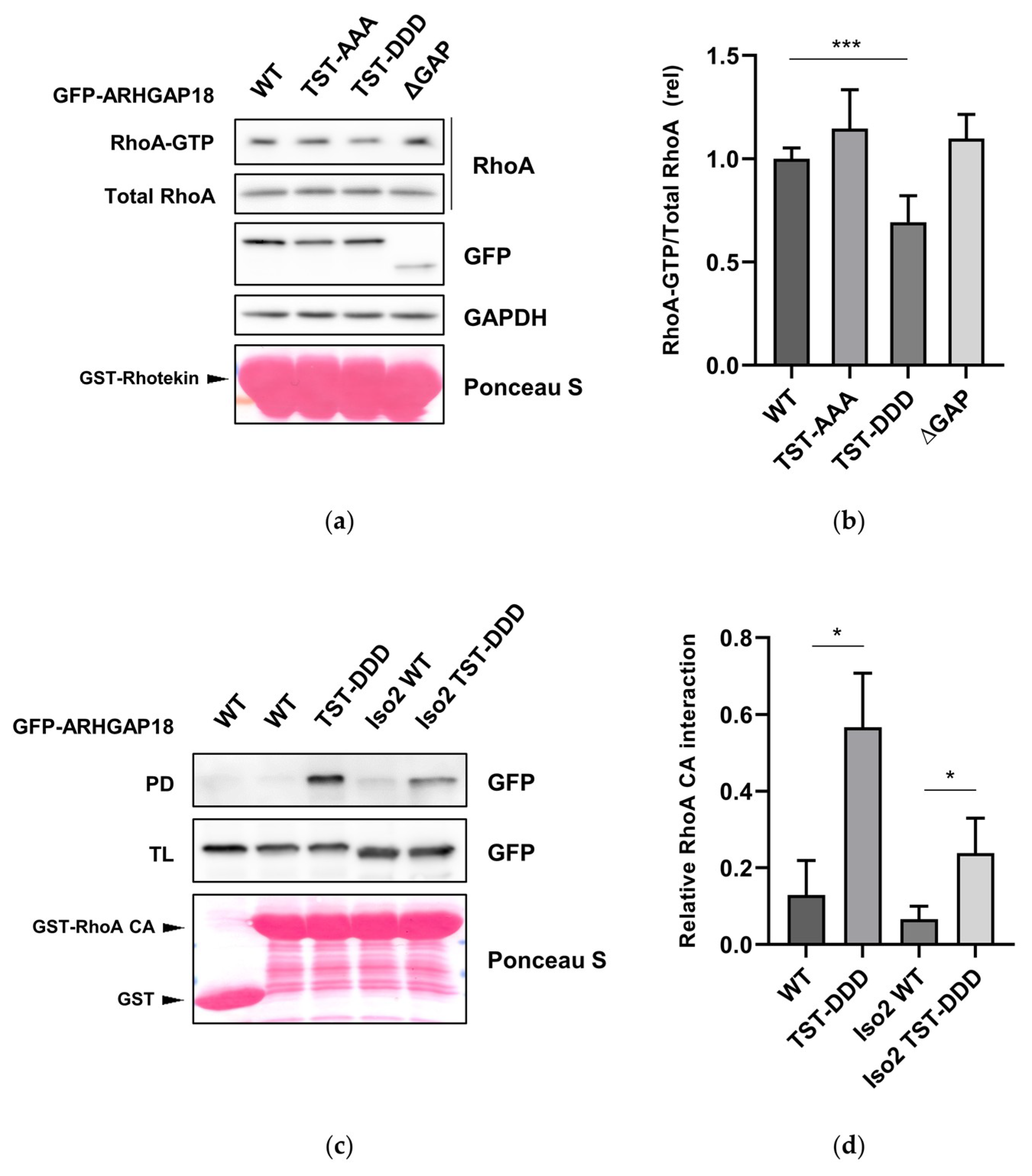
2.5. Phosphorylation of ARHGAP18 Leads to Activation of Its GAP Domain
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Cell Cultivation
4.2. Plasmid Construction
4.3. Protein Expression and Purification
4.4. Screen for PKN3 Substrates, Sample Preparation and Data Analysis
4.5. Immunoprecipitation and Immunoblotting
4.6. Antibodies
4.7. Kinase Assays
4.8. RhoA-GTP Pull-Down Assay
4.9. RhoA Pull-Down Assay
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| PKN3 | Protein Kinase N3 |
| ARHGAP18 | Rho GTPase Activating Protein 18 |
| N6-Bn ATPγS | N6-Benzyl ATPγS |
| PNBM | p-nitrobenzyl mesylate |
References
- Hashimoto, T.; Mukai, H.; Kawamata, T. Localization of PKN mRNA in the rat brain. Mol. Brain Res. 1998. [Google Scholar] [CrossRef]
- Mukai, H.; Ono, Y. A novel protein kinase with leucine zipper-like sequences: Its catalytic domain is highly homologous to that of protein kinase C. Biochem. Biophys. Res. Commun. 1994. [Google Scholar] [CrossRef] [PubMed]
- Quilliam, L.A.; Lambert, Q.T.; Mickelson-Young, L.A.; Westwick, J.K.; Sparks, A.B.; Kay, B.K.; Jenkins, N.A.; Gilbert, D.J.; Copeland, N.G.; Der, C.J. Isolation of a NCK-associated kinase, PRK2, an SH3-binding protein and potential effector of Rho protein signaling. J. Biol. Chem. 1996, 271, 28772–28776. [Google Scholar] [CrossRef] [PubMed]
- Aleku, M.; Schulz, P.; Keil, O.; Santel, A.; Schaeper, U.; Dieckhoff, B.; Janke, O.; Endruschat, J.; Durieux, B.; Röder, N.; et al. Atu027, a liposomal small interfering RNA formulation targeting protein kinase N3, inhibits cancer progression. Cancer Res. 2008, 68, 9788–9798. [Google Scholar] [CrossRef] [PubMed]
- Uehara, S.; Udagawa, N.; Mukai, H.; Ishihara, A.; Maeda, K.; Yamashita, T.; Murakami, K.; Nishita, M.; Nakamura, T.; Kato, S.; et al. Protein kinase N3 promotes bone resorption by osteoclasts in response to Wnt5a-Ror2 signaling. Sci. Signal. 2017, 10, 1–12. [Google Scholar] [CrossRef]
- Uehara, S.; Udagawa, N.; Kobayashi, Y. Regulation of osteoclast function via Rho-Pkn3-c-Src pathways. J. Oral Biosci. 2019, 61, 135–140. [Google Scholar] [CrossRef]
- Oishi, K.; Mukai, H.; Shibata, H.; Takahashi, M.; Ona, Y. Identification and characterization of PKNβ, a novel isoform of protein kinase PKN: Expression and arachidonic acid dependency are different from those of PKNα. Biochem. Biophys. Res. 1999, 814, 808–814. [Google Scholar] [CrossRef]
