Light-Induced Oxidase Activity of DNAzyme-Modified Quantum Dots
Abstract
:1. Introduction
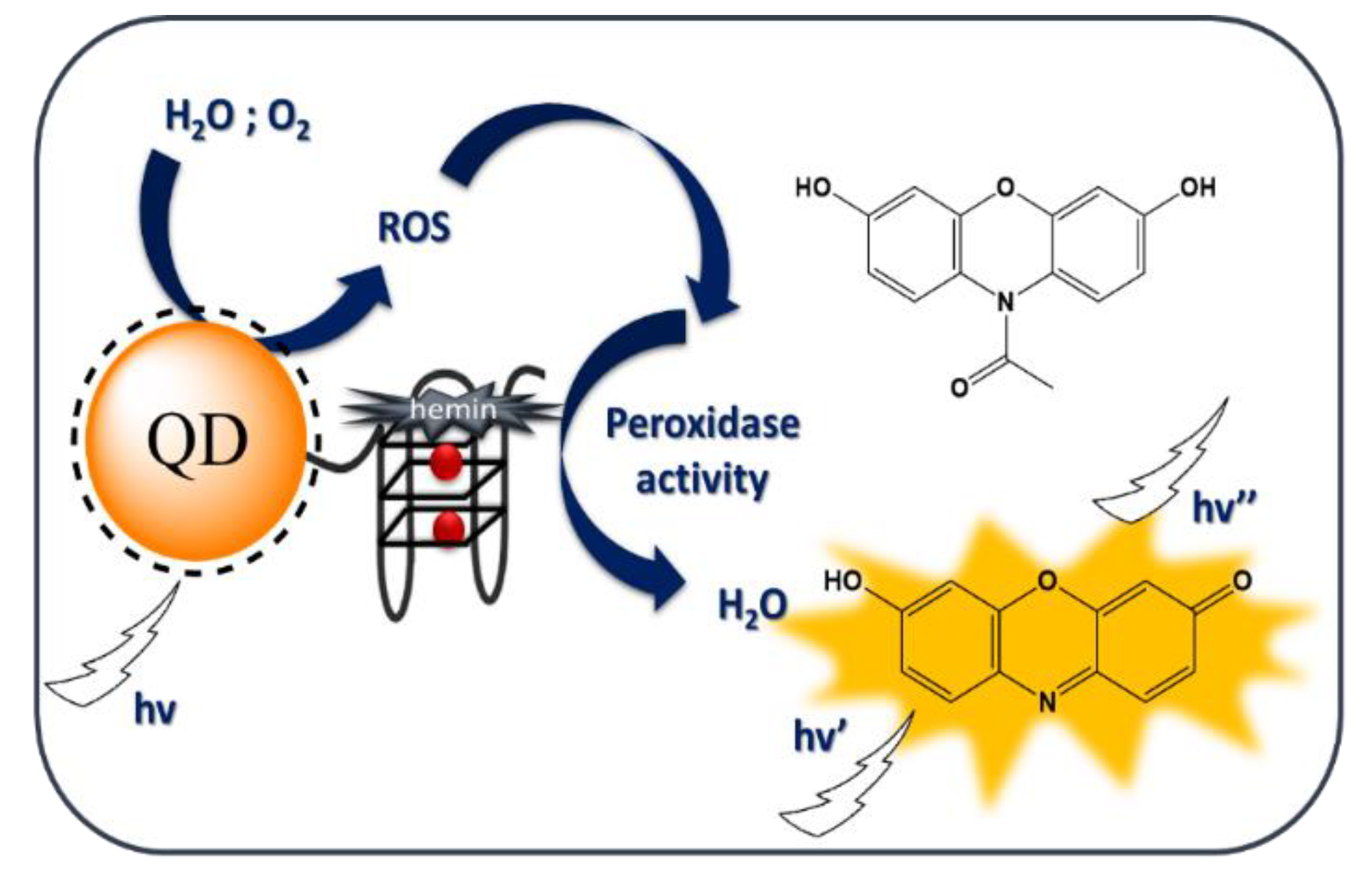
2. Results and Discussion
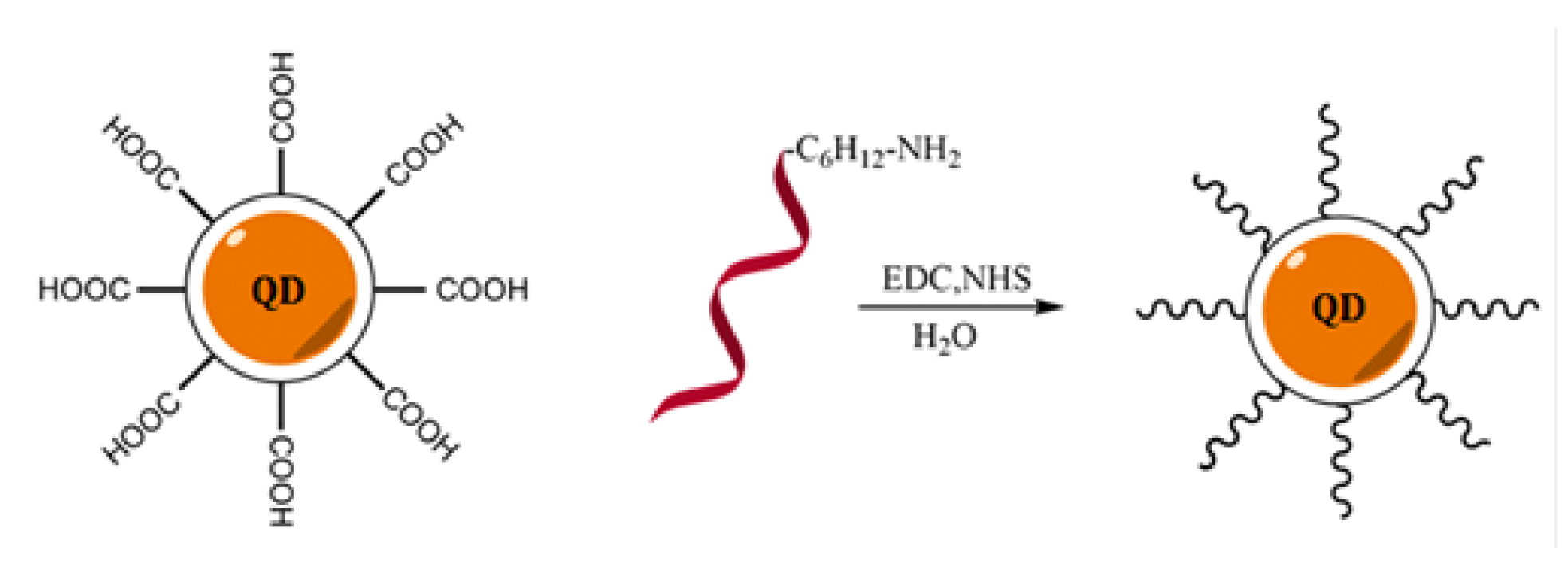
2.1. Design and Synthesis of the QD-DNA Conjugate
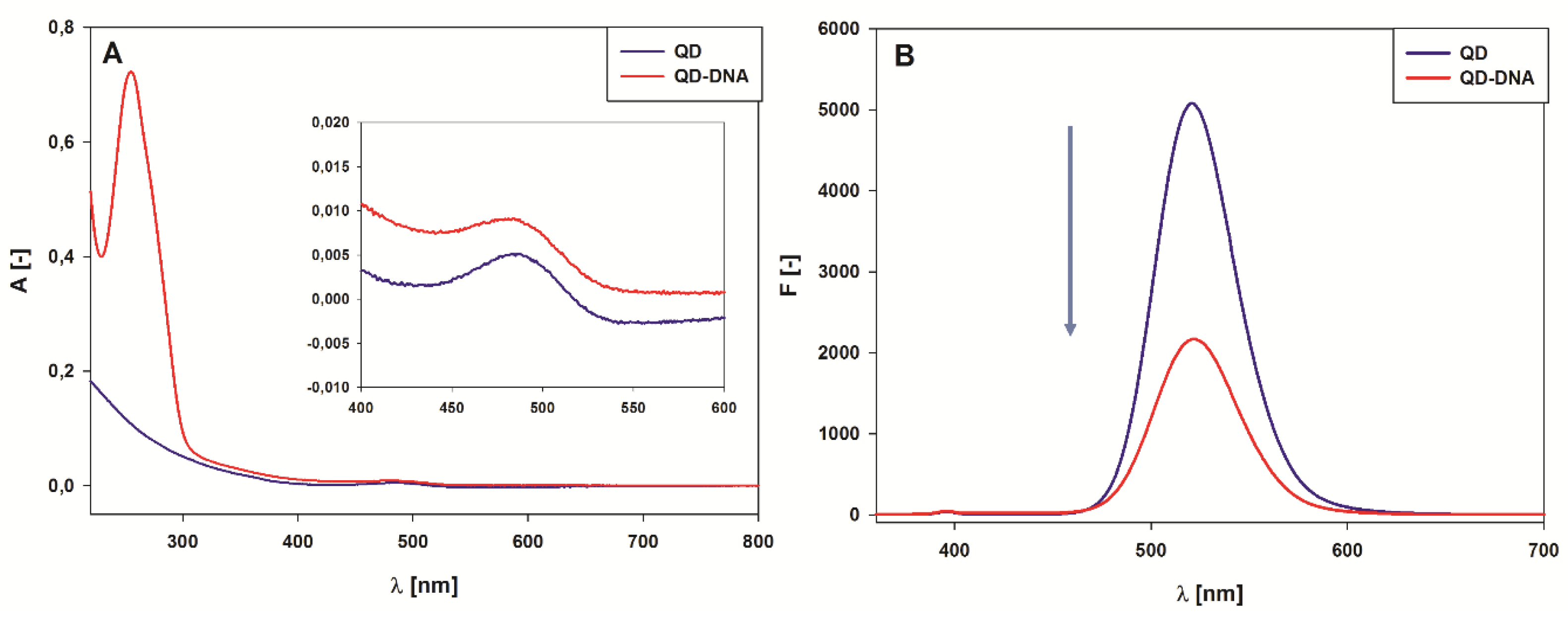
2.2. Spectroscopic Characterization of the QD-DNA Conjugate
2.3. Peroxidase Activity of the QD-DNA Conjugate
3. Materials and Methods
3.1. Materials
3.2. Synthesis and Purification of QD-DNA Conjugates
3.3. Determination of the Size of Nanoparticles and their Zeta Potential
3.4. Spectroscopic Characterization of QD and QD-DNA Systems
3.5. Gel Electrophoresis
3.6. Activity Measurements
3.7. Light-Induced Switching of Peroxidase Activity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Halliwell, B. Reactive Species and Antioxidants. Redox Biology Is a Fundamental Theme of Aerobic Life. Plant Physiol. 2006, 141, 312–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halliwell, B.; Gutteridge, J.M.C. Oxygen Toxicity, Oxygen Radicals, Transition Metals and Disease. Biochem. J. 1984, 219, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Hayyan, M.; Hashim, M.A.; AlNashef, I.M. Superoxide Ion: Generation and Chemical Implications. Chem. Rev. 2016, 116, 3029–3085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kroemer, G.; Zamzami, N.; Susin, S.A. Mitochondrial Control of Apoptosis. Immunol. Today 1997, 18, 44–51. [Google Scholar] [CrossRef]
