Pathogenic Single Nucleotide Polymorphisms on Autophagy-Related Genes
Abstract
:1. Introduction
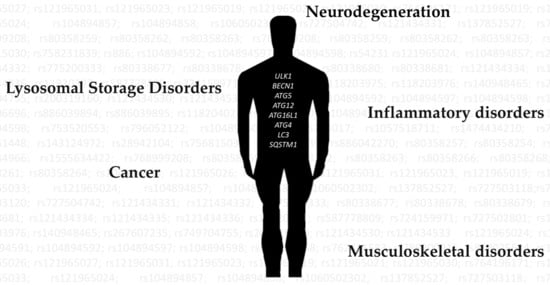
2. Autophagy Dysregulation in Disease
3. Relevant Variants on Autophagy-Related Genes
3.1. The ULK1/2 Kinase Complex
3.2. The Class III Phosphatidylinositol 3-Kinase (PI3KC3) Complexes
3.3. PI(3)P-Binding Proteins and the ATG9-Containing Membranes
3.4. The ATG12 and ATG8 Conjugation Systems
3.5. Selective Autophagy Receptors
3.6. Cellular Machineries Involved in Autophagosome-Lysosome Fusion
4. Concluding Remarks
Supplementary Materials
Funding
Conflicts of Interest
Abbreviations
| ALFY | Autophagy-linked FYVE protein |
| AMBRA1 | Activating molecule in BECN1-regulated autophagy protein 1 |
| AMPK | Adenosine monophosphate-activated protein kinase |
| ARSB | Arylsulfatase B |
| ATG10 | Autophagy-related protein 10 |
| ATG101 | Autophagy-related protein 101 |
| ATG12 | Autophagy-related protein 12 |
| ATG13 | Autophagy-related protein 13 |
| ATG14 | Autophagy-related protein 14 |
| ATG16L1 | Autophagy-related protein 16 like 1 |
| ATG2A | Autophagy-related protein 2 homolog A |
| ATG2B | Autophagy-related protein 2 homolog B |
| ATG4A | Autophagy related 4A cysteine peptidase |
| ATG4B | Autophagy related 4B cysteine peptidase |
| ATG4C | Autophagy related 4C cysteine peptidase |
| ATG4D | Autophagy related 4D cysteine peptidase |
| ATG5 | Autophagy-related protein 5 |
| ATG7 | Autophagy-related protein 7 |
| ATG9A | Autophagy-related protein 9A |
| ATG9B | Autophagy-related protein 9B |
| ATGL | Adipose triglyceride lipase |
| ATL3 | Atlastin-3 |
| Barkor | Beclin 1-associated autophagy-related key regulator |
| BCL-2 | Apoptosis regulator Bcl-2 |
| BECN1 | Beclin-1 |
| BIRC6 | Baculoviral IAP repeat-containing protein 6 |
| BNIP3 | BCL2/adenovirus E1B 19 kDa protein-interacting protein 3 |
| BNIP3L | BCL2/adenovirus E1B 19 kDa protein-interacting protein 3-like |
| CCPG1 | Cell cycle progression protein 1 |
| CHMP2B | Charged multivesicular body protein 2b |
| CHMP4B | Charged multivesicular body protein 4b |
| CTNS | Cystinosin |
| CTSD | Cathepsin D |
| CTSF | Cathepsin F |
| DFCP1 | Double FYVE-containing protein 1 |
| DNAJC5 | DnaJ homolog subfamily C member 5 |
| EPG5 | Ectopic P granules protein 5 homolog |
| FAM134B | Family With Sequence Similarity 134, Member B |
| FKBP8 | Peptidyl-prolyl cis-trans isomerase FKBP8 |
| FUCA1 | Tissue alpha-L-fucosidase |
| FUNDC1 | FUN14 domain-containing protein 1 |
| FYCO1 | FYVE and coiled-coil domain-containing protein 1 |
| GAA | Lysosomal alpha-glucosidase |
| GABARAP | Gamma-aminobutyric acid receptor-associated protein |
| GABARAPL1 | Gamma-aminobutyric acid receptor-associated protein-like 1 |
| GALNS | N-acetylgalactosamine-6-sulfatase |
| GARARAPL2 | Gamma-aminobutyric acid receptor-associated protein-like 2 |
| GLB1 | Beta-galactosidase |
| GNS | N-acetylglucosamine-6-sulfatase |
| GRASP55 | Golgi reassembly-stacking protein of 55 kDa |
| GRN | Progranulin |
| GUSB | Beta-glucuronidase |
| HGS | Hepatocyte growth factor-regulated tyrosine kinase substrate |
| HGSNAT | Heparan-alpha-glucosaminide N-acetyltransferase |
| HOPS | Hepatocyte odd Ppotein shuttling protein |
| HSC70 | Heat shock cognate 71 kDa protein |
| HSL | Hormone-sensitive lipase |
| HTT | Huntingtin |
| HYAL1 | Hyaluronidase-1 |
| IDS | Iduronate 2-sulfatase |
| IDUA | Alpha-L-iduronidase |
| KIF5B | Kinesin-1 heavy chain |
| LAMP2 | Lysosome-associated membrane protein 2 |
| MAN2B1 | Lysosomal alpha-mannosidase |
| MANBA | Beta-mannosidase |
| MAP1-LC3A | Microtubule-associated proteins 1A/1B light chain 3A |
| MAP1-LC3B | Microtubule-associated proteins 1A/1B light chain 3B |
| MAP1-LC3C | Microtubule-associated proteins 1A/1B light chain 3C |
| MFSD8 | Major facilitator superfamily domain-containing protein 8 |
| mTOR | Mechanistic/mammalian target of rapamycin |
| NAGLU | Alpha-N-acetylglucosaminidase |
| NBR1 | Next to BRCA1 gene 1 protein |
| NCOA4 | Nuclear receptor coactivator 4 |
| NDP52 | Nuclear domain 10 protein 52 |
| NPC1 | NPC intracellular cholesterol transporter 1 |
| NPC2 | NPC intracellular cholesterol transporter 2 |
| NUFIP1 | Nuclear fragile X mental retardation-interacting protein 1 |
| OPTN | Optineurin |
| PI3KC3 | Phosphatidylinositol 3-kinase catalytic subunit type 3 |
| PIK3R4 | Phosphoinositide 3-kinase regulatory subunit 4 |
| PLEKHM1 | Pleckstrin homology domain-containing family M member 1 |
| RB1CC1 | RB1-inducible coiled-coil protein 1 |
| RTN3 | Reticulon-3 |
| SEC62 | Translocation protein SEC62 |
| SGSH | N-sulphoglucosamine sulphohydrolase |
| SLC17A5 | Sialin |
| SNAP29 | Synaptosomal-associated protein 29 |
| SQSTM1 | Sequestosome-1 |
| STK36 | Serine/threonine-protein kinase 36 |
| STX17 | Syntaxin-17 |
| TAX1BP1 | Tax1-binding protein 1 |
| TEX264 | Testis-expressed protein 264 |
| TOLLIP | Toll-interacting protein |
| ULK1 | Unc-51 like autophagy activating kinase |
| UVRAG | UV radiation resistance-associated gene protein |
| VAMP7 | Vesicle-associated membrane protein 7 |
| VAMP8 | Vesicle-associated membrane protein 8 |
| VPS4B | Vacuolar protein sorting-associated protein 4B |
| WIPI1 | WD repeat domain phosphoinositide-interacting protein 1 |
| WIPI2 | WD repeat domain phosphoinositide-interacting protein 2 |
| WIPI3 | WD repeat domain phosphoinositide-interacting protein 3 |
| WIPI4 | WD repeat domain phosphoinositide-interacting protein 4 |
| YKT6 | Synaptobrevin homolog YKT6 |
References
- Kroemer, G.; Mariño, G.; Levine, B. Autophagy and the Integrated Stress Response. Mol. Cell 2010, 40, 280–293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galluzzi, L.; Baehrecke, E.H.; Ballabio, A.; Boya, P.; Bravo-San Pedro, J.M.; Cecconi, F.; Choi, A.M.; Chu, C.T.; Codogno, P.; Colombo, M.I.; et al. Molecular definitions of autophagy and related processes. Embo J. 2017, 36, 1811–1836. [Google Scholar] [CrossRef] [PubMed]
- Parzych, K.R.; Klionsky, D.J. An overview of autophagy: Morphology, mechanism, and regulation. Antioxid. Redox Signal. 2014, 20, 460–473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mijaljica, D.; Prescott, M.; Devenish, R.J. Microautophagy in Mammalian Cells: Revisiting a 40-Year-Old Conundrum. Autophagy 2011, 7, 673–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaushik, S.; Cuervo, A.M. The coming of age of chaperone-mediated autophagy. Nat. Rev. Mol. Cell Biol. 2018, 19, 365–381. [Google Scholar] [CrossRef] [PubMed]
- Kawabata, T.; Yoshimori, T. Autophagosome Biogenesis and Human Health. Cell Discov. 2020, 6, 33. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Cuervo, A.M. Autophagy in the cellular energetic balance. Cell Metab. 2020, 6, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levine, B.; Kroemer, G. Biological Functions of Autophagy Genes: A Disease Perspective. Cell 2019, 176, 11–42. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.G.; Zhang, H. Core autophagy genes and human diseases. Curr. Opin. Cell Biol. 2019, 61, 117–125. [Google Scholar] [CrossRef]
- Choi, A.M.K.; Ryter, S.W.; Levine, B. Autophagy in Human Health and Disease. N. Engl. J. Med. 2013, 368, 651–662. [Google Scholar] [CrossRef]
- Yang, Y.; Klionsky, D.J. Autophagy and disease: Unanswered questions. Cell Death Differ. 2020, 27, 858–871. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Pietrocola, F.; Bravo-San Pedro, J.M.; Amaravadi, R.K.; Baehrecke, E.H.; Cecconi, F.; Codogno, P.; Debnath, J.; Gewirtz, D.A.; Karantza, V.; et al. Autophagy in malignant transformation and cancer progression. Embo J. 2015, 34, 856–880. [Google Scholar] [CrossRef]
- Lai, J.; Chen, B.; Mok, H.; Zhang, G.; Ren, C.; Liao, N. Comprehensive analysis of autophagy-related prognostic genes in breast cancer. J. Cell. Mol. Med. 2020, 24, 9145–9153. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, R.; Chen, W.; Chen, Q.; Zhou, J. Construction of a prognosis-predicting model based on autophagy-related genes for hepatocellular carcinoma (HCC) patients. Aging 2020, 12, 14582. [Google Scholar] [CrossRef]
- Feng, H.; Zhong, L.; Yang, X.; Wan, Q.; Pei, X.; Wang, J. Development and validation of prognostic index based on autophagy-related genes in patient with head and neck squamous cell carcinoma. Cell Death Discov. 2020, 6, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.; Jiang, L.; Luo, S.; Zhao, X.; Hu, H.; Zhao, G.; Tang, W. Development of an autophagy-related gene expression signature for prognosis prediction in prostate cancer patients. J. Transl. Med. 2020, 18, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Sun, Z.; Jing, C.; Xiao, C.; Li, T. An autophagy-related long non-coding RNA prognostic signature accurately predicts survival outcomes in bladder urothelial carcinoma patients. Aging 2020, 12, 15624–15637. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Zhang, S.; Nie, Z.; Wen, X.; Gao, Y. Identification of an Autophagy-Related Prognostic Signature for Clear Cell Renal Cell Carcinoma. Front. Oncol. 2020, 10, 873. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, S.; Cao, H.; Wang, J.; Wen, T.; Hu, X.; Li, H. Prognostic significance of autophagy-related genes within esophageal carcinoma. BMC Cancer 2020, 20, 1–11. [Google Scholar] [CrossRef]
- Xing, Q.; Ji, C.; Zhu, B.; Cong, R.; Wang, Y. Identification of small molecule drugs and development of a novel autophagy-related prognostic signature for kidney renal clear cell carcinoma. Cancer Med. 2020, 9, 7034–7051. [Google Scholar] [CrossRef]
- Huo, X.; Qi, J.; Huang, K.; Bu, S.; Yao, W.; Chen, Y.; Nie, J. Identification of an autophagy-related gene signature that can improve prognosis of hepatocellular carcinoma patients. BMC Cancer 2020, 20, 771. [Google Scholar] [CrossRef] [PubMed]
- Du, J.-X.; Chen, C.; Luo, Y.-H.; Cai, J.-L.; Cai, C.-Z.; Xu, J.; Ni, X.-J.; Zhu, W. Establishment and validation of a novel autophagy-related gene signature for patients with breast cancer. Gene 2020, 762, 144974. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Ma, X.; Liu, J.; Wan, Y.; Jiang, Y.; Xia, Y.; Cheng, W. Prognostic value of an autophagy-related gene expression signature for endometrial cancer patients. Cancer Cell Int. 2020, 20, 306. [Google Scholar] [CrossRef]
- Yang, H.; Han, M.; Li, H. Construction and Validation of an Autophagy-Related Prognostic Risk Signature for Survival Predicting in Clear Cell Renal Cell Carcinoma Patients. Front. Oncol. 2020, 10, 707. [Google Scholar] [CrossRef] [PubMed]
- Shibutani, S.T.; Saitoh, T.; Nowag, H.; Münz, C.; Yoshimori, T. Autophagy and Autophagy-Related Proteins in the Immune System. Nat. Immunol. 2015, 16, 1014–1024. [Google Scholar] [CrossRef]
- Clarke, A.J.; Simon, A.K. Autophagy in the Renewal, Differentiation and Homeostasis of Immune Cells. Nat. Rev. Immunol. 2019, 19, 170–183. [Google Scholar] [CrossRef]
- Münz, C. Autophagy proteins in antigen processing for presentation on MHC molecules. Immunol. Rev. 2016, 272, 17–27. [Google Scholar] [CrossRef]
- Cui, B.; Lin, H.; Yu, J.; Yu, J.; Hu, Z. Autophagy and the Immune Response. Adv. Exp. Med. Biol. 2019, 1206, 595–634. [Google Scholar]
- Deretic, V.; Levine, B. Autophagy Balances Inflammation in Innate Immunity. Autophagy 2018, 14, 243–251. [Google Scholar] [CrossRef] [Green Version]