- Leenders, F.; Möpert, K.; Schmiedeknecht, A.; Santel, A.; Czauderna, F.; Aleku, M.; Penschuck, S.; Dames, S.; Sternberger, M.; Röhl, T.; et al. PKN3 is required for malignant prostate cell growth downstream of activated PI 3-kinase. EMBO J. 2004, 23, 3303–3313. [Google Scholar] [CrossRef]
- Unsal-Kacmaz, K.; Ragunathan, S.; Rosfjord, E.; Dann, S.; Upeslacis, E.; Grillo, M.; Hernandez, R.; Mack, F.; Klippel, A. The interaction of PKN3 with RhoC promotes malignant growth. Mol. Oncol. 2012, 6, 284–298. [Google Scholar] [CrossRef]
- Möpert, K.; Löffler, K.; Röder, N.; Kaufmann, J.; Santel, A. Depletion of protein kinase N3 (PKN3) impairs actin and adherens junctions dynamics and attenuates endothelial cell activation. Eur. J. Cell Biol. 2012, 91, 694–705. [Google Scholar] [CrossRef]
- Santel, A.; Aleku, M.; Roder, N.; Mopert, K.; Durieux, B.; Janke, O.; Keil, O.; Endruschat, J.; Dames, S.; Lange, C.; et al. Atu027 Prevents Pulmonary Metastasis in Experimental and Spontaneous Mouse Metastasis Models. Clin. Cancer Res. 2010, 16, 5469–5480. [Google Scholar] [CrossRef] [PubMed]
- Gemperle, J.; Hexnerová, R.; Lepšík, M.; Tesina, P.; Dibus, M.; Novotný, M.; Brábek, J.; Veverka, V.; Rosel, D. Structural characterization of CAS SH3 domain selectivity and regulation reveals new CAS interaction partners. Sci. Rep. 2017, 7, 8057. [Google Scholar] [CrossRef] [PubMed]
- Gemperle, J.; Dibus, M.; Koudelková, L.; Rosel, D.; Brábek, J. The interaction of p130Cas with PKN3 promotes malignant growth. Mol. Oncol. 2019, 13, 264–289. [Google Scholar] [CrossRef] [PubMed]
- Kraus, M.; Dolinski, B.; Rosahl, T.W.; Magee, J.A. Protein kinase N3 deficiency impedes PI3-kinase pathway-driven leukemogenesis without affecting normal hematopoiesis. Leukemia 2014, 29, 255–258. [Google Scholar] [CrossRef] [PubMed]
- Mukai, H.; Muramatsu, A.; Mashud, R.; Kubouchi, K.; Tsujimoto, S.; Hongu, T.; Kanaho, Y.; Tsubaki, M.; Nishida, S.; Shioi, G.; et al. PKN3 is the major regulator of angiogenesis and tumor metastasis in mice. Sci. Rep. 2016, 6, 18979. [Google Scholar] [CrossRef]
- Allen, J.J.; Li, M.; Brinkworth, C.S.; Paulson, J.L.; Wang, D.; Hübner, A.; Chou, W.-H.; Davis, R.J.; Burlingame, A.L.; Messing, R.O.; et al. A semisynthetic epitope for kinase substrates. Nat. Methods 2007, 4, 511–516. [Google Scholar] [CrossRef]
- Hertz, N.T.; Wang, B.T.; Allen, J.J.; Zhang, C.; Dar, A.C.; Burlingame, A.L.; Shokat, K.M. Chemical Genetic Approach for Kinase-Substrate Mapping by Covalent Capture of Thiophosphopeptides and Analysis by Mass Spectrometry. Curr. Protoc. Chem. Biol. 2010, 2, 15–36. [Google Scholar] [CrossRef]