- Marzo, I. Bax and Adenine Nucleotide Translocator Cooperate in the Mitochondrial Control of Apoptosis. Science 1998, 281, 2027–2031. [Google Scholar] [CrossRef]
- Murphy, M.P. How Mitochondria Produce Reactive Oxygen Species. Biochem. J. 2009, 417, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Qu, X.; Kim, E.; Lei, M.; Dai, K.; Tan, X.; Xu, M.; Li, J.; Liu, Y.; Shi, X.; et al. Bio-Inspired Redox-Cycling Antimicrobial Film for Sustained Generation of Reactive Oxygen Species. Biomaterials 2018, 162, 109–122. [Google Scholar] [CrossRef]
- Del Río, L.A.; López-Huertas, E. ROS Generation in Peroxisomes and Its Role in Cell Signaling. Plant Cell Physiol. 2016, 57, 1364–1376. [Google Scholar] [CrossRef]
- Cai, P.-J.; Xiao, X.; He, Y.-R.; Li, W.-W.; Zang, G.-L.; Sheng, G.-P.; Hon-Wah Lam, M.; Yu, L.; Yu, H.-Q. Reactive Oxygen Species (ROS) Generated by Cyanobacteria Act as an Electron Acceptor in the Biocathode of a Bio-Electrochemical System. Biosens. Bioelectron. 2013, 39, 306–310. [Google Scholar] [CrossRef]
- Walling, C. Fenton’s Reagent Revisited. Acc. Chem. Res. 1975, 8, 125–131. [Google Scholar] [CrossRef]
- Zepp, R.G.; Faust, B.C.; Hoign, J. Hydroxyl Radical Formation in Aqueous Reactions (PH 3-8) of Iron(I1) with Hydrogen Peroxide: The Photo-Fenton Reaction. Environ. Sci. Technol. 1992, 26, 313–319. [Google Scholar] [CrossRef]