- Cadwell, K. Crosstalk between Autophagy and Inflammatory Signalling Pathways: Balancing Defence and Homeostasis. Nat. Rev. Immunol. 2016, 16, 661–675. [Google Scholar] [CrossRef]
- Yin, H.; Wu, H.; Chen, Y.; Zhang, J.; Zheng, M.; Chen, G.; Li, L.; Lu, Q. The Therapeutic and Pathogenic Role of Autophagy in Autoimmune Diseases. Front. Immunol. 2018, 9, 1512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.; Eun, H.; Jo, E.-K. Roles of Autophagy-Related Genes in the Pathogenesis of Inflammatory Bowel Disease. Cells 2019, 8, 77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levine, B.; Mizushima, N.; Virgin, H.W. Autophagy in Immunity and Inflammation. Nature 2011, 469, 323–335. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Bowman, J.W.; Jung, J.U. Autophagy during Viral infection A Double-Edged Sword. Nat. Rev. Microbiol. 2018, 16, 341–354. [Google Scholar] [CrossRef]
- Menzies, F.M.; Fleming, A.; Caricasole, A.; Bento, C.F.; Andrews, S.P.; Ashkenazi, A.; Füllgrabe, J.; Jackson, A.; Jimenez Sanchez, M.; Karabiyik, C.; et al. Autophagy and Neurodegeneration: Pathogenic Mechanisms and Therapeutic Opportunities. Neuron 2017, 93, 1015–1034. [Google Scholar] [CrossRef] [Green Version]
- Park, H.; Kang, J.H.; Lee, S. Autophagy in Neurodegenerative Diseases: A Hunter for Aggregates. Int. J. Mol. Sci. 2020, 21, 3369. [Google Scholar] [CrossRef]
- Vicencio, E.; Beltrán, S.; Labrador, L.; Manque, P.; Nassif, M.; Woehlbier, U. Implications of Selective Autophagy Dysfunction for ALS Pathology. Cells 2020, 9, 381. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Runwal, G.; Obrocki, P.; Rubinsztein, D.C. Autophagy in childhood neurological disorders. Dev. Med. Child Neurol. 2019, 61, 639–645. [Google Scholar] [CrossRef]
- Margeta, M. Autophagy Defects in Skeletal Myopathies. Annu. Rev. Pathol. Mech. Dis. 2020, 15, 261–285. [Google Scholar] [CrossRef] [Green Version]
- Pierrefite-Carle, V.; Santucci-Darmanin, S.; Breuil, V.; Camuzard, O.; Carle, G.F. Autophagy in bone: Self-eating to stay in balance. Ageing Res. Rev. 2015, 24, 206–217. [Google Scholar] [CrossRef]
- Vinatier, C.; Domínguez, E.; Guicheux, J.; Caramés, B. Role of the Inflammation-Autophagy-Senescence Integrative Network in Osteoarthritis. Front. Physiol. 2018, 9, 706. [Google Scholar] [CrossRef] [PubMed]
- Ballabio, A.; Bonifacino, J.S. Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat. Rev. Mol. Cell Biol. 2020, 21, 101–118. [Google Scholar] [CrossRef] [PubMed]
- Sun, A. Lysosomal storage disease overview. Ann. Transl. Med. 2018, 6, 476. [Google Scholar] [CrossRef] [PubMed]
- Palhegyi, A.M.; Seranova, E.; Dimova, S.; Hoque, S.; Sarkar, S. Biomedical Implications of Autophagy in Macromolecule Storage Disorders. Front. Cell Dev. Biol. 2019, 7, 179. [Google Scholar] [CrossRef] [Green Version]
- Parenti, G.; Andria, G.; Ballabio, A. Lysosomal storage diseases: From pathophysiology to therapy. Annu. Rev. Med. 2015, 66, 471–486. [Google Scholar] [CrossRef]
- Ariosa, A.R.; Klionsky, D.J. Autophagy core machinery: Overcoming spatial barriers in neurons. J. Mol. Med. 2016, 94, 1217–1227. [Google Scholar] [CrossRef] [Green Version]
- Koyama-Honda, I.; Itakura, E.; Fujiwara, T.K.; Mizushima, N. Temporal analysis of recruitment of mammalian ATG proteins to the autophagosome formation site. Autophagy 2013, 9, 1491–1499. [Google Scholar] [CrossRef] [Green Version]
- Wesselborg, S.; Stork, B. Autophagy Signal Transduction by ATG Proteins: From Hierarchies to Networks. Cell Mol Life Sci. 2015, 72, 4721–4757. [Google Scholar] [CrossRef] [Green Version]
- Nakatogawa, H. Mechanisms governing autophagosome biogenesis. Nat. Rev. Mol. Cell Biol. 2020, 21, 439–458. [Google Scholar] [CrossRef]
- Zachari, M.; Ganley, I.G. The Mammalian ULK1 Complex and Autophagy Initiation. Esssays Biochem. 2017, 61, 585–596. [Google Scholar]
- Tamargo-Gómez, I.; Mariño, G. AMPK: Regulation of Metabolic Dynamics in the Context of Autophagy. Int. J. Mol. Sci. 2018, 19, 3812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Zhang, H. Regulation of Autophagy by mTOR Signaling Pathway; Springer: Berlin/Heidelberg, Germany, 2019; Volume 1206, pp. 67–83. [Google Scholar]
- Morgan, A.R.; Lam, W.J.; Han, D.Y.; Fraser, A.G.; Ferguson, L.R. Association analysis of ULK1 with Crohn‘s disease in a New Zealand population. Gastroenterol. Res. Pract. 2012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henckaerts, L.; Cleynen, I.; Brinar, M.; John, J.M.; Van Steen, K.; Rutgeerts, P.; Vermeire, S. Genetic variation in the autophagy gene ULK1 and risk of Crohn’s disease. Inflamm. Bowel Dis. 2011, 17, 1392–1397. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.R.; Liang, L.; Chen, W.W.; Wen, C.; Wan, B.S.; Luo, L.L.; Zhao, Y.L.; Chen, J.; Yue, J. ULK1 polymorphisms confer susceptibility to pulmonary tuberculosis in a Chinese population. Int. J. Tuberc. Lung Dis. 2019, 23, 265–271. [Google Scholar] [CrossRef]
- Horne, D.J.; Graustein, A.D.; Shah, J.A.; Peterson, G.; Savlov, M.; Steele, S.; Narita, M.; Hawn, T.R. Human ULK1 Variation and Susceptibility to Mycobacterium tuberculosis Infection. J. Infect. Dis. 2016, 214, 1260–1267. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Han, R.; Wang, M.; Li, X.; Yang, X.; Xia, Q.; Liu, R.; Yuan, Y.; Hu, X.; Chen, M.; et al. Association between the autophagy-related gene ULK1 and ankylosing spondylitis susceptibility in the Chinese Han population: A case-control study. Postgrad. Med. J. 2017, 93, 752–757. [Google Scholar] [CrossRef]
- Wolthers, B.O.; Frandsen, T.L.; Abrahamsson, J.; Albertsen, B.K.; Helt, L.R.; Heyman, M.; Jónsson, G.; Kõrgvee, L.T.; Lund, B.; Raja, R.A.; et al. Asparaginase-associated pancreatitis: A study on phenotype and genotype in the NOPHO ALL2008 protocol. Leukemia 2017, 31, 325–332. [Google Scholar] [CrossRef]
- Bronson, P.G.; Chang, D.; Bhangale, T.; Seldin, M.F.; Ortmann, W.; Ferreira, R.C.; Urcelay, E.; Pereira, L.F.; Martin, J.; Plebani, A.; et al. Common variants at PVT1, ATG13-AMBRA1, AHI1 and CLEC16A are associated with selective IgA deficiency. Nat. Genet. 2016, 48, 1425–1429. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; An, T.; Li, M.; Yi, Z.; Li, C.; Sun, X.; Guan, X.; Li, L.; Wang, Y.; Zhang, Y.; et al. The association between early-onset cardiac events caused by neoadjuvant or adjuvant chemotherapy in triple-negative breast cancer patients and some novel autophagy-related polymorphisms in their genomic DNA: A real-world study. Cancer Commun. 2018, 38, 71. [Google Scholar] [CrossRef] [Green Version]
- Berger, M.D.; Yamauchi, S.; Cao, S.; Hanna, D.L.; Sunakawa, Y.; Schirripa, M.; Matsusaka, S.; Yang, D.; Groshen, S.; Zhang, W.; et al. Autophagy-related polymorphisms predict hypertension in patients with metastatic colorectal cancer treated with FOLFIRI and bevacizumab: Results from TRIBE and FIRE-3 trials. Eur. J. Cancer 2017, 77, 13–20. [Google Scholar] [CrossRef]
- Hurley, J.H.; Young, L.N. Mechanisms of autophagy initiation. Annu. Rev. Biochem. 2017, 86, 225–244. [Google Scholar] [CrossRef] [PubMed]
- Kang, R.; Zeh, H.J.; Lotze, M.T.; Tang, D. The Beclin 1 Network Regulates Autophagy and Apoptosis. Cell Death Differ. 2011, 18, 571–580. [Google Scholar] [CrossRef] [PubMed]
- Cianfanelli, V.; De Zio, D.; Di Bartolomeo, S.; Nazio, F.; Strappazzon, F.; Cecconi, F. Ambra1 at a glance. J. Cell Sci. 2015, 128, 2003–2008. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.M.; Jung, C.H.; Seo, M.; Kim, E.K.; Park, J.M.; Bae, S.S.; Kim, D.H. MTORC1 phosphorylates UVRAG to negatively regulate autophagosome and endosome maturation. Mol. Cell 2015, 57, 207–218. [Google Scholar] [CrossRef] [Green Version]
- Zhong, Y.; Wang, Q.J.; Li, X.; Yan, Y.; Backer, J.M.; Chait, B.T.; Heintz, N.; Yue, Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat. Cell Biol. 2009, 11, 468–476. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, K.; Saitoh, T.; Tabata, K.; Omori, H.; Satoh, T.; Kurotori, N.; Maejima, I.; Shirahama-Noda, K.; Ichimura, T.; Isobe, T.; et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat. Cell Biol. 2009, 11, 385–396. [Google Scholar] [CrossRef]
- Cheng, X.; Ma, X.; Ding, X.; Li, L.; Jiang, X.; Shen, Z.; Chen, S.; Liu, W.; Gong, W.; Sun, Q. Pacer Mediates the Function of Class III PI3K and HOPS Complexes in Autophagosome Maturation by Engaging Stx17. Mol. Cell 2017, 65, 1029–1043.e5. [Google Scholar] [CrossRef] [Green Version]
- Hamet, P.; Haloui, M.; Harvey, F.; Marois-Blanchet, F.-C.; Sylvestre, M.-P.; Tahir, M.-R.; Simon, P.H.G.; Kanzki, B.S.; Raelson, J.; Long, C.; et al. PROX1 gene CC genotype as a major determinant of early onset of type 2 diabetes in slavic study participants from Action in Diabetes and Vascular Disease. J. Hypertens. 2017, 35, S24–S32. [Google Scholar] [CrossRef] [Green Version]
- Kazachkova, N.; Raposo, M.; Ramos, A.; Montiel, R.; Lima, M. Promoter Variant Alters Expression of the Autophagic BECN1 Gene: Implications for Clinical Manifestations of Machado-Joseph Disease. Cerebellum 2017, 16, 957–963. [Google Scholar] [CrossRef]
- Zhao, L.-L.; Liu, H.-L.; Luo, S.; Walsh, K.M.; Li, W.; Wei, Q. Associations of novel variants in PIK3C3, INSR and MAP3K4 of the ATM pathway genes with pancreatic cancer risk. Am. J. Cancer Res. 2020, 10, 2128–2144. [Google Scholar]
- Ng, D.; Hu, N.; Hu, Y.; Wang, C.; Giffen, C.; Tang, Z.-Z.; Han, X.-Y.; Yang, H.H.; Lee, M.P.; Goldstein, A.M.; et al. Replication of a genome-wide case-control study of esophageal squamous cell carcinoma. Int. J. Cancer 2008, 123, 1610–1615. [Google Scholar] [CrossRef] [Green Version]
- Hu, N.; Wang, C.; Hu, Y.; Yang, H.H.; Giffen, C.; Tang, Z.Z.; Han, X.Y.; Goldstein, A.M.; Emmert-Buck, M.R.; Buetow, K.H.; et al. Genome-wide association study in esophageal cancer using GeneChip mapping 10K array. Cancer Res. 2005, 65, 2542–2546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, N.; Zheng, Y.; Liu, J.; Lei, T.; Xu, Y.; Yang, M. Genetic variations associated with telomere length confer risk of gastric cardia adenocarcinoma. Gastric Cancer 2019, 22, 1089–1099. [Google Scholar] [CrossRef] [Green Version]
- Niewold, T.B.; Kariuki, S.N.; Franek, B.S.; Mikolaitis, R.A.; Utset, T.O.; Jolly, M.; Skol, A.D. Promoter variant of PIK3C3 is associated with autoimmunity against Ro and Sm epitopes in African-American lupus patients. J. Biomed. Biotechnol. 2010, 2010. [Google Scholar]
- Stopkova, P.; Saito, T.; Papolos, D.F.; Vevera, J.; Paclt, I.; Zukov, I.; Bersson, Y.B.; Margolis, B.A.; Strous, R.D.; Lachman, H.M. Identification of PIK3C3 promoter variant associated with bipolar disorder and schizophrenia. Biol. Psychiatry 2004, 55, 981–988. [Google Scholar] [CrossRef] [PubMed]
- Rietschel, M.; Mattheisen, M.; Degenhardt, F.; Mühleisen, T.W.; Kirsch, P.; Esslinger, C.; Herms, S.; Demontis, D.; Steffens, M.; Strohmaier, J.; et al. Association between genetic variation in a region on chromosome 11 and schizophrenia in large samples from Europe. Mol. Psychiatry 2012, 17, 906–917. [Google Scholar] [CrossRef] [Green Version]
- Mitjans, M.; Begemann, M.; Ju, A.; Dere, E.; Wüstefeld, L.; Hofer, S.; Hassouna, I.; Balkenhol, J.; Oliveira, B.; van der Auwera, S.; et al. Sexual dimorphism of AMBRA1-related autistic features in human and mouse. Transl. Psychiatry 2017, 7, e1247. [Google Scholar] [CrossRef]
