The Loss of HLA-F/KIR3DS1 Ligation Is Mediated by Hemoglobin Peptides
Abstract
1. Introduction
2. Results
2.1. Hemoglobin Subunits were Upregulated in CD4+/HIV+ T Cells
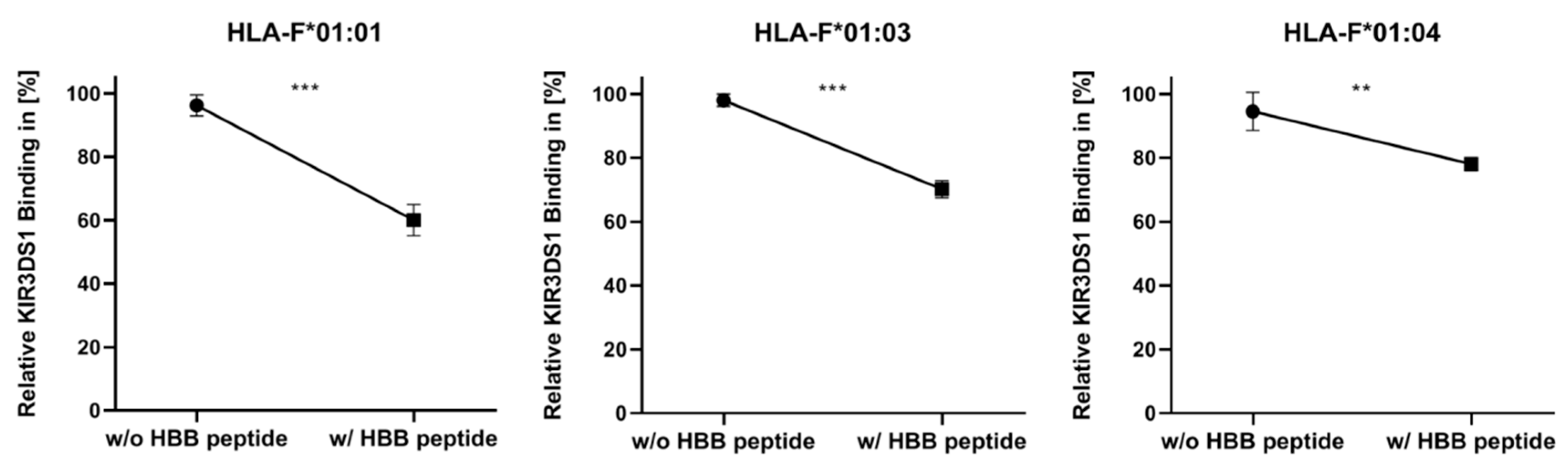
2.2. Presentation of HBB Peptide Reduced the Recognition of HLA-F by KIR3DS1
3. Discussion
4. Materials and Methods
4.1. Proteome Analysis of CD4+ T Cells of HIV-Infected Person
4.2. Maintenance of Cell Lines
4.3. Cloning and Transduction of HLA-F Constructs
4.4. Detection of HLA-F on Transduced Cells via KIR3DS1-Binding Assay
4.5. Peptide Binding Assay for HLA-F
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Lee, N.; Ishitani, A.; Geraghty, D.E. HLA-F is a surface marker on activated lymphocytes. Eur. J. Immunol. 2010, 40, 2308–2318. [Google Scholar] [CrossRef] [PubMed]
- Goodridge, J.P.; Burian, A.; Lee, N.; Geraghty, D.E. HLA-F complex without peptide binds to MHC class I protein in the open conformer form. J. Immunol. 2010, 184, 6199–6208. [Google Scholar] [CrossRef] [PubMed]
- Wainwright, S.D.; Biro, P.A.; Holmes, C.H. HLA-F is a predominantly empty, intracellular, TAP-associated MHC class Ib protein with a restricted expression pattern. J. Immunol. 2000, 164, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Goodridge, J.P.; Burian, A.; Lee, N.; Geraghty, D.E. HLA-F and MHC class I open conformers are ligands for NK cell Ig-like receptors. J. Immunol. 2013, 191, 3553–3562. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Beltran, W.F.; Holzemer, A.; Martrus, G.; Chung, A.W.; Pacheco, Y.; Simoneau, C.R.; Rucevic, M.; Lamothe-Molina, P.A.; Pertel, T.; Kim, T.E.; et al. Open conformers of HLA-F are high-affinity ligands of the activating NK-cell receptor KIR3DS1. Nat. Immunol. 2016, 17, 1067–1074. [Google Scholar] [CrossRef] [PubMed]
- Dulberger, C.L.; McMurtrey, C.P.; Holzemer, A.; Neu, K.E.; Liu, V.; Steinbach, A.M.; Garcia-Beltran, W.F.; Sulak, M.; Jabri, B.; Lynch, V.J.; et al. Human Leukocyte Antigen F Presents Peptides and Regulates Immunity through Interactions with NK Cell Receptors. Immunity 2017, 46, 1018. [Google Scholar] [CrossRef]
- Ho, G.T.; Heinen, F.J.; Huyton, T.; Blasczyk, R.; Bade-Doding, C. HLA-F*01:01 presents peptides with N-terminal flexibility and a preferred length of 16 residues. Immunogenetics 2019, 71, 353–360. [Google Scholar] [CrossRef]