- Katsounaros, I.; Cherevko, S.; Zeradjanin, A.R.; Mayrhofer, K.J.J. Oxygen Electrochemistry as a Cornerstone for Sustainable Energy Conversion. Angew. Chem. Int. Ed. 2014, 53, 102–121. [Google Scholar] [CrossRef] [PubMed]
- Katsounaros, I.; Schneider, W.B.; Meier, J.C.; Benedikt, U.; Biedermann, P.U.; Cuesta, A.; Auer, A.A.; Mayrhofer, K.J.J. The Impact of Spectator Species on the Interaction of H2O2 with Platinum—Implications for the Oxygen Reduction Reaction Pathways. Phys. Chem. Chem. Phys. 2013, 15, 8058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Surib, N.A.; Kuila, A.; Saravanan, P.; Sim, L.C.; Leong, K.H. A Ligand Strategic Approach with Cu-MOF for Enhanced Solar Light Photocatalysis. New J. Chem. 2018, 42, 11124–11130. [Google Scholar] [CrossRef]
- Liao, Y.; Brame, J.; Que, W.; Xiu, Z.; Xie, H.; Li, Q.; Fabian, M.; Alvarez, P.J. Photocatalytic Generation of Multiple ROS Types Using Low-Temperature Crystallized Anodic TiO2 Nanotube Arrays. J. Hazard. Mater. 2013, 260, 434–441. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Casey, P.S.; McCall, M.J.; Fenech, M. Effects of Surface Chemistry on Cytotoxicity, Genotoxicity, and the Generation of Reactive Oxygen Species Induced by ZnO Nanoparticles. Langmuir 2010, 26, 15399–15408. [Google Scholar] [CrossRef]
- Lipovsky, A.; Tzitrinovich, Z.; Friedmann, H.; Applerot, G.; Gedanken, A.; Lubart, R. EPR Study of Visible Light-Induced ROS Generation by Nanoparticles of ZnO. J. Phys. Chem. C 2009, 113, 15997–16001. [Google Scholar] [CrossRef]