- Chang, G.H.; Lay, A.J.; Ting, K.K.; Zhao, Y.; Coleman, P.R.; Powter, E.E.; Formaz-Preston, A.; Jolly, C.J.; Bower, N.I.; Hogan, B.M.; et al. Arhgap18: An endogenous inhibitor of angiogenesis, limiting tip formation and stabilizing junctions. Small GTPases 2014, 5. [Google Scholar] [CrossRef]
- Maeda, M.; Hasegawa, H.; Hyodo, T.; Ito, S.; Asano, E.; Yuang, H.; Funasaka, K.; Shimokata, K.; Hasegawa, Y.; Hamaguchi, M.; et al. ARHGAP18, a GTPase-activating protein for RhoA, controls cell shape, spreading and motility. Mol. Biol. Cell 2011, 22, 3840–3852. [Google Scholar] [CrossRef]
- Li, X.; Tao, Y.; Wang, X.; Wang, T.; Liu, J. Advanced glycosylation end products (AGEs) controls proliferation, invasion and permeability through orchestrating ARHGAP18/RhoA pathway in human umbilical vein endothelial cells. Glycoconj. J. 2020, 37, 209–219. [Google Scholar] [CrossRef]
- Porazinski, S.; Wang, H.; Asaoka, Y.; Behrndt, M.; Miyamoto, T.; Morita, H.; Hata, S.; Sasaki, T.; Krens, S.F.G.; Osada, Y.; et al. YAP is essential for tissue tension to ensure vertebrate 3D body shape. Nature 2015, 521, 217–221. [Google Scholar] [CrossRef] [PubMed]
- Vautrin-Glabik, A.; Botia, B.; Kischel, P.; Ouadid-Ahidouch, H.; Rodat-Despoix, L. IP3R3 silencing induced actin cytoskeletal reorganization through ARHGAP18/RhoA/mDia1/FAK pathway in breast cancer cell lines. Biochim. Biophys. Acta - Mol. Cell Res. 2018, 1865, 945–958. [Google Scholar] [CrossRef] [PubMed]
- Lovelace, M.D.; Powter, E.E.; Coleman, P.R.; Zhao, Y.; Parker, A.; Chang, G.H.; Lay, A.J.; Hunter, J.; McGrath, A.P.; Jormakka, M.; et al. The RhoGAP protein ARHGAP18/SENEX localizes to microtubules and regulates their stability in endothelial cells. Mol. Biol. Cell 2017, 28, 1066–1078. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ji, S.; Fu, L.; Jiang, T.; Wu, D.; Meng, F. Over-expression of ARHGAP18 suppressed cell proliferation, migration, invasion and tumor growth in gastric cancer by restraining over-activation of MAPK signaling pathways. Onco. Targets. Ther. 2018, 11, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Humphries, B.; Wang, Z.; Li, Y.; Jhan, J.R.; Jiang, Y.; Yang, C. ARHGAP18 downregulation by miR-200b suppresses metastasis of triple-negative breast cancer by enhancing activation of RhoA. Cancer Res. 2017, 77, 4051–4064. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Rojas, A.; Maya-Núñez, G.; Huerta-Reyes, M.; Pérez-Solis, M.A.; Silva-García, R.; Guillén, N.; Olivo-Marin, J.C. Activation of human gonadotropin-releasing hormone receptor promotes down regulation of ARHGAP18 and regulates the cell invasion of MDA-MB-231 cells. Mol. Cell. Endocrinol. 2018, 460, 94–103. [Google Scholar] [CrossRef] [PubMed]