- Ho, G.T.; Heinen, F.J.; Blasczyk, R.; Pich, A.; Bade-Doeding, C. HLA-F Allele-Specific Peptide Restriction Represents an Exceptional Proteomic Footprint. Int. J. Mol. Sci. 2019, 20, 5572. [Google Scholar] [CrossRef]
- Lepin, E.J.; Bastin, J.M.; Allan, D.S.; Roncador, G.; Braud, V.M.; Mason, D.Y.; van der Merwe, P.A.; McMichael, A.J.; Bell, J.I.; Powis, S.H.; et al. Functional characterization of HLA-F and binding of HLA-F tetramers to ILT2 and ILT4 receptors. Eur. J. Immunol. 2000, 30, 3552–3561. [Google Scholar] [CrossRef]
- Manandhar, T.; Hò, G.-G.T.; Pump, W.C.; Blasczyk, R.; Bade-Doeding, C. Battle between Host Immune Cellular Responses and HCMV Immune Evasion. Int. J. Mol. Sci. 2019, 20, 3626. [Google Scholar] [CrossRef]
- Guha, D.; Ayyavoo, V. Innate immune evasion strategies by human immunodeficiency virus type 1. ISRN Aids 2013, 2013, 954806. [Google Scholar] [CrossRef] [PubMed]
- Pappworth, I.Y.; Wang, E.C.; Rowe, M. The switch from latent to productive infection in epstein-barr virus-infected B cells is associated with sensitization to NK cell killing. J. Virol. 2007, 81, 474–482. [Google Scholar] [CrossRef] [PubMed]
- Tomasec, P.; Braud, V.M.; Rickards, C.; Powell, M.B.; McSharry, B.P.; Gadola, S.; Cerundolo, V.; Borysiewicz, L.K.; McMichael, A.J.; Wilkinson, G.W. Surface expression of HLA-E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science 2000, 287, 1031. [Google Scholar] [CrossRef] [PubMed]
- Nattermann, J.; Nischalke, H.D.; Hofmeister, V.; Ahlenstiel, G.; Zimmermann, H.; Leifeld, L.; Weiss, E.H.; Sauerbruch, T.; Spengler, U. The HLA-A2 restricted T cell epitope HCV core 35-44 stabilizes HLA-E expression and inhibits cytolysis mediated by natural killer cells. Am. J. Pathol. 2005, 166, 443–453. [Google Scholar] [CrossRef]
- Nattermann, J.; Nischalke, H.D.; Hofmeister, V.; Kupfer, B.; Ahlenstiel, G.; Feldmann, G.; Rockstroh, J.; Weiss, E.H.; Sauerbruch, T.; Spengler, U. HIV-1 infection leads to increased HLA-E expression resulting in impaired function of natural killer cells. Antivir. Ther. 2005, 10, 95–107. [Google Scholar]