- Litchfield, K.; Levy, M.; Orlando, G.; Loveday, C.; Law, P.J.; Migliorini, G.; Holroyd, A.; Broderick, P.; Karlsson, R.; Haugen, T.B.; et al. Identification of 19 new risk loci and potential regulatory mechanisms influencing susceptibility to testicular germ cell tumor. Nat. Genet. 2017, 49, 1133–1140. [Google Scholar] [CrossRef]
- Ross, C.J.; Towfic, F.; Shankar, J.; Laifenfeld, D.; Thoma, M.; Davis, M.; Weiner, B.; Kusko, R.; Zeskind, B.; Knappertz, V.; et al. A pharmacogenetic signature of high response to Copaxone in late-phase clinical-trial cohorts of multiple sclerosis. Genome Med. 2017, 9, 50. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.-K.; Lee, W.-Y.; Kwon, J.-T.; Sohn, D.-R.; Hong, S.-J.; Kim, H.-J. Association of ultraviolet radiation resistance-associated gene polymorphisms with rheumatoid arthritis. Biomed. Rep. 2014, 2, 117–121. [Google Scholar] [CrossRef]
- Jeong, T.-J.; Shin, M.-K.; Uhm, Y.-K.; Kim, H.-J.; Chung, J.-H.; Lee, M.-H. Association of UVRAG polymorphisms with susceptibility to non-segmental vitiligo in a Korean sample. Exp. Dermatol. 2010, 19, e323–e325. [Google Scholar] [CrossRef]
- Mercer, T.J.; Gubas, A.; Tooze, S.A. A Molecular Perspective of Mammalian Autophagosome Biogenesis. Biol. Chem. 2018, 293, 5386–5395. [Google Scholar] [CrossRef] [Green Version]
- Axe, E.L.; Walker, S.A.; Manifava, M.; Chandra, P.; Roderick, H.L.; Habermann, A.; Griffiths, G.; Ktistakis, N.T. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 2008, 182, 685–701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Proikas-Cezanne, T.; Takacs, Z.; Dönnes, P.; Kohlbacher, O. WIPI Proteins: Essential PtdIns3P Effectors at the Nascent Autophagosome. J. Cell Sci. 2015, 128, 207–217. [Google Scholar] [CrossRef] [Green Version]
- Dooley, H.C.; Razi, M.; Polson, H.E.J.; Girardin, S.E.; Wilson, M.I.; Tooze, S.A. WIPI2 Links LC3 Conjugation with PI3P, Autophagosome Formation, and Pathogen Clearance by Recruiting Atg12-5-16L1. Mol. Cell 2014, 55, 238–252. [Google Scholar] [CrossRef] [Green Version]
- Bakula, D.; Müller, A.J.; Zuleger, T.; Takacs, Z.; Franz-Wachtel, M.; Thost, A.K.; Brigger, D.; Tschan, M.P.; Frickey, T.; Robenek, H.; et al. WIPI3 and WIPI4 β-propellers are scaffolds for LKB1-AMPK-TSC signalling circuits in the control of autophagy. Nat. Commun. 2017, 8, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Osawa, T.; Kotani, T.; Kawaoka, T.; Hirata, E.; Suzuki, K.; Nakatogawa, H.; Ohsumi, Y.; Noda, N.N. Atg2 mediates direct lipid transfer between membranes for autophagosome formation. Nat. Struct. Mol. Biol. 2019, 26, 281–288. [Google Scholar] [CrossRef]
- Osawa, T.; Noda, N.N. Atg2: A novel phospholipid transfer protein that mediates de novo autophagosome biogenesis. Protein Sci. A Publ. Protein Soc. 2019, 28, 1005–1012. [Google Scholar] [CrossRef] [Green Version]
- Filimonenko, M.; Isakson, P.; Finley, K.D.; Anderson, M.; Jeong, H.; Melia, T.J.; Bartlett, B.J.; Myers, K.M.; Birkeland, H.C.G.; Lamark, T.; et al. The Selective Macroautophagic Degradation of Aggregated Proteins Requires the PI3P-Binding Protein Alfy. Mol. Cell 2010, 38, 265–279. [Google Scholar] [CrossRef] [Green Version]
- Noda, T. Autophagy in the Context of the Cellular Membrane-Trafficking System: The Enigma of Atg9 Vesicles. Biochem. Soc. Trans. 2017, 45, 1323–1331. [Google Scholar] [CrossRef] [Green Version]
- Orsi, A.; Razi, M.; Dooley, H.C.; Robinson, D.; Weston, A.E.; Collinson, L.M.; Tooze, S.A. Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol. Biol. Cell 2012, 23, 1860–1873. [Google Scholar] [CrossRef] [PubMed]
- Sobota, R.S.; Stein, C.M.; Kodaman, N.; Maro, I.; Wieland-Alter, W.; Igo, R.P.; Magohe, A.; Malone, L.L.; Chervenak, K.; Hall, N.B.; et al. A chromosome 5q31.1 locus associates with tuberculin skin test reactivity in HIV-positive individuals from tuberculosis hyper-endemic regions in east Africa. PLoS Genet. 2017, 13, e1006710. [Google Scholar] [CrossRef] [PubMed]
- Kadir, R.; Harel, T.; Markus, B.; Perez, Y.; Bakhrat, A.; Cohen, I.; Volodarsky, M.; Feintsein-Linial, M.; Chervinski, E.; Zlotogora, J.; et al. ALFY-Controlled DVL3 Autophagy Regulates Wnt Signaling, Determining Human Brain Size. PLoS Genet. 2016, 12, e1005919. [Google Scholar] [CrossRef] [Green Version]
- Lesseur, C.; Diergaarde, B.; Olshan, A.F.; Wünsch-Filho, V.; Ness, A.R.; Liu, G.; Lacko, M.; Eluf-Neto, J.; Franceschi, S.; Lagiou, P.; et al. Genome-wide association analyses identify new susceptibility loci for oral cavity and pharyngeal cancer. Nat. Genet. 2016, 48, 1544–1550. [Google Scholar] [CrossRef]
- Ohba, C.; Nabatame, S.; Iijima, Y.; Nishiyama, K.; Tsurusaki, Y.; Nakashima, M.; Miyake, N.; Tanaka, F.; Ozono, K.; Saitsu, H.; et al. De novo WDR45 mutation in a patient showing clinically Rett syndrome with childhood iron deposition in brain. J. Hum. Genet. 2014, 59, 292–295. [Google Scholar] [CrossRef] [PubMed]
- Tschentscher, A.; Dekomien, G.; Ross, S.; Cremer, K.; Kukuk, G.M.; Epplen, J.T.; Hoffjan, S. Analysis of the C19orf12 and WDR45 genes in patients with neurodegeneration with brain iron accumulation. J. Neurol. Sci. 2015, 349, 105–109. [Google Scholar] [CrossRef]
- Saitsu, H.; Nishimura, T.; Muramatsu, K.; Kodera, H.; Kumada, S.; Sugai, K.; Kasai-Yoshida, E.; Sawaura, N.; Nishida, H.; Hoshino, A.; et al. De novo mutations in the autophagy gene WDR45 cause static encephalopathy of childhood with neurodegeneration in adulthood. Nat. Genet. 2013, 45, 445–449. [Google Scholar]
- Lee, H.-S.; Park, T. Nuclear receptor and VEGF pathways for gene-blood lead interactions, on bone mineral density, in Korean smokers. PLoS ONE 2018, 13, e0193323. [Google Scholar] [CrossRef] [Green Version]
- Suleiman, J.; Allingham-Hawkins, D.; Hashem, M.; Shamseldin, H.E.; Alkuraya, F.S.; El-Hattab, A.W. WDR45B-related intellectual disability, spastic quadriplegia, epilepsy, and cerebral hypoplasia: A consistent neurodevelopmental syndrome. Clin. Genet. 2018, 93, 360–364. [Google Scholar] [CrossRef]
- Brinar, M.; Vermeire, S.; Cleynen, I.; Lemmens, B.; Sagaert, X.; Henckaerts, L.; Van Assche, G.; Geboes, K.; Rutgeerts, P.; De Hertogh, G. Genetic variants in autophagy-related genes and granuloma formation in a cohort of surgically treated Crohn’s disease patients. J. Crohn’s Colitis 2012, 6, 43–50. [Google Scholar] [CrossRef] [Green Version]
- Yamada, Y.; Sakuma, J.; Takeuchi, I.; Yasukochi, Y.; Kato, K.; Oguri, M.; Fujimaki, T.; Horibe, H.; Muramatsu, M.; Sawabe, M.; et al. Identification of C21orf59 and ATG2A as novel determinants of renal function-related traits in Japanese by exome-wide association studies. Oncotarget 2017, 8, 45259–45273. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Mateos, J.; Seijas-Tamayo, R.; Klain, J.C.A.; Borgonõn, M.P.; Pérez-Ruiz, E.; Mesiá, R.; Del Barco, E.; Coloma, C.S.; Dominguez, A.R.; Daroqui, J.C.; et al. Analysis of autophagy gene polymorphisms in Spanish patients with head and neck squamous cell carcinoma. Sci. Rep. 2017, 7, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buffen, K.; Oosting, M.; Quintin, J.; Ng, A.; Kleinnijenhuis, J.; Kumar, V.; van de Vosse, E.; Wijmenga, C.; van Crevel, R.; Oosterwijk, E.; et al. Autophagy Controls BCG-Induced Trained Immunity and the Response to Intravesical BCG Therapy for Bladder Cancer. PLoS Pathog. 2014, 10, e1004485. [Google Scholar] [CrossRef] [PubMed]
- Mehrabi Pour, M.; Nasiri, M.; Kamfiroozie, H.; Zibaeenezhad, M.J. Association of the ATG9B gene polymorphisms with coronary artery disease susceptibility: A case-control study. J. Cardiovasc. Thorac. Res. 2019, 11, 109–115. [Google Scholar] [CrossRef]
- Mizushima, N. The ATG conjugation systems in autophagy. Curr. Opin. Cell Biol. 2020, 63, 1–10. [Google Scholar] [CrossRef]
- Hanada, T.; Noda, N.N.; Satomi, Y.; Ichimura, Y.; Fujioka, Y.; Takao, T.; Inagaki, F.; Ohsumi, Y. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J. Biol. Chem. 2007, 282, 37298–37302. [Google Scholar] [CrossRef] [Green Version]
- Khor, B.; Conway, K.L.; Omar, A.S.; Biton, M.; Haber, A.L.; Rogel, N.; Baxt, L.A.; Begun, J.; Kuballa, P.; Gagnon, J.D.; et al. Distinct Tissue-Specific Roles for the Disease-Associated Autophagy Genes ATG16L2 and ATG16L1. J. Immunol. 2019, 203, 1820–1829. [Google Scholar] [CrossRef]
- Ishibashi, K.; Fujita, N.; Kanno, E.; Omori, H.; Yoshimori, T.; Itoh, T.; Fukuda, M. Atg16L2, a novel isoform of mammalian Atg16L that is not essential for canonical autophagy despite forming an Atg12-5-16L2 complex. Autophagy 2011, 7, 1500–1513. [Google Scholar] [CrossRef]
- Wesch, N.; Kirkin, V.; Rogov, V.V. Atg8-Family Proteins-Structural Features and Molecular Interactions in Autophagy and Beyond. Cells 2020, 9, 2008. [Google Scholar] [CrossRef]
- Fernandez, A.F.; Lopez-Otin, C. The functional and pathologic relevance of autophagy proteases. J. Clin. Investig. 2015, 125, 33–41. [Google Scholar] [CrossRef] [Green Version]
- Nair, U.; Yen, W.L.; Mari, M.; Cao, Y.; Xie, Z.; Baba, M.; Reggiori, F.; Klionsky, D.J. A role for Atg8-PE deconjugation in autophagosome biogenesis. Autophagy 2012, 8, 780–793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Z.Q.; Ni, T.; Hong, B.; Wang, H.Y.; Jiang, F.J.; Zou, S.; Chen, Y.; Zheng, X.L.; Klionsky, D.J.; Liang, Y.; et al. Dual roles of Atg8 - PE deconjugation by Atg4 in autophagy. Autophagy 2012, 8, 883–892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z.; Liu, Z. Potentially functional variants of autophagy-related genes are associated with the efficacy and toxicity of radiotherapy in patients with nasopharyngeal carcinoma. Mol. Genet. Genom. Med. 2019, 7, 1–8. [Google Scholar] [CrossRef]
- Song, X.; Yuan, Z.; Yuan, H.; Wang, L.; Ji, P.; Jin, G.; Dai, J.; Ma, H. ATG12 expression quantitative trait loci associated with head and neck squamous cell carcinoma risk in a Chinese Han population. Mol. Carcinog. 2018, 57, 1030–1037. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.W.; Hsieh, M.S.; Chang, Y.H.; Huang, P.M.; Lee, J.M. Genetic polymorphisms of ATG5 predict survival and recurrence in patients with early-stage esophageal squamous cell carcinoma. Oncotarget 2017, 8, 91494–91504. [Google Scholar] [CrossRef]
- Shen, M.; Lin, L. Functional variants of autophagy-related genes are associated with the development of hepatocellular carcinoma. Life Sci. 2019, 235, 116675. [Google Scholar] [CrossRef]
- Yuan, J.; Zhang, N.; Yin, L.; Zhu, H.; Zhang, L.; Zhou, L.; Yang, M. Clinical Implications of the Autophagy Core Gene Variations in Advanced Lung Adenocarcinoma Treated with Gefitinib. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, K.A.M.; Luo, L.; Thompson, T.A.; Torres, S.; Hu, C.A.A.; Thomas, N.E.; Lilyquist, J.; Anton-Culver, H.; Gruber, S.B.; From, L.; et al. Variants in autophagy-related genes and clinical characteristics in melanoma: A population-based study. Cancer Med. 2016, 5, 3336–3345. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.X.; Zhou, X.; Huang, T.T.; Tang, Y.; Liu, B.; Peng, P.; Sun, L.; Wang, Y.H.; Yuan, X.L. The Thr300Ala variant of ATG16L1 is associated with decreased risk of brain metastasis in patients with non-small cell lung cancer. Autophagy 2017, 13, 1053–1063. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Ma, F.; Wang, J.; Li, Q.; Zhang, P.; Yuan, P.; Luo, Y.; Cai, R.; Fan, Y.; Chen, S.; et al. Genetic polymorphisms of autophagy-related gene 5 (ATG5) rs473543 predict different disease-free survivals of triple-negative breast cancer patients receiving anthracycline- and/or taxane-based adjuvant chemotherapy. Chin. J. Cancer 2018, 37, 4. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Hang, D.; Jiang, Y.; Chen, J.; Han, J.; Zhou, W.; Jin, G.; Ma, H.; Dai, J. Evaluation of genetic variants in autophagy pathway genes as prognostic biomarkers for breast cancer. Gene 2017, 627, 549–555. [Google Scholar] [CrossRef] [PubMed]