- Aleskandarany, M.A.; Sonbul, S.; Surridge, R.; Mukherjee, A.; Caldas, C.; Diez-Rodriguez, M.; Ashankyty, I.; Albrahim, K.I.; Elmouna, A.M.; Aneja, R.; et al. Rho-GTPase activating-protein 18: A biomarker associated with good prognosis in invasive breast cancer. Br. J. Cancer 2017, 117, 1176–1184. [Google Scholar] [CrossRef][Green Version]
- Coleman, P.R.; Hahn, C.N.; Grimshaw, M.; Lu, Y.; Li, X.; Brautigan, P.J.; Beck, K.; Stocker, R.; Vadas, M.A.; Gamble, J.R. Stress-induced premature senescence mediated by a novel gene, SENEX, results in an anti-inflammatory phenotype in endothelial cells. Blood 2010, 116, 4016–4024. [Google Scholar] [CrossRef]
- Lay, A.J.; Coleman, P.R.; Formaz-Preston, A.; Ka Ting, K.; Roediger, B.; Weninger, W.; Schwartz, M.A.; Vadas, M.A.; Gamble, J.R. ARHGAP18: A flow-responsive gene that regulates endothelial cell alignment and protects against atherosclerosis. J. Am. Heart Assoc. 2019, 8. [Google Scholar] [CrossRef]
- Coleman, P.R.; Lay, A.J.; Ting, K.K.; Zhao, Y.; Li, J.; Jarrah, S.; Vadas, M.A.; Gamble, J.R. YAP and the RhoC regulator ARHGAP18, are required to mediate flow-dependent endothelial cell alignment. Cell Commun. Signal. 2020, 18. [Google Scholar] [CrossRef]
- Cibrián Uhalte, E.; Kirchner, M.; Hellwig, N.; Allen, J.J.; Donat, S.; Shokat, K.M.; Selbach, M.; Abdelilah-Seyfried, S. In Vivo Conditions to Identify Prkci Phosphorylation Targets Using the Analog-Sensitive Kinase Method in Zebrafish. PLoS ONE 2012, 7, e40000. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.X.; Jung, D.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef] [PubMed]
- Shibata, H.; Oishi, K.; Yamagiwa, A.; Matsumoto, M.; Mukai, H.; Ono, Y. PKNbeta interacts with the SH3 Domains of Graf and a Novel Graf Related Protein, Graf 2, Which Are GTPase Activating Proteins for Rho. J. Biochem. 2001, 31, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Collazos, A.; Michael, N.; Whelan, R.D.H.; Kelly, G.; Mellor, H.; Pang, L.C.H.; Totty, N.; Parker, P.J. Site recognition and substrate screens for PKN family proteins. Biochem. J. 2011, 438, 535–543. [Google Scholar] [CrossRef]
- Browne, C.M.; Jiang, B.; Ficarro, S.B.; Doctor, Z.M.; Johnson, J.L.; Card, J.D.; Sivakumaren, S.C.; Alexander, W.M.; Yaron, T.M.; Murphy, C.J.; et al. A Chemoproteomic Strategy for Direct and Proteome-Wide Covalent Inhibitor Target-Site Identification. J. Am. Chem. Soc. 2019, 141, 191–203. [Google Scholar] [CrossRef]