- Carosella, E.D.; Rouas-Freiss, N.; Tronik-Le Roux, D.; Moreau, P.; LeMaoult, J. HLA-G: An Immune Checkpoint Molecule. Adv. Immunol. 2015, 127, 33–144. [Google Scholar] [CrossRef]
- Celik, A.A.; Simper, G.S.; Hiemisch, W.; Blasczyk, R.; Bade-Döding, C. HLA-G peptide preferences change in transformed cells: Impact on the binding motif. Immunogenetics 2018, 70, 485–494. [Google Scholar] [CrossRef]
- Hofstetter, A.R.; Sullivan, L.C.; Lukacher, A.E.; Brooks, A.G. Diverse roles of non-diverse molecules: MHC class Ib molecules in host defense and control of autoimmunity. Curr. Opin. Immunol. 2011, 23, 104–110. [Google Scholar] [CrossRef]
- Cohen, G.B.; Gandhi, R.T.; Davis, D.M.; Mandelboim, O.; Chen, B.K.; Strominger, J.L.; Baltimore, D. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity 1999, 10, 661–671. [Google Scholar] [CrossRef]
- Korner, C.; Altfeld, M. Role of KIR3DS1 in human diseases. Front. Immunol. 2012, 3, 326. [Google Scholar] [CrossRef]
- Kiani, Z.; Bruneau, J.; Geraghty, D.E.; Bernard, N.F. HLA-F on Autologous HIV-Infected Cells Activates Primary NK Cells Expressing the Activating Killer Immunoglobulin-Like Receptor KIR3DS1. J. Virol. 2019, 93. [Google Scholar] [CrossRef] [PubMed]
- Celik, A.A.; Simper, G.S.; Huyton, T.; Blasczyk, R.; Bade-Doding, C. HLA-G mediated immune regulation is impaired by a single amino acid exchange in the alpha 2 domain. Hum. Immunol. 2018, 79, 453–462. [Google Scholar] [CrossRef] [PubMed]
- Celik, A.A.; Kraemer, T.; Huyton, T.; Blasczyk, R.; Bade-Doding, C. The diversity of the HLA-E-restricted peptide repertoire explains the immunological impact of the Arg107Gly mismatch. Immunogenetics 2016, 68, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Kraemer, T.; Celik, A.A.; Huyton, T.; Kunze-Schumacher, H.; Blasczyk, R.; Bade-Doding, C. HLA-E: Presentation of a Broader Peptide Repertoire Impacts the Cellular Immune Response-Implications on HSCT Outcome. Stem Cells Int. 2015, 2015, 346714. [Google Scholar] [CrossRef]
- Chan, K.F.; Gully, B.S.; Gras, S.; Beringer, D.X.; Kjer-Nielsen, L.; Cebon, J.; McCluskey, J.; Chen, W.; Rossjohn, J. Divergent T-cell receptor recognition modes of a HLA-I restricted extended tumour-associated peptide. Nat. Commun. 2018, 9, 1026. [Google Scholar] [CrossRef]