- Michailidou, K.; Beesley, J.; Lindstrom, S.; Canisius, S.; Dennis, J.; Lush, M.J.; Maranian, M.J.; Bolla, M.K.; Wang, Q.; Shah, M.; et al. Genome-wide association analysis of more than 120,000 individuals identifies 15 new susceptibility loci for breast cancer. Nat. Genet. 2015, 47, 373–380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, Z.; Xue, J.; He, Y.; Ma, H.; Jin, G.; Chen, J.; Hu, Z.; Liu, X.a.; Shen, H. Potentially functional polymorphisms in ATG10 are associated with risk of breast cancer in a Chinese population. Gene 2013, 527, 491–495. [Google Scholar] [CrossRef] [PubMed]
- Santoni, M.; Piva, F.; De Giorgi, U.; Mosca, A.; Basso, U.; Santini, D.; Buti, S.; Lolli, C.; Terrone, C.; Maruzzo, M.; et al. Autophagic gene polymorphisms in liquid biopsies and outcome of patients with metastatic clear cell renal cell carcinoma. Anticancer Res. 2018, 38, 5773–5782. [Google Scholar] [CrossRef]
- Nikseresht, M.; Shahverdi, M.; Dehghani, M.; Abidi, H.; Mahmoudi, R.; Ghalamfarsa, G.; Manzouri, L.; Ghavami, S. Association of single nucleotide autophagy-related protein 5 gene polymorphism rs2245214 with susceptibility to non–small cell lung cancer. J. Cell. Biochem. 2019, 120, 1924–1931. [Google Scholar] [CrossRef]
- Xie, K.; Liang, C.; Li, Q.; Yan, C.; Wang, C.; Gu, Y.; Zhu, M.; Du, F.; Wang, H.; Dai, J.; et al. Role of ATG10 expression quantitative trait loci in non-small cell lung cancer survival. Int. J. Cancer 2016, 139, 1564–1573. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, J.S.; Li, N.; Weinhold, N.; Försti, A.; Ali, M.; Van Duin, M.; Thorleifsson, G.; Johnson, D.C.; Chen, B.; Halvarsson, B.M.; et al. Genome-wide association study identifies multiple susceptibility loci for multiple myeloma. Nat. Commun. 2016, 7, 22. [Google Scholar] [CrossRef]
- Plantinga, T.S.; van de Vosse, E.; Huijbers, A.; Netea, M.G.; Joosten, L.A.B.; Smit, J.W.A.; Netea-Maier, R.T. Role of Genetic Variants of Autophagy Genes in Susceptibility for Non-Medullary Thyroid Cancer and Patients Outcome. PLoS ONE 2014, 9, e94086. [Google Scholar] [CrossRef] [Green Version]
- Huijbers, A.; Plantinga, T.S.; Joosten, L.A.B.; Aben, K.K.H.; Gudmundsson, J.; den Heijer, M.; Kiemeney, L.A.L.M.; Netea, M.G.; Hermus, A.R.M.M.; Netea-Maier, R.T. The effect of the ATG16L1 Thr300Ala Polymorphism on Susceptibility and Outcome of Patients with Epithelial Cell-Derived Thyroid Carcinoma. Endocr. Relat. Cancer 2012, 19, L15–L18. [Google Scholar] [CrossRef]
- Grimm, W.A.; Messer, J.S.; Murphy, S.F.; Nero, T.; Lodolce, J.P.; Weber, C.R.; Logsdon, M.F.; Bartulis, S.; Sylvester, B.E.; Springer, A.; et al. The Thr300Ala variant in ATG16L1 is associated with improved survival in human colorectal cancer and enhanced production of type I interferon. Gut 2016, 65, 456–464. [Google Scholar] [CrossRef] [Green Version]
- Burada, F.; Ciurea, M.E.; Nicoli, R.; Streata, I.; Vilcea, I.D.; Rogoveanu, I.; Ioana, M. ATG16L1 T300A Polymorphism is Correlated with Gastric Cancer Susceptibility. Pathol. Oncol. Res. 2016, 22, 317–322. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.Y.; Huang, S.P.; Lin, V.C.; Yu, C.C.; Chang, T.Y.; Lu, T.L.; Chiang, H.C.; Bao, B.Y. Genetic variants of the autophagy pathway as prognostic indicators for prostate cancer. Sci. Rep. 2015, 5, 14045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, L.; Xu, J.; Song, J.; Xu, Y.; Zhang, B.; Gao, C.; Zhu, D.; Zhou, C.; Bi, D.; Wang, Y.; et al. Autophagy-Related Gene 7 Polymorphisms and Cerebral Palsy in Chinese Infants. Front. Cell. Neurosci. 2019, 13, 494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, J.; Xia, L.; Shang, Q.; Du, J.; Zhu, D.; Wang, Y.; Bi, D.; Song, J.; Ma, C.; Gao, C.; et al. A Variant of the Autophagy-Related 5 Gene Is Associated with Child Cerebral Palsy. Cell. Neurosci. 2017, 11, 407. [Google Scholar] [CrossRef] [Green Version]
- Metzger, S.; Saukko, M.; Van Che, H.; Tong, L.; Puder, Y.; Riess, O.; Nguyen, H.P. Age at onset in Huntington’s disease is modified by the autophagy pathway: Implication of the V471A polymorphism in Atg7. Hum. Genet. 2010, 128, 453–459. [Google Scholar] [CrossRef]
- Metzger, S.; Walter, C.; Riess, O.; Roos, R.A.C.; Nielsen, J.E.; Craufurd, D.; Nguyen, H.P. The V471A Polymorphism in Autophagy-Related Gene ATG7 Modifies Age at Onset Specifically in Italian Huntington Disease Patients. PLoS ONE 2013, 8, e68951. [Google Scholar] [CrossRef]
- Kim, M.; Sandford, E.; Gatica, D.; Qiu, Y.; Liu, X.; Zheng, Y.; Schulman, B.A.; Xu, J.; Semple, I.; Ro, S.H.; et al. Mutation in ATG5 reduces autophagy and leads to ataxia with developmental delay. eLife 2016, 5, e12245. [Google Scholar] [CrossRef] [Green Version]
- Chen, D.; Zhu, C.; Wang, X.; Feng, X.; Pang, S.; Huang, W.; Hawley, R.G.; Yan, B. A novel and functional variant within the ATG5 gene promoter in sporadic Parkinson’s disease. Neurosci. Lett. 2013, 538, 49–53. [Google Scholar] [CrossRef]
- Yuan, J.; Han, R.; Esther, A.; Wu, Q.; Yang, J.; Yan, W.; Ji, X.; Liu, Y.; Li, Y.; Yao, W.; et al. Polymorphisms in autophagy related genes and the coal workers’ pneumoconiosis in a Chinese population. Gene 2017, 632, 36–42. [Google Scholar] [CrossRef]
- Lee, T.H.; Ko, T.M.; Chen, C.H.; Chang, Y.J.; Lu, L.S.; Chang, C.H.; Huang, K.L.; Chang, T.Y.; Lee, J.D.; Chang, K.C.; et al. A genome-wide association study links small-vessel ischemic stroke to autophagy. Sci. Rep. 2017, 7, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Hampe, J.; Franke, A.; Rosenstiel, P.; Till, A.; Teuber, M.; Huse, K.; Albrecht, M.; Mayr, G.; De La Vega, F.M.; Briggs, J.; et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat. Genet. 2007, 39, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, S.; Mandell, M.A.; Deretic, V. IRGM governs the core autophagy machinery to conduct antimicrobial defense. Mol. Cell 2015, 58, 507–521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bekpen, C.; Xavier, R.J.; Eichler, E.E. Human IRGM gene “to be or not to be”. Semin. Immunopathol. 2010, 32, 437–444. [Google Scholar] [CrossRef] [PubMed]
- Dezelak, M.; Repnik, K.; Koder, S.; Ferkolj, I.; Potočnik, U. A Prospective Pharmacogenomic Study of Crohn’s Disease Patients during Routine Therapy with Anti-TNF-α Drug Adalimumab: Contribution of ATG5, NFKB1, and CRP Genes to Pharmacodynamic Variability. Omics A J. Integr. Biol. 2016, 20, 296–309. [Google Scholar] [CrossRef] [PubMed]
- Usategui-Martín, R.; García-Aparicio, J.; Corral-Gudino, L.; Calero-Paniagua, I.; Del Pino-Montes, J.; González Sarmiento, R. Polymorphisms in Autophagy Genes Are Associated with Paget Disease of Bone. PLoS ONE 2015, 10, e0128984. [Google Scholar] [CrossRef] [PubMed]
- Kamel, A.M.; Badary, M.S.; Mohamed, W.A.; Ahmed, G.H.; El-Feky, M.A. Evaluation of autophagy-related genes in Egyptian systemic lupus erythematosus patients. Int. J. Rheum. Dis. 2020, 5, 1–7. [Google Scholar] [CrossRef]
- Ciccacci, C.; Perricone, C.; Alessandri, C.; Latini, A.; Politi, C.; Delunardo, F.; Pierdominici, M.; Conti, F.; Novelli, G.; Ortona, E.; et al. Evaluation of ATG5 polymorphisms in Italian patients with systemic lupus erythematosus: Contribution to disease susceptibility and clinical phenotypes. Lupus 2018, 27, 1464–1469. [Google Scholar] [CrossRef]
- Dang, J.; Li, J.; Xin, Q.; Shan, S.; Bian, X.; Yuan, Q.; Liu, N.; Ma, X.; Li, Y.; Liu, Q. Gene–gene interaction of ATG5, ATG7, BLK and BANK1 in systemic lupus erythematosus. Int. J. Rheum. Dis. 2016, 19, 1284–1293. [Google Scholar] [CrossRef]
- López, P.; Alonso-Pérez, E.; Rodríguez-Carrio, J.; Suárez, A. Influence of Atg5 Mutation in SLE Depends on Functional IL-10 Genotype. PLoS ONE 2013, 8, e78756. [Google Scholar] [CrossRef] [Green Version]
- Alonso-Perez, E.; Suarez-Gestal, M.; Calaza, M.; Ordi-Ros, J.; Balada, E.; Bijl, M.; Papasteriades, C.; Carreira, P.; Skopouli, F.N.; Witte, T.; et al. Further Evidence of Subphenotype Association with Systemic Lupus Erythematosus Susceptibility Loci: A European Cases Only Study. PLoS ONE 2012, 7, e45356. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.J.; Lu, X.L.; Lv, J.C.; Yang, H.Z.; Qin, L.X.; Zhao, M.H.; Su, Y.; Li, Z.G.; Zhang, H. Genetic association of PRDM1-ATG5 intergenic region and autophagy with systemic lupus erythematosus in a Chinese population. Ann. Rheum. Dis. 2011, 70, 1330–1337. [Google Scholar] [CrossRef] [PubMed]
- Harley, J.B.; Alarcón-Riquelme, M.E.; Criswell, L.A.; Jacob, C.O.; Kimberly, R.P.; Moser, K.L.; Tsao, B.P.; Vyse, T.J.; Langefeld, C.D.; Nath, S.K.; et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat. Genet. 2008, 40, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Yu, H.; Zhang, L.; Li, H.; Liu, Y.; Kijlstra, A.; Yang, P. Association of ATG5 gene polymorphisms with behçet’s disease and ATG10 gene polymorphisms with VKH syndrome in a chinese han population. Investig. Ophthalmol. Vis. Sci. 2015, 56, 8280–8287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, P.P.; Wang, H.X.; Zhuang, J.C.; Liu, Q.B.; Zhao, G.X.; Li, Z.X.; Wu, Z.Y. Variants of autophagy-related gene 5 are associated with neuromyelitis optica in the Southern Han Chinese population. Autoimmunity 2014, 47, 563–566. [Google Scholar] [CrossRef] [PubMed]
- Mayes, M.D.; Bossini-Castillo, L.; Gorlova, O.; Martin, J.E.; Zhou, X.; Chen, W.V.; Assassi, S.; Ying, J.; Tan, F.K.; Arnett, F.C.; et al. Immunochip analysis identifies multiple susceptibility loci for systemic sclerosis. Am. J. Hum. Genet. 2014, 94, 47–61. [Google Scholar] [CrossRef] [Green Version]
- Martin, J.E.; Assassi, S.; Diaz-Gallo, L.M.; Broen, J.C.; Simeon, C.P.; Castellvi, I.; Vicente-Rabaneda, E.; Fonollosa, V.; Ortego-Centeno, N.; González-Gay, M.A.; et al. A systemic sclerosis and systemic lupus erythematosus pan-meta-GWAS reveals new shared susceptibility loci. Hum. Mol. Genet. 2013, 22, 4021–4029. [Google Scholar] [CrossRef] [Green Version]
- You, Y.; Huo, J.; Huang, J.; Wang, M.; Shao, Y.; Ge, M.; Li, X.; Huang, Z.; Zhang, J.; Nie, N.; et al. Contribution of autophagy-related gene 5 variants to acquired aplastic anemia in Han-Chinese population. J. Cell. Biochem. 2019, 120, 11409–11417. [Google Scholar] [CrossRef]
- Martin, L.J.; Gupta, J.; Jyothula, S.S.S.K.; Butsch Kovacic, M.; Biagini Myers, J.M.; Patterson, T.L.; Ericksen, M.B.; He, H.; Gibson, A.M.; Baye, T.M.; et al. Functional Variant in the Autophagy-Related 5 Gene Promotor is Associated with Childhood Asthma. PLoS ONE 2012, 7, e33454. [Google Scholar] [CrossRef]
- Poon, A.H.; Chouiali, F.; Tse, S.M.; Litonjua, A.A.; Hussain, S.N.A.; Baglole, C.J.; Eidelman, D.H.; Olivenstein, R.; Martin, J.G.; Weiss, S.T.; et al. Genetic and histologic evidence for autophagy in asthma pathogenesis. J. Allergy Clin. Immunol. 2012, 129, 569–571. [Google Scholar] [CrossRef] [Green Version]