- Linardopoulou, E.V.; Parghi, S.S.; Friedman, C.; Osborn, G.E.; Parkhurst, S.M.; Trask, B.J. Human Subtelomeric WASH Genes Encode a New Subclass of the WASP Family. PLoS Genet. 2007, 3, e237. [Google Scholar] [CrossRef]
- Yang, C.; Pring, M.; Wear, M.A.; Huang, M.; Cooper, J.A.; Svitkina, T.M.; Zigmond, S.H. Mammalian CARMIL inhibits actin filament capping by capping protein. Dev. Cell 2005, 9, 209–221. [Google Scholar] [CrossRef]
- Liang, Y.; Niederstrasser, H.; Edwards, M.; Jackson, C.E.; Cooper, J.A. Distinct roles for CARMIL isoforms in cell migration. Mol. Biol. Cell 2009, 20, 5290–5305. [Google Scholar] [CrossRef]
- Mori, K.; Amano, M.; Takefuji, M.; Kato, K.; Morita, Y.; Nishioka, T.; Matsuura, Y.; Murohara, T.; Kaibuchi, K. Rho-kinase contributes to sustained RhoA activation through phosphorylation of p190A RhoGAP. J. Biol. Chem. 2009, 284, 5067–5076. [Google Scholar] [CrossRef]
- Jiang, W.; Betson, M.; Mulloy, R.; Foster, R.; Lévay, M.; Ligeti, E.; Settleman, J. P190A RhoGAP is a glycogen synthase kinase-3-β substrate required for polarized cell migration. J. Biol. Chem. 2008, 283, 20978–20988. [Google Scholar] [CrossRef]
- Minoshima, Y.; Kawashima, T.; Hirose, K.; Tonozuka, Y.; Kawajiri, A.; Bao, Y.C.; Deng, X.; Tatsuka, M.; Narumiya, S.; May, W.S.; et al. Phosphorylation by Aurora B converts MgcRacGAP to a RhoGAP during cytokinesis. Dev. Cell 2003, 4, 549–560. [Google Scholar] [CrossRef]
- Luo, W.; Janoštiak, R.; Tolde, O.; Ryzhova, L.M.; Koudelková, L.; Dibus, M.; Brábek, J.; Hanks, S.K.; Rosel, D. ARHGAP42 is activated by Src-mediated tyrosine phosphorylation to promote cell motility. J. Cell Sci. 2017, 130, 2382–2393. [Google Scholar] [CrossRef] [PubMed]
- Lusis, A.J. Atherosclerosis. Nature 2000, 407, 233–241. [Google Scholar] [CrossRef]
- Hägg, S.; Skogsberg, J.; Lundström, J.; Noori, P.; Nilsson, R.; Zhong, H.; Maleki, S.; Shang, M.M.; Brinne, B.R.; Bradshaw, M.; et al. Multi-organ expression profiling uncovers a gene module in coronary artery disease involving transendothelial migration of leukocytes and LIM domain binding 2: The Stockholm Atherosclerosis Gene Expression (STAGE) study. PLoS Genet. 2009, 5, e1000754. [Google Scholar] [CrossRef]
- Zhang, C.J.; Zhu, N.; Liu, Z.; Shi, Z.; Long, J.; Zu, X.Y.; Tang, Z.W.; Hu, Z.Y.; Liao, D.F.; Qin, L. Wnt5a/Ror2 pathway contributes to the regulation of cholesterol homeostasis and inflammatory response in atherosclerosis. Biochim. Biophys. Acta - Mol. Cell Biol. Lipids 2020, 1865, 158547. [Google Scholar] [CrossRef]