- Beringer, D.X.; Kleijwegt, F.S.; Wiede, F.; van der Slik, A.R.; Loh, K.L.; Petersen, J.; Dudek, N.L.; Duinkerken, G.; Laban, S.; Joosten, A.; et al. T cell receptor reversed polarity recognition of a self-antigen major histocompatibility complex. Nat. Immunol. 2015, 16, 1153–1161. [Google Scholar] [CrossRef]
- Liepe, J.; Marino, F.; Sidney, J.; Jeko, A.; Bunting, D.E.; Sette, A.; Kloetzel, P.M.; Stumpf, M.P.; Heck, A.J.; Mishto, M. A large fraction of HLA class I ligands are proteasome-generated spliced peptides. Science 2016, 354, 354–358. [Google Scholar] [CrossRef]
- Supuran, C.T. Carbonic anhydrases: Novel therapeutic applications for inhibitors and activators. Nat. Rev. Drug Discov. 2008, 7, 168–181. [Google Scholar] [CrossRef]
- Saha, D.; Patgaonkar, M.; Shroff, A.; Ayyar, K.; Bashir, T.; Reddy, K.V. Hemoglobin expression in nonerythroid cells: Novel or ubiquitous? Int. J. Inflamm. 2014, 2014, 803237. [Google Scholar] [CrossRef]
- Lampe, J.B.; Schneider-Schaulies, S.; Aguzzi, A. Expression of the interferon-induced MxA protein in viral encephalitis. Neuropathol. Appl. Neurobiol. 2003, 29, 273–279. [Google Scholar] [CrossRef]
- Betancor, G.; Dicks, M.D.J.; Jimenez-Guardeno, J.M.; Ali, N.H.; Apolonia, L.; Malim, M.H. The GTPase Domain of MX2 Interacts with the HIV-1 Capsid, Enabling Its Short Isoform to Moderate Antiviral Restriction. Cell Rep. 2019, 29, 1923–1933.e1923. [Google Scholar] [CrossRef]
- Guo, T.; Zuo, Y.; Qian, L.; Liu, J.; Yuan, Y.; Xu, K.; Miao, Y.; Feng, Q.; Chen, X.; Jin, L.; et al. ADP-ribosyltransferase PARP11 modulates the interferon antiviral response by mono-ADP-ribosylating the ubiquitin E3 ligase beta-TrCP. Nat. Microbiol. 2019, 4, 1872–1884. [Google Scholar] [CrossRef] [PubMed]
- Gia-Gia Toni, H.; Funmilola, H.; Florian, S.; Rainer, B.; Christina, B.-D. Dynamic Interaction between Immune Escape Mechanism and HLA-Ib Regulation. Immunogenetics 2019, 179–182. [Google Scholar] [CrossRef]
- Pump, W.C.; Kraemer, T.; Huyton, T.; Ho, G.T.; Blasczyk, R.; Bade-Doeding, C. Between Innate and Adaptive Immune Responses: NKG2A, NKG2C, and CD8(+) T Cell Recognition of HLA-E Restricted Self-Peptides Acquired in the Absence of HLA-Ia. Int. J. Mol. Sci. 2019, 20, 1454. [Google Scholar] [CrossRef] [PubMed]