- Jansen, A.F.M.; Schoffelen, T.; Bleeker-Rovers, C.P.; Wever, P.C.; Jaeger, M.; Oosting, M.; Adriaans, A.; Joosten, L.A.B.; Netea, M.G.; van Deuren, M.; et al. Genetic variations in innate immunity genes affect response to Coxiella burnetii and are associated with susceptibility to chronic Q fever. Clin. Microbiol. Infect. 2019, 25, e11–e631. [Google Scholar] [CrossRef]
- Shao, Y.; Chen, F.; Chen, Y.; Zhang, W.; Lin, Y.; Cai, Y.; Yin, Z.; Tao, S.; Liao, Q.; Zhao, J.; et al. Association between genetic polymorphisms in the autophagy-related 5 gene promoter and the risk of sepsis. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Fan, X.; Wang, X.; Zhang, X.; Zhang, K.; Han, Q.; Lv, Y.; Liu, Z. Genetic association of polymorphisms at the intergenic region between PRDM1 and ATG5 with hepatitis B virus infection in Han Chinese patients. J. Med Virol. 2020, 92, 1198–1205. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Fan, X.; Wang, X.; Deng, H.; Zhang, K.; Zhang, X.; Han, Q.; Lv, Y.; Liu, Z. Autophagy-Related 5 Gene rs510432 Polymorphism Is Associated with Hepatocellular Carcinoma in Patients with Chronic Hepatitis B Virus Infection. Immunol. Investig. 2019, 48, 378–391. [Google Scholar] [CrossRef]
- Tanaka, S.; Nagashima, H.; Uotani, T.; Graham, D.Y.; Yamaoka, Y. Autophagy-related genes in Helicobacter pylori infection. Helicobacter 2017, 22, 1–10. [Google Scholar] [CrossRef]
- Castaño-Rodríguez, N.; Kaakoush, N.O.; Goh, K.L.; Fock, K.M.; Mitchell, H.M. Autophagy in Helicobacter pylori Infection and Related Gastric Cancer. Helicobacter 2015, 20, 353–369. [Google Scholar] [CrossRef] [PubMed]
- Raju, D.; Hussey, S.; Ang, M.; Terebiznik, M.R.; Sibony, M.; Galindo-Mata, E.; Gupta, V.; Blanke, S.R.; Delgado, A.; Romero-Gallo, J.; et al. Vacuolating cytotoxin and variants in Atg16L1 that disrupt autophagy promote helicobacter pylori infection in humans. Gastroenterology 2012, 142, 1160–1171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Douroudis, K.; Kingo, K.; Traks, T.; Rätsep, R.; Silm, H.; Vasar, E.; Kõks, S. ATG16L1 gene polymorphisms are associated with palmoplantar pustulosis. Hum. Immunol. 2011, 72, 613–615. [Google Scholar] [CrossRef] [PubMed]
- Douroudis, K.; Kingo, K.; Traks, T.; Reimann, E.; Raud, K.; Rätsep, R.; Mössner, R.; Silm, H.; Vasar, E.; Kõks, S. Polymorphisms in the ATG16L1 Gene are Associated with Psoriasis Vulgaris. Acta Derm. Venereol. 2012, 92, 85–87. [Google Scholar] [CrossRef] [Green Version]
- Mao, J.J.; Wu, L.X.; Wang, W.; Ye, Y.Y.; Yang, J.; Chen, H.; Yang, Q.F.; Zhang, X.Y.; Wang, B.; Chen, W.X. Nucleotide variation in ATG4A and susceptibility to cervical cancer in southwestern chinese women. Oncol. Lett. 2018, 15, 2992–3000. [Google Scholar] [CrossRef]
- He, Q.; Lu, Y.; Hu, S.; Huang, Q.; Li, S.; Huang, Y.; Hu, Q.; Wu, L.; Chen, W. An intron SNP rs807185 in ATG4A decreases the risk of lung cancer in a southwest Chinese population. Eur. J. Cancer Prev. 2016, 25, 255–258. [Google Scholar] [CrossRef]
- Turcot, V.; Lu, Y.; Highland, H.M.; Schurmann, C.; Justice, A.E.; Fine, R.S.; Bradfield, J.P.; Esko, T.; Giri, A.; Graff, M.; et al. Protein-altering variants associated with body mass index implicate pathways that control energy intake and expenditure in obesity. Nat. Genet. 2018, 50, 26–41. [Google Scholar] [CrossRef] [Green Version]
- Franceschini, N.; Giambartolomei, C.; de Vries, P.S.; Finan, C.; Bis, J.C.; Huntley, R.P.; Lovering, R.C.; Tajuddin, S.M.; Winkler, T.W.; Graff, M.; et al. GWAS and colocalization analyses implicate carotid intima-media thickness and carotid plaque loci in cardiovascular outcomes. Nat. Commun. 2018, 9, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, C.; Wen, Y.; Guo, X.; Yang, T.; Shen, H.; Chen, X.; Tian, Q.; Tan, L.; Deng, H.W.; Zhang, F. Genetic association, mRNA and protein expression analysis identify ATG4C as a susceptibility gene for Kashin–Beck disease. Osteoarthr. Cartil. 2017, 25, 281–286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Portilla-Fernandez, E.; Ghanbari, M.; van Meurs, J.B.J.; Danser, A.H.J.; Franco, O.H.; Muka, T.; Roks, A.; Dehghan, A. Dissecting the association of autophagy-related genes with cardiovascular diseases and intermediate vascular traits: A population-based approach. PLoS ONE 2019, 14, e0214137. [Google Scholar] [CrossRef] [Green Version]
- Hysi, P.G.; Choquet, H.; Khawaja, A.P.; Wojciechowski, R.; Tedja, M.S.; Yin, J.; Simcoe, M.J.; Patasova, K.; Mahroo, O.A.; Thai, K.K.; et al. Meta-analysis of 542,934 subjects of European ancestry identifies new genes and mechanisms predisposing to refractive error and myopia. Nat. Genet. 2020, 52, 401–407. [Google Scholar] [CrossRef]
- Qi, Y.Y.; Zhou, X.J.; Nath, S.K.; Sun, C.; Wang, Y.N.; Hou, P.; Mu, R.; Li, C.; Guo, J.P.; Li, Z.G.; et al. A Rare Variant (rs933717) at FBXO31-MAP1LC3B in Chinese Is Associated With Systemic Lupus Erythematosus. Arthritis Rheumatol. 2018, 70, 287–297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johansen, T.; Lamark, T. Selective Autophagy: ATG8 Family Proteins, LIR Motifs and Cargo Receptors. J. Mol. Biol. 2020, 432, 80–103. [Google Scholar] [CrossRef]
- Khaminets, A.; Behl, C.; Dikic, I. Ubiquitin-Dependent and Independent Signals In Selective Autophagy. Trends Cell Biol. 2016, 26, 6–16. [Google Scholar] [CrossRef]
- Katsuragi, Y.; Ichimura, Y.; Komatsu, M. P62/SQSTM1 Functions as a Signaling Hub and an Autophagy Adaptor. FEBS J. 2015, 82, 4672–4678. [Google Scholar] [CrossRef] [Green Version]
- Kirkin, V.; McEwan, D.G.; Novak, I.; Dikic, I. A Role for Ubiquitin in Selective Autophagy; Elsevier: Amsterdam, The Netherlands, 2009; Volume 34, pp. 259–269. [Google Scholar]
- Korac, J.; Schaeffer, V.; Kovacevic, I.; Clement, A.M.; Jungblut, B.; Behl, C.; Terzic, J.; Dikic, I. Ubiquitin-independent function of optineurin in autophagic clearance of protein aggregates. J. Cell Sci. 2013, 126, 580–592. [Google Scholar] [CrossRef] [Green Version]
- Lazarou, M.; Sliter, D.A.; Kane, L.A.; Sarraf, S.A.; Wang, C.; Burman, J.L.; Sideris, D.P.; Fogel, A.I.; Youle, R.J. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 2015, 524, 309–314. [Google Scholar] [CrossRef] [Green Version]
- Wild, P.; Farhan, H.; McEwan, D.G.; Wagner, S.; Rogov, V.V.; Brady, N.R.; Richter, B.; Korac, J.; Waidmann, O.; Choudhary, C.; et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 2011, 333, 228–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vargas, J.N.S.; Wang, C.; Bunker, E.; Hao, L.; Maric, D.; Schiavo, G.; Randow, F.; Youle, R.J. Spatiotemporal Control of ULK1 Activation by NDP52 and TBK1 during Selective Autophagy. Mol. Cell 2019, 74, 347–362.e6. [Google Scholar] [CrossRef] [Green Version]
- Ravenhill, B.J.; Boyle, K.B.; von Muhlinen, N.; Ellison, C.J.; Masson, G.R.; Otten, E.G.; Foeglein, A.; Williams, R.; Randow, F. The Cargo Receptor NDP52 Initiates Selective Autophagy by Recruiting the ULK Complex to Cytosol-Invading Bacteria. Mol. Cell 2019, 74, 320–329.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tumbarello, D.A.; Manna, P.T.; Allen, M.; Bycroft, M.; Arden, S.D.; Kendrick-Jones, J.; Buss, F. The Autophagy Receptor TAX1BP1 and the Molecular Motor Myosin VI Are Required for Clearance of Salmonella Typhimurium by Autophagy. PLoS Pathog. 2015, 11, e1005174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, K.; Psakhye, I.; Jentsch, S. Autophagic clearance of PolyQ proteins mediated by ubiquitin-Atg8 adaptors of the conserved CUET protein family. Cell 2014, 158, 549–563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gatica, D.; Lahiri, V.; Klionsky, D.J. Cargo recognition and degradation by selective autophagy. Nat. Cell Biol. 2018, 20, 233–242. [Google Scholar] [CrossRef]
- Yoo, S.M.; Jung, Y.K. A Molecular Approach to Mitophagy and Mitochondrial Dynamics. Mol. Cells 2018, 41, 18–26. [Google Scholar]
- Wei, Y.; Chiang, W.C.; Sumpter, R.; Mishra, P.; Levine, B. Prohibitin 2 Is an Inner Mitochondrial Membrane Mitophagy Receptor. Cell 2017, 168, 224–238.e10. [Google Scholar] [CrossRef] [Green Version]
- Princely Abudu, Y.; Pankiv, S.; Mathai, B.J.; Håkon Lystad, A.; Bindesbøll, C.; Brenne, H.B.; Yoke Wui Ng, M.; Thiede, B.; Yamamoto, A.; Mutugi Nthiga, T.; et al. NIPSNAP1 and NIPSNAP2 Act as “Eat Me” Signals for Mitophagy. Dev. Cell 2019, 49, 509–525.e12. [Google Scholar] [CrossRef] [Green Version]
- Kirkin, V.; Rogov, V.V. A Diversity of Selective Autophagy Receptors Determines the Specificity of the Autophagy Pathway. Mol. Cell 2019, 76, 268–285. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Lopez, N.; Garcia-Macia, M.; Sahu, S.; Athonvarangkul, D.; Liebling, E.; Merlo, P.; Cecconi, F.; Schwartz, G.J.; Singh, R. Autophagy in the CNS and Periphery Coordinate Lipophagy and Lipolysis in the Brown Adipose Tissue and Liver. Cell Metab. 2016, 23, 113–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, S.; Wells, C.D.; Roach, P.J. Starch-binding domain-containing protein 1 (Stbd1) and glycogen metabolism: Identification of the Atg8 family interacting motif (AIM) in Stbd1 required for interaction with GABARAPL1. Biochem. Biophys. Res. Commun. 2011, 413, 420–425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wyant, G.A.; Abu-Remaileh, M.; Frenkel, E.M.; Laqtom, N.N.; Dharamdasani, V.; Lewis, C.A.; Chan, S.H.; Heinze, I.; Ori, A.; Sabatini, D.M. Nufip1 is a ribosome receptor for starvation-induced ribophagy. Science 2018, 360, 751–758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dowdle, W.E.; Nyfeler, B.; Nagel, J.; Elling, R.A.; Liu, S.; Triantafellow, E.; Menon, S.; Wang, Z.; Honda, A.; Pardee, G.; et al. Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo. Nat. Cell Biol. 2014, 16, 1069–1079. [Google Scholar] [CrossRef]
- Mancias, J.D.; Wang, X.; Gygi, S.P.; Harper, J.W.; Kimmelman, A.C. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature 2014, 508, 105–109. [Google Scholar] [CrossRef]
- Haack, T.B.; Ignatius, E.; Calvo-Garrido, J.; Iuso, A.; Isohanni, P.; Maffezzini, C.; Lönnqvist, T.; Suomalainen, A.; Gorza, M.; Kremer, L.S.; et al. Absence of the Autophagy Adaptor SQSTM1/p62 Causes Childhood-Onset Neurodegeneration with Ataxia, Dystonia, and Gaze Palsy. Am. J. Hum. Genet. 2016, 99, 735–743. [Google Scholar] [CrossRef]
- Teyssou, E.; Takeda, T.; Lebon, V.; Boillée, S.; Doukouré, B.; Bataillon, G.; Sazdovitch, V.; Cazeneuve, C.; Meininger, V.; Leguern, E.; et al. Mutations in SQSTM1 encoding p62 in amyotrophic lateral sclerosis: Genetics and neuropathology. Acta Neuropathol. 2013, 125, 511–522. [Google Scholar] [CrossRef]
- Fecto, F.; Yan, J.; Vemula, S.P.; Liu, E.; Yang, Y.; Chen, W.; Zheng, J.G.; Shi, Y.; Siddique, N.; Arrat, H.; et al. SQSTM1 mutations in familial and sporadic amyotrophic lateral sclerosis. Arch. Neurol. 2011, 68, 1440–1446. [Google Scholar] [CrossRef]
- Le Ber, I.; Camuzat, A.; Guerreiro, R.; Bouya-Ahmed, K.; Bras, J.; Nicolas, G.; Gabelle, A.; Didic, M.; De Septenville, A.; Millecamps, S.; et al. SQSTM1 Mutations in french patients with frontotemporal dementia or frontotemporal dementia with amyotrophic lateral sclerosis. JAMA Neurol. 2013, 70, 1403–1410. [Google Scholar]
- Boutoleau-Bretonnière, C.; Camuzat, A.; Le Ber, I.; Bouya-Ahmed, K.; Guerreiro, R.; Deruet, A.L.; Evrard, C.; Bras, J.; Lamy, E.; Auffray-Calvier, E.; et al. A phenotype of atypical apraxia of speech in a family carrying SQSTM1 mutation. J. Alzheimer’s Dis. 2015, 43, 625–630. [Google Scholar] [CrossRef] [Green Version]