- Sirelkhatim, A.; Mahmud, S.; Seeni, A.; Kaus, N.H.M.; Ann, L.C.; Bakhori, S.K.M.; Hasan, H.; Mohamad, D. Review on Zinc Oxide Nanoparticles: Antibacterial Activity and Toxicity Mechanism. Nano-Micro Lett. 2015, 7, 219–242. [Google Scholar] [CrossRef] [Green Version]
- Brzicova, T.; Sikorova, J.; Milcova, A.; Vrbova, K.; Klema, J.; Pikal, P.; Lubovska, Z.; Philimonenko, V.; Franco, F.; Topinka, J.; et al. Nano-TiO2 Stability in Medium and Size as Important Factors of Toxicity in Macrophage-like Cells. Toxicol. In Vitro 2019, 54, 178–188. [Google Scholar] [CrossRef]
- Chen, L.; Hu, C.; Guo, Y.; Shi, Q.; Zhou, B. TiO2 Nanoparticles and BPA Are Combined to Impair the Development of Offspring Zebrafish after Parental Coexposure. Chemosphere 2019, 217, 732–741. [Google Scholar] [CrossRef]
- Aliakbari, F.; Haji Hosseinali, S.; KhaliliSarokhalil, Z.; Shahpasand, K.; Akbar Saboury, A.; Akhtari, K.; Falahati, M. Reactive Oxygen Species Generated by Titanium Oxide Nanoparticles Stimulate the Hemoglobin Denaturation and Cytotoxicity against Human Lymphocyte Cell. J. Biomol. Struct. Dyn. 2019, 37, 4875–4881. [Google Scholar] [CrossRef] [PubMed]