- Nemeth, J.; Vongrad, V.; Metzner, K.J.; Strouvelle, V.P.; Weber, R.; Pedrioli, P.; Aebersold, R.; Gunthard, H.F.; Collins, B.C. In vivo and in vitro Proteome Analysis of Human Immunodeficiency Virus (HIV)-1-infected, Human CD4(+) T Cells. Mol. Cell. Proteom. 2017, 16, S108–S123. [Google Scholar] [CrossRef]
- Berge, T.; Eriksson, A.; Brorson, I.S.; Hogestol, E.A.; Berg-Hansen, P.; Doskeland, A.; Mjaavatten, O.; Bos, S.D.; Harbo, H.F.; Berven, F. Quantitative proteomic analyses of CD4(+) and CD8(+) T cells reveal differentially expressed proteins in multiple sclerosis patients and healthy controls. Clin. Proteom. 2019, 16, 19. [Google Scholar] [CrossRef]
- Perutz, M.F.; Rossmann, M.G.; Cullis, A.F.; Muirhead, H.; Will, G.; North, A.C. Structure of haemoglobin: A three-dimensional Fourier synthesis at 5.5-A. resolution, obtained by X-ray analysis. Nature 1960, 185, 416–422. [Google Scholar] [CrossRef]
- Liu, L.L.; Zhu, S.G.; Jiang, X.Y.; Ren, J.; Lin, Y.; Zhang, N.N.; Tong, M.L.; Zhang, H.L.; Zheng, W.H.; Fu, H.J.; et al. LncRNA Expression in CD4+ T Cells in Neurosyphilis Patients. Front. Cell. Infect. Microbiol. 2017, 7, 461. [Google Scholar] [CrossRef]
- Basu, A.; Chakrabarti, A. Hemoglobin interacting proteins and implications of spectrin hemoglobin interaction. J. Proteom. 2015, 128, 469–475. [Google Scholar] [CrossRef]
- Gomes, I.; Dale, C.S.; Casten, K.; Geigner, M.A.; Gozzo, F.C.; Ferro, E.S.; Heimann, A.S.; Devi, L.A. Hemoglobin-derived peptides as novel type of bioactive signaling molecules. AAPS J. 2010, 12, 658–669. [Google Scholar] [CrossRef]
- Huyton, T.; Ladas, N.; Schumacher, H.; Blasczyk, R.; Bade-Doeding, C. Pocketcheck: Updating the HLA class I peptide specificity roadmap. Tissue Antigens 2012, 80, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Elsner, H.A.; DeLuca, D.; Strub, J.; Blasczyk, R. HistoCheck: Rating of HLA class I and II mismatches by an internet-based software tool. Bone Marrow Transplant. 2004, 33, 165–169. [Google Scholar] [CrossRef] [PubMed]
- Kramer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]
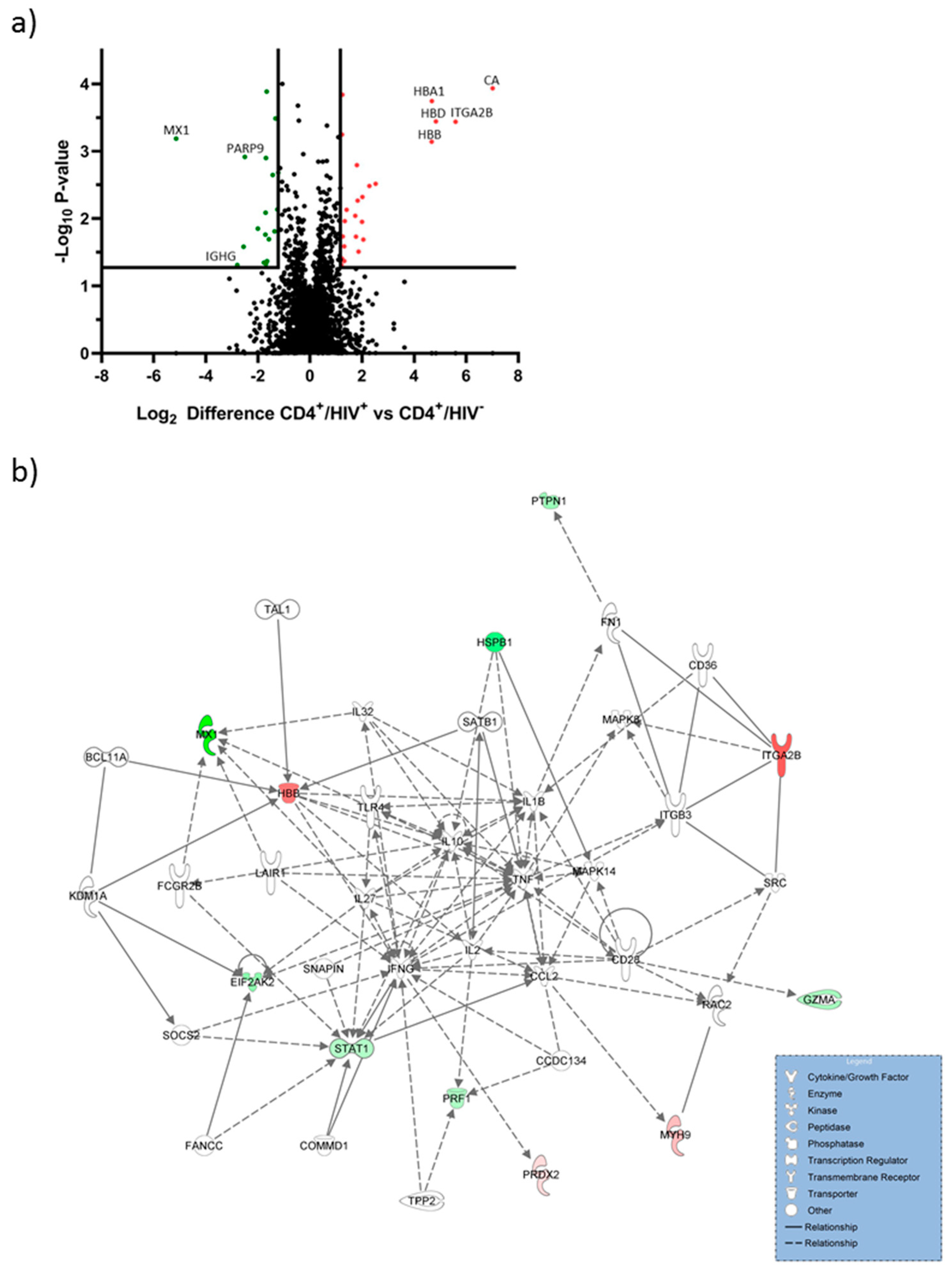
| Protein Name | Gene Code | log2 Regulation | p Value |
|---|---|---|---|
| Carbonic anhydrase 1 | CA1 | 7.01 | <0.001 |