- Gang, Q.; Bettencourt, C.; Machado, P.M.; Brady, S.; Holton, J.L.; Pittman, A.M.; Hughes, D.; Healy, E.; Parton, M.; Hilton-Jones, D.; et al. Rare variants in SQSTM1 and VCP genes and risk of sporadic inclusion body myositis. Neurobiol. Aging 2016, 47, e1–e218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rea, S.L.; Walsh, J.P.; Ward, L.; Yip, K.; Ward, B.K.; Kent, G.N.; Steer, J.H.; Xu, J.; Ratajczak, T. A novel mutation (K378X) in the Sequestosome 1 gene associated with increased NF-κB signaling and Paget’s disease of bone with a severe phenotype. J. Bone Miner. Res. 2006, 21, 1136–1145. [Google Scholar] [CrossRef]
- Hocking, L.J.; Lucas, G.J.A.; Daroszewska, A.; Mangion, J.; Olavesen, M.; Cundy, T.; Nicholson, G.C.; Ward, L.; Bennett, S.T.; Wuyts, W.; et al. Domain-specific mutations in sequestosome 1 (SQSTM1) cause familial and sporadic Paget’s diease. Hum. Mol. Genet. 2002, 11, 2735–2739. [Google Scholar] [CrossRef] [PubMed]
- Ellinghaus, D.; Zhang, H.; Zeissig, S.; Lipinski, S.; Till, A.; Jiang, T.; Stade, B.; Bromberg, Y.; Ellinghaus, E.; Keller, A.; et al. Association between variants of PRDM1 and NDP52 and crohn’s disease, based on exome sequencing and functional studies. Gastroenterology 2013, 145, 339–347. [Google Scholar] [CrossRef] [Green Version]
- Pap, É.M.; Farkas, K.; Széll, M.; Németh, G.; Rajan, N.; Nagy, N. Identification of putative phenotype-modifying genetic factors associated with phenotypic diversity in Brooke-Spiegler syndrome. Exp. Dermatol. 2020. [Google Scholar]
- Maruyama, H.; Morino, H.; Ito, H.; Izumi, Y.; Kato, H.; Watanabe, Y.; Kinoshita, Y.; Kamada, M.; Nodera, H.; Suzuki, H.; et al. Mutations of optineurin in amyotrophic lateral sclerosis. Nature 2010, 465, 223–226. [Google Scholar] [CrossRef]
- Silva, I.A.L.; Conceição, N.; Gagnon, É.; Caiado, H.; Brown, J.P.; Gianfrancesco, F.; Michou, L.; Cancela, M.L. Effect of genetic variants of OPTN in the pathophysiology of Paget’s disease of bone. Biochim. Et Biophys. Acta Mol. Basis Dis. 2018, 1864, 143–151. [Google Scholar] [CrossRef]
- Albagha, O.M.E.; Visconti, M.R.; Alonso, N.; Langston, A.L.; Cundy, T.; Dargie, R.; Dunlop, M.G.; Fraser, W.D.; Hooper, M.J.; Isaia, G.; et al. Genome-wide association study identifies variants at CSF1, OPTN and TNFRSF11A as genetic risk factors for Paget’s disease of bone. Nat. Genet. 2010, 42, 520–524. [Google Scholar] [CrossRef] [Green Version]
- Rezaie, T.; Child, A.; Hitchings, R.; Brice, G.; Miller, L.; Coca-Prados, M.; Héon, E.; Krupin, T.; Ritch, R.; Kreutzer, D.; et al. Adult-onset primary open-angle glaucoma caused by mutations in optineurin. Sci. N.Y. 2002, 295, 1077–1079. [Google Scholar] [CrossRef] [PubMed]
- Newton, C.A.; Oldham, J.M.; Ley, B.; Anand, V.; Adegunsoye, A.; Liu, G.; Batra, K.; Torrealba, J.; Kozlitina, J.; Glazer, C.; et al. Telomere length and genetic variant associations with interstitial lung disease progression and survival. Eur. Respir. J. 2019, 53, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Araujo, F.J.d.; Silva, L.D.O.d.; Mesquita, T.G.; Pinheiro, S.K.; Vital, W.d.S.; Chrusciak-Talhari, A.; Guerra, J.A.d.O.; Talhari, S.; Ramasawmy, R. Polymorphisms in the TOLLIP Gene Influence Susceptibility to Cutaneous Leishmaniasis Caused by Leishmania guyanensis in the Amazonas State of Brazil. PLoS Negl. Trop. Dis. 2015, 9, e0003875. [Google Scholar] [CrossRef]
- Montoya-Buelna, M.; Fafutis-Morris, M.; Tovar-Cuevas, A.J.; Alvarado-Navarro, A.; Valle, Y.; Padilla-Gutierrez, J.R.; Muñoz-Valle, J.F.; Figuera-Villanueva, L.E. Role of toll-interacting protein gene polymorphisms in leprosy Mexican patients. Biomed Res. Int. 2013, 2013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shah, J.A.; Berrington, W.R.; Vary, J.C.; Wells, R.D.; Peterson, G.J.; Kunwar, C.B.; Khadge, S.; Hagge, D.A.; Hawn, T.R. Genetic variation in toll-interacting protein is associated with leprosy susceptibility and cutaneous expression of interleukin 1 receptor antagonist. J. Infect. Dis. 2016, 213, 1189–1197. [Google Scholar] [CrossRef] [Green Version]
- Brasil, L.W.; Barbosa, L.R.A.; De Araujo, F.J.; Da Costa, A.G.; Da Silva, L.D.O.; Pinheiro, S.K.; De Almeida, A.C.G.; Kuhn, A.; Vitor-Silva, S.; De Melo, G.C.; et al. TOLLIP gene variant is associated with Plasmodium vivax malaria in the Brazilian Amazon. Malar. J. 2017, 16, 116. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.; Huang, W.; Wang, D.; Wang, Y.; Wang, M.; Zhang, M.; He, J.Q. Evaluation of TLR2, TLR4, and TOLLIP polymorphisms for their role in tuberculosis susceptibility. Apmis 2018, 126, 501–508. [Google Scholar] [CrossRef]
- Song, Z.; Yin, J.; Yao, C.; Sun, Z.; Shao, M.; Zhang, Y.; Tao, Z.; Huang, P.; Tong, C. Variants in the Toll-interacting protein gene are associated with susceptibility to sepsis in the Chinese Han population. Crit. Care 2011, 15, R12. [Google Scholar] [CrossRef] [Green Version]
- Ruiz, M.T.; Balachi, J.F.; Fernandes, R.A.; Galbiatti, A.L.S.; Manigua, J.V.; Pavarino-Bertelli, É.C.; Goloni-Bertollo, E.M. Analysis of the TAX1BP1 gene in head and neck cancer patients. Braz. J. Otorhinolaryngol. 2010, 76, 193–198. [Google Scholar] [CrossRef] [Green Version]
- Geller, F.; Feenstra, B.; Carstensen, L.; Pers, T.H.; Van Rooij, I.A.L.M.; Körberg, I.B.; Choudhry, S.; Karjalainen, J.M.; Schnack, T.H.; Hollegaard, M.V.; et al. Genome-wide association analyses identify variants in developmental genes associated with hypospadias. Nat. Genet. 2014, 46, 957–963. [Google Scholar] [CrossRef]
- Lin, E.; Kuo, P.-H.; Liu, Y.-L.; Yu, Y.W.Y.; Yang, A.C.; Tsai, S.-J. A Deep Learning Approach for Predicting Antidepressant Response in Major Depression Using Clinical and Genetic Biomarkers. Front. Psychiatry 2018, 9, 290. [Google Scholar] [CrossRef] [Green Version]
- Ikeda, M.; Takahashi, A.; Kamatani, Y.; Momozawa, Y.; Saito, T.; Kondo, K.; Shimasaki, A.; Kawase, K.; Sakusabe, T.; Iwayama, Y.; et al. Genome-wide association study detected novel susceptibility genes for schizophrenia and shared trans-populations/diseases genetic effect. Schizophr. Bull. 2019, 45, 824–834. [Google Scholar] [CrossRef] [PubMed]
- Nicoletti, P.; Cartsos, V.M.; Palaska, P.K.; Shen, Y.; Floratos, A.; Zavras, A.I. Genomewide Pharmacogenetics of Bisphosphonate-Induced Osteonecrosis of the Jaw: The Role of RBMS3. Oncologist 2012, 17, 279–287. [Google Scholar] [CrossRef] [Green Version]
- Ilgaz Aydinlar, E.; Rolfs, A.; Serteser, M.; Parman, Y. Mutation in FAM134B causing hereditary sensory neuropathy with spasticity in a Turkish family. Muscle Nerve 2014, 49, 774–775. [Google Scholar] [CrossRef] [PubMed]
- Murphy, S.M.; Davidson, G.L.; Brandner, S.; Houlden, H.; Reilly, M.M. Mutation in FAM134B causing severe hereditary sensory neuropathy. J. Neurol. Neurosurg. Psychiatry 2012, 83, 119–120. [Google Scholar] [CrossRef]
- Fischer, D.; Schabhüttl, M.; Wieland, T.; Windhager, R.; Strom, T.M.; Auer-Grumbach, M. A Novel Missense Mutation Confirms ATL3 as a Gene for Hereditary Sensory Neuropathy Type 1. Brain 2014, 137, 286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kornak, U.; Mademan, I.; Schinke, M.; Voigt, M.; Krawitz, P.; Hecht, J.; Barvencik, F.; Schinke, T.; Gießelmann, S.; Beil, F.T.; et al. Sensory neuropathy with bone destruction due to a mutation in the membrane-shaping atlastin GTPase 3. Brain 2014, 137, 683–692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manjurano, A.; Clark, T.G.; Nadjm, B.; Mtove, G.; Wangai, H.; Sepulveda, N.; Campino, S.G.; Maxwell, C.; Olomi, R.; Rockett, K.R.; et al. Candidate Human Genetic Polymorphisms and Severe Malaria in a Tanzanian Population. PLoS ONE 2012, 7, e47463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Apinjoh, T.O.; Anchang-Kimbi, J.K.; Njua-Yafi, C.; Ngwai, A.N.; Mugri, R.N.; Clark, T.G.; Rockett, K.A.; Kwiatkowski, D.P.; Achidi, E.A. Association of candidate gene polymorphisms and TGF-beta/IL-10 levels with malaria in three regions of Cameroon: A case-control study. Malar. J. 2014, 13, 236. [Google Scholar] [CrossRef] [Green Version]
- Tavian, D.; Missaglia, S.; Redaelli, C.; Pennisi, E.M.; Invernici, G.; Wessalowski, R.; Maiwald, R.; Arca, M.; Coleman, R.A. Contribution of novel ATGL missense mutations to the clinical phenotype of NLSD-M: A strikingly low amount of lipase activity may preserve cardiac function. Hum. Mol. Genet. 2012, 21, 5318–5328. [Google Scholar] [CrossRef]
- Zolotov, S.; Xing, C.; Mahamid, R.; Shalata, A.; Sheikh-Ahmad, M.; Garg, A. Homozygous LIPE mutation in siblings with multiple symmetric lipomatosis, partial lipodystrophy, and myopathy. Am. J. Med Genet. Part A 2017, 173, 190–194. [Google Scholar] [CrossRef] [Green Version]
- Farhan, S.M.K.; Robinson, J.F.; McIntyre, A.D.; Marrosu, M.G.; Ticca, A.F.; Loddo, S.; Carboni, N.; Brancati, F.; Hegele, R.A. A Novel LIPE Nonsense Mutation Found Using Exome Sequencing in Siblings With Late-Onset Familial PartialLipodystrophy. Can. J. Cardiol. 2014, 30, 1649–1654. [Google Scholar] [CrossRef] [PubMed]
- Jansen, I.E.; Gibbs, J.R.; Nalls, M.A.; Price, T.R.; Lubbe, S.; van Rooij, J.; Uitterlinden, A.G.; Kraaij, R.; Williams, N.M.; Brice, A.; et al. Establishing the role of rare coding variants in known Parkinson’s disease risk loci. Neurobiol. Aging 2017, 59, e11–e220. [Google Scholar] [CrossRef] [PubMed]
- International Parkinson’s Disease Genomics, C.; Wellcome Trust Case Control, C. A two-stage meta-analysis identifies several new loci for Parkinson’s disease. PLoS Genet. 2011, 7, e1002142. [Google Scholar]
- Yucesoy, B.; Kaufman, K.M.; Lummus, Z.L.; Weirauch, M.T.; Zhang, G.; Cartier, A.; Boulet, L.P.; Sastre, J.; Quirce, S.; Tarlo, S.M.; et al. Genome-wide association study identifies novel loci associated with diisocyanate-induced occupational asthma. Toxicol. Sci. 2015, 146, 192–201. [Google Scholar] [CrossRef]
- Wang, Y.; Ray, A.M.; Johnson, E.K.; Zuhlke, K.A.; Cooney, K.A.; Lange, E.M. Evidence for an association between prostate cancer and chromosome 8q24 and 10q11 genetic variants in African American men: The flint men’s health study. Prostate 2011, 71, 225–231. [Google Scholar] [CrossRef] [Green Version]
- Sheu, S.Y.; Schwertheim, S.; Worm, K.; Grabellus, F.; Schmid, K.W. Diffuse sclerosing variant of papillary thyroid carcinoma: Lack of BRAF mutation but occurrence of RET/PTC rearrangements. Mod. Pathol. 2007, 20, 779–787. [Google Scholar] [CrossRef]
- Lee, J.M.; Gillis, T.; Mysore, J.S.; Ramos, E.M.; Myers, R.H.; Hayden, M.R.; Morrison, P.J.; Nance, M.; Ross, C.A.; Margolis, R.L.; et al. Common SNP-based haplotype analysis of the 4p16.3 Huntington disease gene region. Am. J. Hum. Genet. 2012, 90, 434–444. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Vicente, M.; Talloczy, Z.; Wong, E.; Tang, G.; Koga, H.; Kaushik, S.; de Vries, R.; Arias, E.; Harris, S.; Sulzer, D.; et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease. Nat. Neurosci. 2010, 13, 567–576. [Google Scholar] [CrossRef] [Green Version]
- Rui, Y.N.; Xu, Z.; Patel, B.; Chen, Z.; Chen, D.; Tito, A.; David, G.; Sun, Y.; Stimming, E.F.; Bellen, H.J.; et al. Huntingtin functions as a scaffold for selective macroautophagy. Nat. Cell Biol. 2015, 17, 262–275. [Google Scholar] [CrossRef] [Green Version]