- García-Mata, R.; Wennerberg, K.; Arthur, W.T.; Noren, N.K.; Ellerbroek, S.M.; Burridge, K. Analysis of activated GAPs and GEFs in cell lysates. Methods Enzymol. 2006, 406, 425–437. [Google Scholar] [CrossRef]


| Gene | Site | Surrounding Sequence | PLP | Score |
|---|---|---|---|---|
| ARHGAP18 | T158 | KRVETVSQTLRKKNKQY | 1.00 | 111.46 |
| S156 | VQKRVETVSQTLRKKNK | 1.00 | 111.46 | |
| BRD4 | S601 | SKPPPTYESEEEDKCKP | 1.00 | 101.9 |
| CCDC144A | S805 | EMARKKMNSEISHRHQK | 0.99 | 82.029 |
| CEACAM16 | T402 | NLTDTGRYTLKTVTVQG | 1.00 | 100.25 |
| T398 | VQKLNLTDTGRYTLKTV | 1.00 | 100.25 | |
| DCAF12L1 | S8 | _MAQQQTGSRKRKAPAV | 1.00 | 83.801 |
| DSCAM | S1408 | LPGDNGGSSIRGYILQY | 0.98 | 87.498 |
| IGSF22 | S318 | LSVGDKRMSAELTVLDE | 0.99 | 84.817 |
| KIF27 | S754 | TGNDAKSVSKQYSLKVT | 1.00 | 120.77 |
| T746 | DLIKELIKTGNDAKSVS | 0.99 | 120.77 | |
| NFE2L2 | T137 | KLHHNYKITIYSM____ | 1.00 | 99.139 |
| ODF1 | S10 | AALSCLLDSVRRDIKKV | 1.00 | 103.7 |
| OR4K15 | T158 | ICKPLHYMTVMSRRVCV | 0.99 | 79.906 |
| POLE1 | S891 | VKKPKVTISYPGAMLNI | 1.00 | 82.483 |
| T889 | TNVKKPKVTISYPGAML | 1.00 | 82.483 | |
| PRDM16 | S1218 | DVLNSTLDSEALKHTLC | 0.99 | 96.371 |
| RETSAT | S322 | VLTKATVQSVLLDSAGK | 0.98 | 99.53 |
| SACS | S1222 | GIFTKPSLSAVLKHFKI | 1.00 | 86.512 |
| SPATA31D1 | T1339 | VLGSKSSPTLKTQPPPE | 0.87 | 86.189 |
| USP42 | T786 | PRDPGTPATKEGAWEAM | 1.00 | 93.096 |
| UTP20 | S926 | HLQVFSKFSNPRALYLE | 1.00 | 101.65 |
| VPS33B | S476 | AGKITDAFSSLAKRSNF | 0.78 | 83.206 |
| S477 | GKITDAFSSLAKRSNFR | 0.78 | 83.206 | |
| VWA3B | T99 | EDGRVYNLTAKSELIYQ | 1.00 | 84.244 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dibus, M.; Brábek, J.; Rösel, D. A Screen for PKN3 Substrates Reveals an Activating Phosphorylation of ARHGAP18. Int. J. Mol. Sci. 2020, 21, 7769. https://doi.org/10.3390/ijms21207769
Dibus M, Brábek J, Rösel D. A Screen for PKN3 Substrates Reveals an Activating Phosphorylation of ARHGAP18. International Journal of Molecular Sciences. 2020; 21(20):7769. https://doi.org/10.3390/ijms21207769
Chicago/Turabian StyleDibus, Michal, Jan Brábek, and Daniel Rösel. 2020. "A Screen for PKN3 Substrates Reveals an Activating Phosphorylation of ARHGAP18" International Journal of Molecular Sciences 21, no. 20: 7769. https://doi.org/10.3390/ijms21207769
APA StyleDibus, M., Brábek, J., & Rösel, D. (2020). A Screen for PKN3 Substrates Reveals an Activating Phosphorylation of ARHGAP18. International Journal of Molecular Sciences, 21(20), 7769. https://doi.org/10.3390/ijms21207769