- Blaškovičová, J.; Sochr, J.; Koutsogiannis, A.; Diamantidou, D.; Kopel, P.; Adam, V.; Labuda, J. Detection of ROS Generated by UV-C Irradiation of CdS Quantum Dots and Their Effect on Damage to Chromosomal and Plasmid DNA. Electroanalysis 2018, 30, 698–704. [Google Scholar] [CrossRef]
- Dou, X.; Zhang, Q.; Shah, S.N.A.; Khan, M.; Uchiyama, K.; Lin, J.-M. MoS2 -Quantum Dot Triggered Reactive Oxygen Species Generation and Depletion: Responsible for Enhanced Chemiluminescence. Chem. Sci. 2019, 10, 497–500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribeiro, D.S.M.; Frigerio, C.; Santos, J.L.M.; Prior, J.A.V. Photoactivation by Visible Light of CdTe Quantum Dots for Inline Generation of Reactive Oxygen Species in an Automated Multipumping Flow System. Anal. Chim. Acta 2012, 735, 69–75. [Google Scholar] [CrossRef]
- Chan, W.C.W.; Maxwell, D.J.; Gao, X.; Bailey, R.E.; Han, M.; Nie, S. Luminescent Quantum Dots for Multiplexed Biological Detection and Imaging. Curr. Opin. Biotechnol. 2002, 13, 40–46. [Google Scholar] [CrossRef]
- Biju, V.; Itoh, T.; Anas, A.; Sujith, A.; Ishikawa, M. Semiconductor Quantum Dots and Metal Nanoparticles: Syntheses, Optical Properties, and Biological Applications. Anal. Bioanal. Chem. 2008, 391, 2469–2495. [Google Scholar] [CrossRef]
- Jamieson, T.; Bakhshi, R.; Petrova, D.; Pocock, R.; Imani, M.; Seifalian, A.M. Biological Applications of Quantum Dots. Biomaterials 2007, 28, 4717–4732. [Google Scholar] [CrossRef]