- Kast, D.J.; Dominguez, R. The Cytoskeleton–Autophagy Connection. Curr Biol. 2017, 27, 318–326. [Google Scholar] [CrossRef] [Green Version]
- Jahreiss, L.; Menzies, F.M.; Rubinsztein, D.C. The itinerary of autophagosomes: From peripheral formation to kiss-and-run fusion with lysosomes. Traffic 2008, 9, 574–587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cardoso, C.M.P.; Groth-Pedersen, L.; Høyer-Hansen, M.; Kirkegaard, T.; Corcelle, E.; Andersen, J.S.; Jäättelä, M.; Nylandsted, J. Depletion of Kinesin 5B Affects Lysosomal Distribution and Stability and Induces Peri-Nuclear Accumulation of Autophagosomes in Cancer Cells. PLoS ONE 2009, 4, e4424. [Google Scholar] [CrossRef] [Green Version]
- Kimura, S.; Noda, T.; Yoshimori, T. Dynein-dependent movement of autophagosomes mediates efficient encounters with lysosomes. Cell Struct. Funct. 2008, 33, 109–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lőrincz, P.; Juhász, G. Autophagosome-Lysosome Fusion. J Mol Biol. 2020, 432, 2462–2482. [Google Scholar] [CrossRef]
- Zhao, Y.G.; Zhang, H. Autophagosome Maturation: An Epic Journey from the ER to Lysosomes. J Cell Biol. 2019, 218, 757–770. [Google Scholar] [CrossRef] [PubMed]
- Takáts, S.; Boda, A.; Csizmadia, T.; Juhász, G. Small GTPases controlling autophagy-related membrane traffic in yeast and metazoans. Small Gtpases 2018, 9, 465–471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szatmári, Z.; Sass, M. The Autophagic Roles of Rab Small GTPases and Their Upstream Regulators. Autophagy 2014, 10, 1154–1166. [Google Scholar] [CrossRef] [Green Version]
- Bröcker, C.; Kuhlee, A.; Gatsogiannis, C.; Kleine Balderhaar, H.J.; Hönscher, C.; Engelbrecht-Vandré, S.; Ungermann, C.; Raunser, S. Molecular architecture of the multisubunit homotypic fusion and vacuole protein sorting (HOPS) tethering complex. Proc. Natl. Acad. Sci. USA 2012, 109, 1991–1996. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Miao, G.; Xue, X.; Guo, X.; Yuan, C.; Wang, Z.; Zhang, G.; Chen, Y.; Feng, D.; Hu, J.; et al. The Vici Syndrome Protein EPG5 Is a Rab7 Effector that Determines the Fusion Specificity of Autophagosomes with Late Endosomes/Lysosomes. Mol. Cell 2016, 63, 781–795. [Google Scholar] [CrossRef] [Green Version]
- McEwan, D.G.; Popovic, D.; Gubas, A.; Terawaki, S.; Suzuki, H.; Stadel, D.; Coxon, F.P.; MirandadeStegmann, D.; Bhogaraju, S.; Maddi, K.; et al. PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol. Cell 2015, 57, 39–54. [Google Scholar] [CrossRef] [Green Version]
- Ding, X.; Jiang, X.; Tian, R.; Zhao, P.; Li, L.; Wang, X.; Chen, S.; Zhu, Y.; Mei, M.; Bao, S.; et al. RAB2 regulates the formation of autophagosome and autolysosome in mammalian cells. Autophagy 2019, 15, 1774–1786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lörincz, P.; Tóth, S.; Benkö, P.; Lakatos, Z.; Boda, A.; Glatz, G.; Zobel, M.; Bisi, S.; Hegedüs, K.; Takáts, S.; et al. Rab2 promotes autophagic and endocytic lysosomal degradation. J. Cell Biol. 2017, 216, 1937–1947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pantoom, S.; Konstantinidis, G.; Voss, S.; Han, H.; Hofnagel, O.; Li, Z.; Wu, Y.W. RAB33B recruits the ATG16L1 complex to the phagophore via a noncanonical RAB binding protein. Autophagy 2020, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Diao, J.; Liu, R.; Rong, Y.; Zhao, M.; Zhang, J.; Lai, Y.; Zhou, Q.; Wilz, L.M.; Li, J.; Vivona, S.; et al. ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature 2015, 520, 563–566. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Wang, L.; Lak, B.; Li, J.; Jokitalo, E.; Wang, Y. GRASP55 Senses Glucose Deprivation through O-GlcNAcylation to Promote Autophagosome-Lysosome Fusion. Dev. Cell 2018, 45, 245–261.e6. [Google Scholar] [CrossRef] [Green Version]
- Ebner, P.; Poetsch, I.; Deszcz, L.; Hoffmann, T.; Zuber, J.; Ikeda, F. The IAP family member BRUCE regulates autophagosome–lysosome fusion. Nat. Commun. 2018, 9, 1–15. [Google Scholar] [CrossRef]
- Nakamura, S.; Yoshimori, T. New Insights into Autophagosome-Lysosome Fusion. J Cell Sci. 2017, 130, 209–1216. [Google Scholar] [CrossRef] [Green Version]
- Lefebvre, C.; Legouis, R.; Culetto, E. ESCRT and autophagies: Endosomal functions and beyond. Semin. Cell Dev. Biol. 2018, 74, 21–28. [Google Scholar] [CrossRef]
- Nara, A.; Mizushima, N.; Yamamoto, A.; Kabeya, Y.; Ohsumi, Y.; Yoshimori, T. SKD1 AAA ATPase-dependent endosomal transport is involved in autolysosome formation. Cell Struct. Funct. 2002, 27, 29–37. [Google Scholar] [CrossRef] [Green Version]
- Tamai, K.; Tanaka, N.; Nara, A.; Yamamoto, A.; Nakagawa, I.; Yoshimori, T.; Ueno, Y.; Shimosegawa, T.; Sugamura, K. Role of Hrs in maturation of autophagosomes in mammalian cells. Biochem. Biophys. Res. Commun. 2007, 360, 721–727. [Google Scholar] [CrossRef]
- Lee, J.A.; Beigneux, A.; Ahmad, S.T.; Young, S.G.; Gao, F.B. ESCRT-III Dysfunction Causes Autophagosome Accumulation and Neurodegeneration. Curr. Biol. 2007, 17, 1561–1567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farg, M.A.; Sundaramoorthy, V.; Sultana, J.M.; Yang, S.; Atkinson, R.A.K.; Levina, V.; Halloran, M.A.; Gleeson, P.A.; Blair, I.P.; Soo, K.Y.; et al. C9ORF72, implicated in amytrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking. Hum. Mol. Genet. 2014, 23, 3579–3595. [Google Scholar] [CrossRef] [PubMed]
- Vantaggiato, C.; Panzeri, E.; Castelli, M.; Citterio, A.; Arnoldi, A.; Santorelli, F.M.; Liguori, R.; Scarlato, M.; Musumeci, O.; Toscano, A.; et al. ZFYVE26/SPASTIZIN and SPG11/SPATACSIN mutations in hereditary spastic paraplegia types AR-SPG15 and AR-SPG11 have different effects on autophagy and endocytosis. Autophagy 2019, 15, 34–57. [Google Scholar] [CrossRef]
- Morgan, N.E.; Cutrona, M.B.; Simpson, J.C. Multitasking Rab Proteins in Autophagy and Membrane Trafficking: A Focus on Rab33b. Int J Mol Sci. 2019, 20, 3916. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Ma, Z.; Jiao, X.; Fariss, R.; Kantorow, W.L.; Kantorow, M.; Pras, E.; Frydman, M.; Pras, E.; Riazuddin, S.; et al. Mutations in FYCO1 cause autosomal-recessive congenital cataracts. Am. J. Hum. Genet. 2011, 88, 827–838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goes, F.S.; Hamshere, M.L.; Seifuddin, F.; Pirooznia, M.; Belmonte-Mahon, P.; Breuer, R.; Schulze, T.; Nöthen, M.; Cichon, S.; Rietschel, M.; et al. Genome-wide association of mood-incongruent psychotic bipolar disorder. Transl. Psychiatry 2012, 2, e180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zillhardt, J.L.; Poirier, K.; Broix, L.; Lebrun, N.; Elmorjani, A.; Martinovic, J.; Saillour, Y.; Muraca, G.; Nectoux, J.; Bessieres, B.; et al. Mosaic parental germline mutations causing recurrent forms of malformations of cortical development. Eur. J. Hum. Genet. 2016, 24, 611–614. [Google Scholar] [CrossRef] [Green Version]
- Petukhova, L.; Duvic, M.; Hordinsky, M.; Norris, D.; Price, V.; Shimomura, Y.; Kim, H.; Singh, P.; Lee, A.; Chen, W.V.; et al. Genome-wide association study in alopecia areata implicates both innate and adaptive immunity. Nature 2010, 466, 113–117. [Google Scholar] [CrossRef] [Green Version]
- Xue, A.; Wu, Y.; Zhu, Z.; Zhang, F.; Kemper, K.E.; Zheng, Z.; Yengo, L.; Lloyd-Jones, L.R.; Sidorenko, J.; Wu, Y.; et al. Genome-wide association analyses identify 143 risk variants and putative regulatory mechanisms for type 2 diabetes. Nat. Commun. 2018, 9, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Horikoshi, M.; Beaumont, R.N.; Day, F.R.; Warrington, N.M.; Kooijman, M.N.; Fernandez-Tajes, J.; Feenstra, B.; Van Zuydam, N.R.; Gaulton, K.J.; Grarup, N.; et al. Genome-wide associations for birth weight and correlations with adult disease. Nature 2016, 538, 248–252. [Google Scholar] [CrossRef]
- Fuchs-Telem, D.; Stewart, H.; Rapaport, D.; Nousbeck, J.; Gat, A.; Gini, M.; Lugassy, Y.; Emmert, S.; Eckl, K.; Hennies, H.C.C.; et al. CEDNIK syndrome results from loss-of-function mutations in SNAP29. Br. J. Dermatol. 2011, 164, 610–616. [Google Scholar] [CrossRef] [PubMed]
- Bare, L.A.; Morrison, A.C.; Rowland, C.M.; Shiffman, D.; Luke, M.M.; Iakoubova, O.A.; Kane, J.P.; Malloy, M.J.; Ellis, S.G.; Pankow, J.S.; et al. Five common gene variants identify elevated genetic risk for coronary heart disease. Genet. Med. 2007, 9, 682–689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luke, M.M.; Lalouschek, W.; Rowland, C.M.; Catanese, J.J.; Bolonick, J.I.; Bui, N.D.; Greisenegger, S.; Endler, G.; Devlin, J.J.; Mannhalter, C. Polymorphisms Associated with Both Noncardioembolic Stroke and Coronary Heart Disease: Vienna Stroke Registry. Cerebrovasc. Dis. 2009, 28, 499–504. [Google Scholar] [CrossRef] [Green Version]
- Cheng, S.; Sun, C.; Lao, W.; Kang, H. Association of VAMP8 rs1010 Polymorphism with Host Susceptibility to Pulmonary Tuberculosis in a Chinese Han Population. Genet. Test. Mol. Biomark. 2019, 23, 299–303. [Google Scholar] [CrossRef]
- Hoffmann, T.J.; Van Den Eeden, S.K.; Sakoda, L.C.; Jorgenson, E.; Habel, L.A.; Graff, R.E.; Passarelli, M.N.; Cario, C.L.; Emami, N.C.; Chao, C.R.; et al. A large multiethnic genome-wide association study of prostate cancer identifies novel risk variants and substantial ethnic differences. Cancer Discov. 2015, 5, 878–891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Houlden, H.; King, R.H.M.; Muddle, J.R.; Warner, T.T.; Reilly, M.M.; Orrell, R.W.; Ginsberg, L. A novel RAB7 mutation associated with ulcero-mutilating neuropathy. Ann. Neurol. 2004, 56, 586–590. [Google Scholar] [CrossRef]
- Meggouh, F.; Bienfait, H.M.E.; Weterman, M.A.J.; De Visser, M.; Baas, F. Charcot-Marie-tooth disease due to a de novo mutation of the RAB7 gene. Neurology 2006, 67, 1476–1478. [Google Scholar] [CrossRef]
- Verhoeven, K.; De Jonghe, P.; Coen, K.; Verpoorten, N.; Auer-Grumbach, M.; Kwon, J.M.; FitzPatrick, D.; Schmedding, E.; De Vriendt, E.; Jacobs, A.; et al. Mutations in the small GTP-ase late endosomal protein RAB7 cause Charcot-Marie-Tooth type 2B neuropathy. Am. J. Hum. Genet. 2003, 72, 722–727. [Google Scholar] [CrossRef] [Green Version]
- Alshammari, M.J.; Al-Otaibi, L.; Alkuraya, F.S. Mutation in RAB33B, which encodes a regulator of retrograde Golgi transport, defines a second Dyggve-Melchior-Clausen locus. J. Med Genet. 2012, 49, 455–461. [Google Scholar] [CrossRef]
- Dupuis, N.; Lebon, S.; Kumar, M.; Drunat, S.; Graul-Neumann, L.M.; Gressens, P.; El Ghouzzi, V. A Novel RAB33B Mutation in Smith-McCort Dysplasia. Hum. Mutat. 2013, 34, 283–286. [Google Scholar] [CrossRef]
- Salian, S.; Cho, T.J.; Phadke, S.R.; Gowrishankar, K.; Bhavani, G.S.L.; Shukla, A.; Jagadeesh, S.; Kim, O.H.; Nishimura, G.; Girisha, K.M. Additional three patients with Smith-McCort dysplasia due to novel RAB33B mutations. Am. J. Med Genet. Part A 2017, 173, 588–595. [Google Scholar] [CrossRef]
- Kondo, H.; Maksimova, N.; Otomo, T.; Kato, H.; Imai, A.; Asano, Y.; Kobayashi, K.; Nojima, S.; Nakaya, A.; Hamada, Y.; et al. Mutation in VPS33A affects metabolism of glycosaminoglycans: A new type of mucopolysaccharidosis with severe systemic symptoms. Hum. Mol. Genet. 2017, 26, 173–183. [Google Scholar] [PubMed] [Green Version]
- Nagel, M.; Jansen, P.R.; Stringer, S.; Watanabe, K.; De Leeuw, C.A.; Bryois, J.; Savage, J.E.; Hammerschlag, A.R.; Skene, N.G.; Muñoz-Manchado, A.B.; et al. Meta-analysis of genome-wide association studies for neuroticism in 449,484 individuals identifies novel genetic loci and pathways. Nat. Genet. 2018, 50, 920–927. [Google Scholar] [CrossRef] [Green Version]