| Integrin alpha-IIb | ITGA2B | 5.59 | <0.001 |
| Hemoglobin subunit delta | HBD | 4.83 | <0.001 |
| Hemoglobin subunit alpha | HBA1 | 4.67 | <0.001 |
| Hemoglobin subunit beta | HBB | 4.67 | <0.001 |
| Flavin reductase (NADPH) | BLVRB | 2.52 | 0.003 |
| Myosin-9 | MYH9 | 2.28 | 0.003 |
| Myosin-14 | MYH14 | 1.99 | 0.011 |
| WD repeat-containing protein 61 | WDR61 | 1.80 | 0.001 |
| Pyridoxal kinase | PDXK | 1.76 | 0.018 |
| Protein Name | Gene Code | log2 Regulation | p Value |
|---|---|---|---|
| Interferon-induced GTP-binding protein Mx1 | MX1 | −5.14 | <0.001 |
| Heat shock protein beta-1 | HSPB1 | −2.79 | 0.048 |
| Ig gamma-1 chain C region | IGHG1 | −2.55 | 0.026 |
| Poly [ADP-ribose] polymerase 9 | PARP9 | −2.50 | 0.001 |
| Myosin 1F | MYO1F | −2.00 | 0.014 |
| Calmodulin Like 5 | CALML5 | −1.71 | 0.017 |
| Retention In Endoplasmic Reticulum Sorting Receptor 1 | RER1 | −1.71 | 0.008 |
| Mannose-P-Dolichol Utilization Defect 1 | MPDU1 | −1.69 | 0.001 |
| Thymidine Phosphorylase | TYMP | −1.66 | <0.001 |
| Protein Tyrosine Phosphatase Non-Receptor Type 1 | PTPN1 | −1.58 | 0.020 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hò, G.-G.T.; Hiemisch, W.; Pich, A.; Behrens, G.M.N.; Blasczyk, R.; Bade-Doeding, C. The Loss of HLA-F/KIR3DS1 Ligation Is Mediated by Hemoglobin Peptides. Int. J. Mol. Sci. 2020, 21, 8012. https://doi.org/10.3390/ijms21218012
Hò G-GT, Hiemisch W, Pich A, Behrens GMN, Blasczyk R, Bade-Doeding C. The Loss of HLA-F/KIR3DS1 Ligation Is Mediated by Hemoglobin Peptides. International Journal of Molecular Sciences. 2020; 21(21):8012. https://doi.org/10.3390/ijms21218012
Chicago/Turabian StyleHò, Gia-Gia T., Wiebke Hiemisch, Andreas Pich, Georg M. N. Behrens, Rainer Blasczyk, and Christina Bade-Doeding. 2020. "The Loss of HLA-F/KIR3DS1 Ligation Is Mediated by Hemoglobin Peptides" International Journal of Molecular Sciences 21, no. 21: 8012. https://doi.org/10.3390/ijms21218012
APA StyleHò, G.-G. T., Hiemisch, W., Pich, A., Behrens, G. M. N., Blasczyk, R., & Bade-Doeding, C. (2020). The Loss of HLA-F/KIR3DS1 Ligation Is Mediated by Hemoglobin Peptides. International Journal of Molecular Sciences, 21(21), 8012. https://doi.org/10.3390/ijms21218012