- Nozik, A.J.; Beard, M.C.; Luther, J.M.; Law, M.; Ellingson, R.J.; Johnson, J.C. Semiconductor Quantum Dots and Quantum Dot Arrays and Applications of Multiple Exciton Generation to Third-Generation Photovoltaic Solar Cells. Chem. Rev. 2010, 110, 6873–6890. [Google Scholar] [CrossRef]
- Fruk, L.; Rajendran, V.; Spengler, M.; Niemeyer, C.M. Light-Induced Triggering of Peroxidase Activity Using Quantum Dots. ChemBioChem 2007, 8, 2195–2198. [Google Scholar] [CrossRef]
- Iñarritu, I.; Torres, E.; Topete, A.; Campos-Terán, J. Immobilization Effects on the Photocatalytic Activity of CdS Quantum Dots-Horseradish Peroxidase Hybrid Nanomaterials. J. Colloid Interface Sci. 2017, 506, 36–45. [Google Scholar] [CrossRef]
- Travascio, P.; Li, Y.; Sen, D. DNA-Enhanced Peroxidase Activity of a DNA Aptamer-Hemin Complex. Chem. Biol. 1998, 5, 505–517. [Google Scholar] [CrossRef] [Green Version]
- Kong, D.-M.; Yang, W.; Wu, J.; Li, C.-X.; Shen, H.-X. Structure–Function Study of Peroxidase-like G-Quadruplex-Hemin Complexes. Analyst 2010, 135, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Kosman, J.; Juskowiak, B. Peroxidase-Mimicking DNAzymes for Biosensing Applications: A Review. Anal. Chim. Acta 2011, 707, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Kosman, J.; Żukowski, K.; Juskowiak, B. Comparison of Characteristics and DNAzyme Activity of G4–Hemin Conjugates Obtained via Two Hemin Attachment Methods. Molecules 2018, 23, 1400. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Lu, C.-H.; Willner, I. From Cascaded Catalytic Nucleic Acids to Enzyme−DNA Nanostructures: Controlling Reactivity, Sensing, Logic Operations, and Assembly of Complex Structures. Chem. Rev. 2014, 114, 2881–2941. [Google Scholar] [CrossRef] [PubMed]
- Goodman, S.M.; Levy, M.; Li, F.-F.; Ding, Y.; Courtney, C.M.; Chowdhury, P.P.; Erbse, A.; Chatterjee, A.; Nagpal, P. Designing Superoxide-Generating Quantum Dots for Selective Light-Activated Nanotherapy. Front. Chem. 2018, 6, 46. [Google Scholar] [CrossRef] [Green Version]
- Pong, B.-K.; Trout, B.L.; Lee, J.-Y. Preparation of DNA-Functionalised CdSe/ZnS Quantum Dots. Available online: https://dspace.mit.edu/handle/1721.1/35870 (accessed on 1 January 2017).
- Karakoti, A.S.; Shukla, R.; Shanker, R.; Singh, S. Surface Functionalization of Quantum Dots for Biological Applications. Adv. Colloid Interface Sci. 2015, 215, 28–45. [Google Scholar] [CrossRef]
- Lišková, M.; Voráčová, I.; Klepárník, K.; Hezinová, V.; Přikryl, J.; Foret, F. Conjugation Reactions in the Preparations of Quantum Dot-Based Immunoluminescent Probes for Analysis of Proteins by Capillary Electrophoresis. Anal. Bioanal. Chem. 2011, 400, 369–379. [Google Scholar] [CrossRef]
- Kubota, Y.; Motosa, Y.; Shigemune, Y.; Fujisaki, Y. Fluorescence quenching of 10-methylacrjdinium chloride by nucleotides. Photochem. Photobiol. 1979, 29, 1099–1106. [Google Scholar] [CrossRef]
- Torimura, M.; Kurata, S.; Yamada, K.; Yokomaku, T.; Kamagata, Y.; Kanagawa, T.; Kurane, R. Fluorescence-Quenching Phenomenon by Photoinduced Electron Transfer between a Fluorescent Dye and a Nucleotide Base. Anal. Sci. 2001, 17, 155–160. [Google Scholar] [CrossRef] [Green Version]
- Fasman, G.D. Handbook of Biochemistry and Molecular Biology, Volume 1: Nucleic Acids, 3rd ed.; CRC Press: Boca Raton, FL, USA, 1975. [Google Scholar]

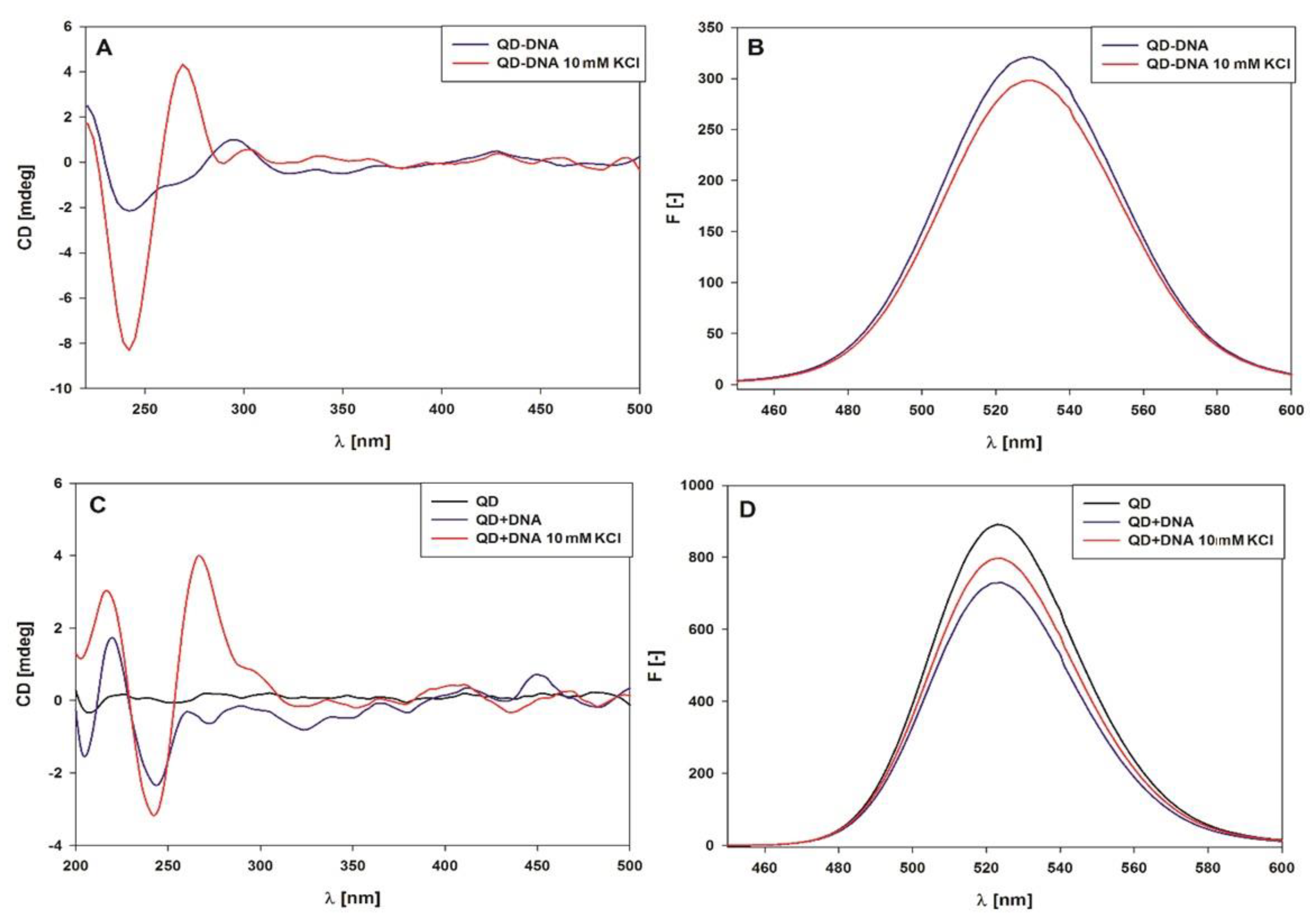
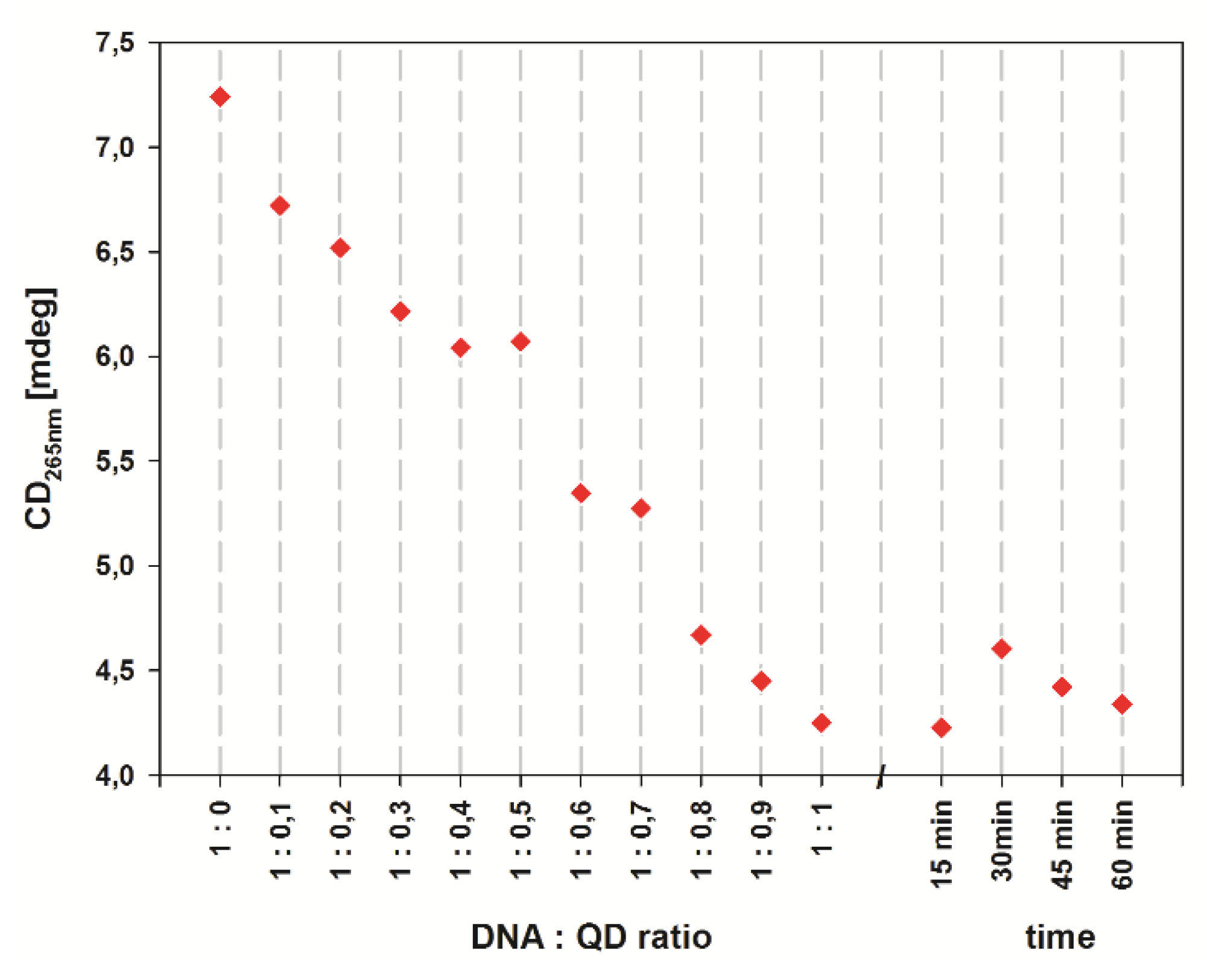
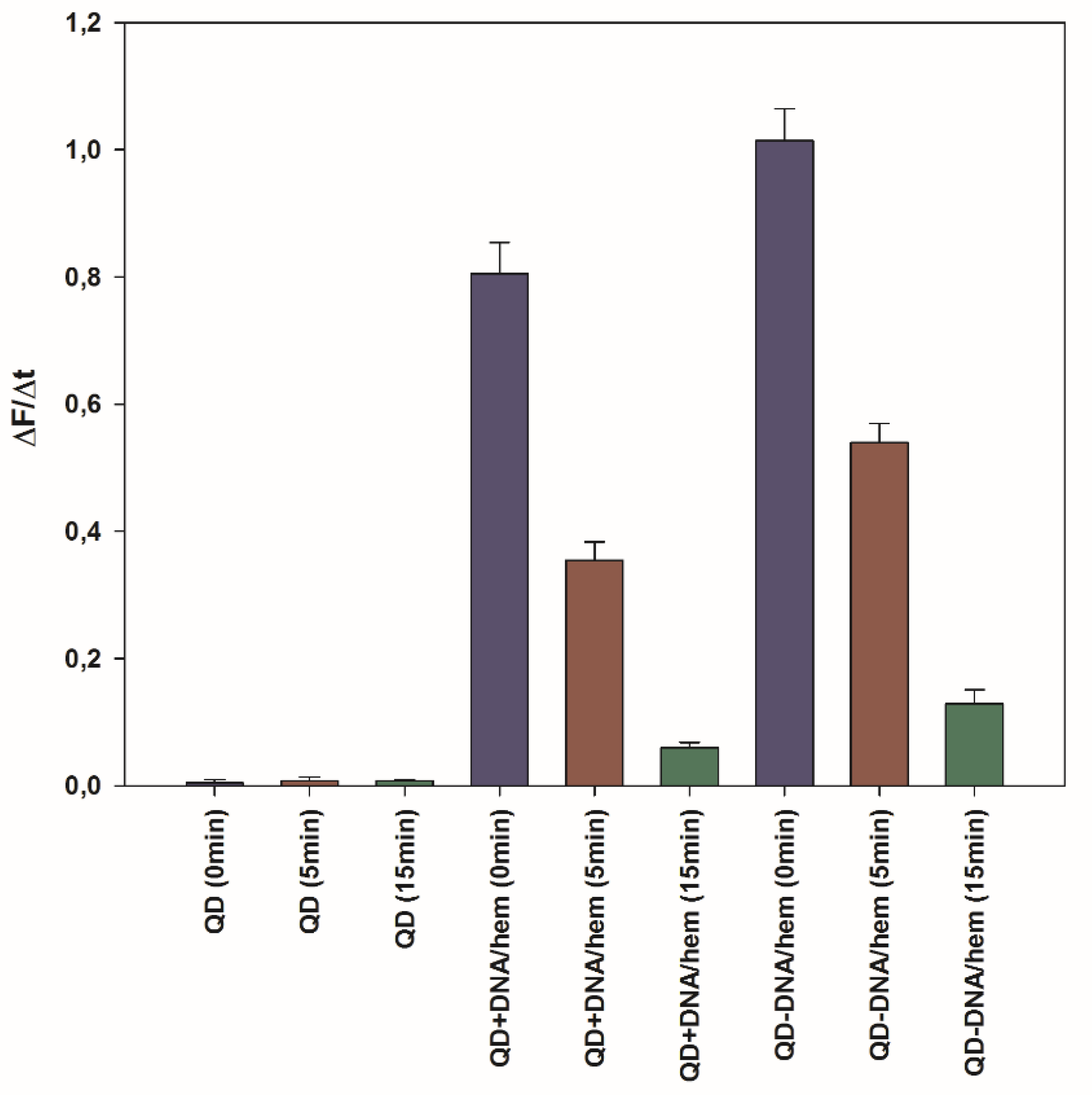
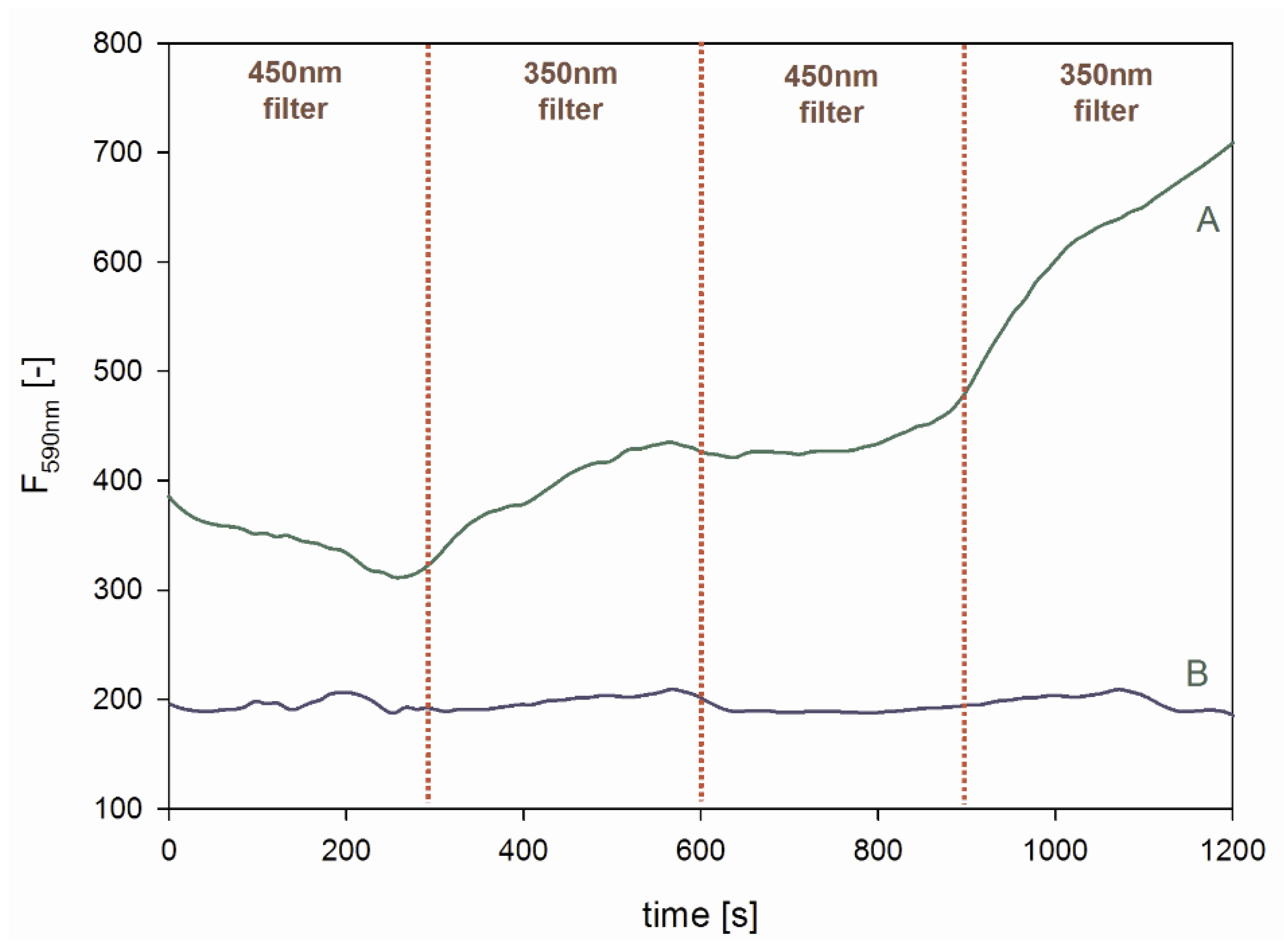
| Z (mV) | ± ΔZ (mV) | |
|---|---|---|
| QD | −41.9 | 3.85 |
| QD + DNA | −55.0 | 1.44 |
| QD − DNA | −47.1 | 4.69 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Żukowski, K.; Kosman, J.; Juskowiak, B. Light-Induced Oxidase Activity of DNAzyme-Modified Quantum Dots. Int. J. Mol. Sci. 2020, 21, 8190. https://doi.org/10.3390/ijms21218190
Żukowski K, Kosman J, Juskowiak B. Light-Induced Oxidase Activity of DNAzyme-Modified Quantum Dots. International Journal of Molecular Sciences. 2020; 21(21):8190. https://doi.org/10.3390/ijms21218190
Chicago/Turabian StyleŻukowski, Krzysztof, Joanna Kosman, and Bernard Juskowiak. 2020. "Light-Induced Oxidase Activity of DNAzyme-Modified Quantum Dots" International Journal of Molecular Sciences 21, no. 21: 8190. https://doi.org/10.3390/ijms21218190