- Van Wesenbeeck, L.; Odgren, P.R.; Coxon, F.P.; Frattini, A.; Moens, P.; Perdu, B.; MacKay, C.A.; Van Hul, E.; Timmermans, J.P.; Vanhoenacker, F.; et al. Involvement of PLEKHM1 in osteoclastic vesicular transport and osteopetrosis in incisors absent rats and humans. J. Clin. Investig. 2007, 117, 919–930. [Google Scholar] [CrossRef] [Green Version]
- Edwards, T.L.; Scott, W.K.; Almonte, C.; Burt, A.; Powell, E.H.; Beecham, G.W.; Wang, L.; Züchner, S.; Konidari, I.; Wang, G.; et al. Genome-Wide association study confirms SNPs in SNCA and the MAPT region as common risk factors for parkinson disease. Ann. Hum. Genet. 2010, 74, 97–109. [Google Scholar] [CrossRef] [Green Version]
- Couch, F.J.; Wang, X.; McGuffog, L.; Lee, A.; Olswold, C.; Kuchenbaecker, K.B.; Soucy, P.; Fredericksen, Z.; Barrowdale, D.; Dennis, J.; et al. Genome-Wide Association Study in BRCA1 Mutation Carriers Identifies Novel Loci Associated with Breast and Ovarian Cancer Risk. PLoS Genet. 2013, 9, e1003212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phelan, C.M.; Kuchenbaecker, K.B.; Tyrer, J.P.; Kar, S.P.; Lawrenson, K.; Winham, S.J.; Dennis, J.; Pirie, A.; Riggan, M.J.; Chornokur, G.; et al. Identification of 12 new susceptibility loci for different histotypes of epithelial ovarian cancer. Nat. Genet. 2017, 49, 680–691. [Google Scholar] [CrossRef] [Green Version]
- Amare, A.T.; Schubert, K.O.; Tekola-Ayele, F.; Hsu, Y.H.; Sangkuhl, K.; Jenkins, G.; Whaley, R.M.; Barman, P.; Batzler, A.; Altman, R.B.; et al. Association of the polygenic scores for personality traits and response to selective serotonin reuptake inhibitors in patients with major depressive disorder. Front. Psychiatry 2018, 9, 16. [Google Scholar] [CrossRef] [Green Version]
- Hagenaars, S.P.; Hill, W.D.; Harris, S.E.; Ritchie, S.J.; Davies, G.; Liewald, D.C.; Gale, C.R.; Porteous, D.J.; Deary, I.J.; Marioni, R.E. Genetic prediction of male pattern baldness. PLoS Genet. 2017, 13, e1006594. [Google Scholar] [CrossRef]
- Piano Mortari, E.; Folgiero, V.; Marcellini, V.; Romania, P.; Bellacchio, E.; D’Alicandro, V.; Bocci, C.; Carrozzo, R.; Martinelli, D.; Petrini, S.; et al. The Vici syndrome protein EPG5 regulates intracellular nucleic acid trafficking linking autophagy to innate and adaptive immunity. Autophagy 2018, 14, 22–37. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.S.; Liu, X.; Xie, C.; Liu, Y.; Xu, C. Non-parametric Survival Analysis of EPG5 Gene with Age at Onset of Alzheimer’s Disease. J. Mol. Neurosci. 2016, 60, 436–444. [Google Scholar] [CrossRef]
- Mekli, K.; Phillips, D.F.; Arpawong, T.E.; Vanhoutte, B.; Tampubolon, G.; Nazroo, J.Y.; Lee, J.; Prescott, C.A.; Stevens, A.; Pendleton, N. Genome-wide scan of depressive symptomatology in two representative cohorts in the United States and the United Kingdom. J. Psychiatr. Res. 2018, 100, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Ayub, H.; Micheal, S.; Akhtar, F.; Khan, M.I.; Bashir, S.; Waheed, N.K.; Ali, M.; Schoenmaker-Koller, F.E.; Shafique, S.; Qamar, R.; et al. Association of a Polymorphism in the BIRC6 Gene with Pseudoexfoliative Glaucoma. PLoS ONE 2014, 9, e105023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van der Zee, J.; Urwin, H.; Engelborghs, S.; Bruyland, M.; Vandenberghe, R.; Dermaut, B.; De Pooter, T.; Peeters, K.; Santens, P.; De Deyn, P.P.; et al. CHMP2B C-truncating mutations in frontotemporal lobar degeneration are associated with an aberrant endosomal phenotype in vitro. Hum. Mol. Genet. 2008, 17, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, R.; Kapogiannis, D.; Huey, E.D.; Grafman, J.; Hardy, J.; Momeni, P. Novel missense mutation in charged multivesicular body protein 2B in a patient with frontotemporal dementia. Alzheimer Dis. Assoc. Disord. 2010, 24, 397–401. [Google Scholar] [CrossRef] [PubMed]
- Skibinski, G.; Parkinson, N.J.; Brown, J.M.; Chakrabarti, L.; Lloyd, S.L.; Hummerich, H.; Nielsen, J.E.; Hodges, J.R.; Spillantini, M.G.; Thusgaard, T.; et al. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat. Genet. 2005, 37, 806–808. [Google Scholar] [CrossRef]
- Cox, L.E.; Ferraiuolo, L.; Goodall, E.F.; Heath, P.R.; Higginbottom, A.; Mortiboys, H.; Hollinger, H.C.; Hartley, J.A.; Brockington, A.; Burness, C.E.; et al. Mutations in CHMP2B in Lower Motor Neuron Predominant Amyotrophic Lateral Sclerosis (ALS). PLoS ONE 2010, 5, e9872. [Google Scholar] [CrossRef]
- Shiels, A.; Bennett, T.M.; Knopf, H.L.S.; Yamada, K.; Yoshiura, K.I.; Niikawa, N.; Shim, S.; Hanson, P.I. CHMP4B, a novel gene for autosomal dominant cataracts linked to chromosome 20q. Am. J. Hum. Genet. 2007, 81, 596–606. [Google Scholar] [CrossRef] [Green Version]
- Raginis-Zborowska, A.; Mekli, K.; Payton, A.; Ollier, W.; Hamdy, S.; Pendleton, N. Genetic determinants of swallowing impairments among community dwelling older population. Exp. Gerontol. 2015, 69, 196–201. [Google Scholar] [CrossRef]
- Kichaev, G.; Bhatia, G.; Loh, P.R.; Gazal, S.; Burch, K.; Freund, M.K.; Schoech, A.; Pasaniuc, B.; Price, A.L. Leveraging Polygenic Functional Enrichment to Improve GWAS Power. Am. J. Hum. Genet. 2019, 104, 65–75. [Google Scholar] [CrossRef] [Green Version]
- Shamim, U.; Ambawat, S.; Singh, J.; Thomas, A.; Pradeep-Chandra-Reddy, C.; Suroliya, V.; Uppilli, B.; Parveen, S.; Sharma, P.; Chanchal, S.; et al. C9orf72 hexanucleotide repeat expansion in Indian patients with ALS: A common founder and its geographical predilection. Neurobiol. Aging 2020, 88, e1–e156. [Google Scholar] [CrossRef]
- Logue, M.W.; Schu, M.; Vardarajan, B.N.; Farrell, J.; Lunetta, K.L.; Jun, G.; Baldwin, C.T.; DeAngelis, M.M.; Farrer, L.A. A search for age-related macular degeneration risk variants in Alzheimer disease genes and pathways. Neurobiol. Aging 2014, 35, e7–e1510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morais, S.; Raymond, L.; Mairey, M.; Coutinho, P.; Brandão, E.; Ribeiro, P.; Loureiro, J.L.; Sequeiros, J.; Brice, A.; Alonso, I.; et al. Massive sequencing of 70 genes reveals a myriad of missing genes or mechanisms to be uncovered in hereditary spastic paraplegias. Eur. J. Hum. Genet. 2017, 25, 1217–1228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bowling, K.M.; Thompson, M.L.; Amaral, M.D.; Finnila, C.R.; Hiatt, S.M.; Engel, K.L.; Cochran, J.N.; Brothers, K.B.; East, K.M.; Gray, D.E.; et al. Genomic diagnosis for children with intellectual disability and/or developmental delay. Genome Med. 2017, 9, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Xie, T.; Deng, L.; Mei, P.; Zhou, Y.; Wang, B.; Zhang, J.; Lin, J.; Wei, Y.; Zhang, X.; Xu, R. A genome-wide association study combining pathway analysis for typical sporadic amyotrophic lateral sclerosis in Chinese Han populations. Neurobiol. Aging 2014, 35, e9–e1778. [Google Scholar] [CrossRef] [PubMed]
- Sagona, A.P.; Nezis, I.P.; Bache, K.G.; Haglund, K.; Bakken, A.C.; Skotheim, R.I.; Stenmark, H. A tumor-associated mutation of FYVE-CENT prevents its interaction with Beclin 1 and interferes with cytokinesis. PLoS ONE 2011, 6, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Settembre, C.; Fraldi, A.; Jahreiss, L.; Spampanato, C.; Venturi, C.; Medina, D.; de Pablo, R.; Tacchetti, C.; Rubinsztein, D.C.; Ballabio, A. A block of autophagy in lysosomal storage disorders. Hum Mol Genet 2008, 17, 119–129. [Google Scholar] [CrossRef]
- Fraldi, A.; Annunziata, F.; Lombardi, A.; Kaiser, H.J.; Medina, D.L.; Spampanato, C.; Fedele, A.O.; Polishchuk, R.; Sorrentino, N.C.; Simons, K.; et al. Lysosomal fusion and SNARE function are impaired by cholesterol accumulation in lysosomal storage disorders. Embo J 2010, 29, 3607–3620. [Google Scholar] [CrossRef] [Green Version]
- Corcelle-Termeau, E.; Vindelov, S.D.; Hamalisto, S.; Mograbi, B.; Keldsbo, A.; Brasen, J.H.; Favaro, E.; Adam, D.; Szyniarowski, P.; Hofman, P.; et al. Excess sphingomyelin disturbs ATG9A trafficking and autophagosome closure. Autophagy 2016, 12, 833–849. [Google Scholar] [CrossRef] [Green Version]
- Elrick, M.J.; Yu, T.; Chung, C.; Lieberman, A.P. Impaired proteolysis underlies autophagic dysfunction in Niemann-Pick type C disease. Hum Mol Genet 2012, 21, 4876–4887. [Google Scholar] [CrossRef] [Green Version]
- Gabande-Rodriguez, E.; Boya, P.; Labrador, V.; Dotti, C.G.; Ledesma, M.D. High sphingomyelin levels induce lysosomal damage and autophagy dysfunction in Niemann Pick disease type A. Cell Death Differ 2014, 21, 864–875. [Google Scholar] [CrossRef]
- Tomanin, R.; Karageorgos, L.; Zanetti, A.; Al-Sayed, M.; Bailey, M.; Miller, N.; Sakuraba, H.; Hopwood, J.J. Mucopolysaccharidosis type VI (MPS VI) and molecular analysis: Review and classification of published variants in the ARSB gene. Hum. Mutat. 2018, 39, 1788–1802. [Google Scholar] [CrossRef] [Green Version]
- Morrone, A.; Caciotti, A.; Atwood, R.; Davidson, K.; Du, C.; Francis-Lyon, P.; Harmatz, P.; Mealiffe, M.; Mooney, S.; Oron, T.R.; et al. Morquio a syndrome-associated mutations: A review of alterations in the GALNS gene and a new locus-specific database. Hum. Mutat. 2014, 35, 1271–1279. [Google Scholar] [CrossRef] [Green Version]
- Brunetti-Pierri, N.; Scaglia, F. GM1 gangliosidosis: Review of clinical, molecular, and therapeutic aspects. Mol. Genet. Metab. 2008, 94, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Zhu, X.; Liu, C.L.; Shi, H.; Shen, S.; Yang, Y.; Hasegawa, K.; Camargo, C.A., Jr.; Liang, L. Shared genetics of asthma and mental health disorders: A large-scale genome-wide cross-trait analysis. Eur. Respir. J. 2019, 54, 1901507. [Google Scholar] [CrossRef]
- Callahan, J.W. Molecular basis of GM1 gangliosidosis and Morquio disease, type B. Structure-function studies of lysosomal β-galactosidase and the non-lysosomal β-galactosidase-like protein. Biochim. Et Biophys. Acta Mol. Basis Dis. 1999, 1455, 85–103. [Google Scholar] [CrossRef] [Green Version]
- Jakobkiewicz-Banecka, J.; Gabig-Ciminska, M.; Kloska, A.; Malinowska, M.; Piotrowska, E.; Banecka-Majkutewicz, Z.; Banecki, B.; Wegrzyn, A.; Wegrzyn, G. Glycosaminoglycans and mucopolysaccharidosis type III. Front Biosci. 2016, 1, 1393–1409. [Google Scholar]
- Tomatsu, S.; Montano, A.M.; Dung, V.C.; Grubb, J.H.; Sly, W.S. Mutations and polymorphisms in GUSB gene in mucopolysaccharidosis VII (sly syndrome). Hum. Mutat. 2009, 30, 511–519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Triggs-Raine, B. Biology of hyaluronan: Insights from genetic disorders of hyaluronan metabolism. World J. Biol. Chem. 2015, 6, 110. [Google Scholar] [CrossRef]
- Froissart, R.; Da Silva, I.M.; Maire, I. Mucopolysaccharidosis type II: An update on mutation spectrum. Acta Paediatr. Int. J. Paediatr. 2007, 96, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Poletto, E.; Pasqualim, G.; Giugliani, R.; Matte, U.; Baldo, G. Worldwide distribution of common IDUA pathogenic variants. Clin. Genet. 2018, 94, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Rowland, T.J.; Sweet, M.E.; Mestroni, L.; Taylor, M.R.G. Danon disease dysregulation of autophagy in a multisystem disorder with cardiomyopathy. J. Cell Sci. 2016, 129, 2135–2143. [Google Scholar] [CrossRef] [Green Version]
- Peruzzo, P.; Pavan, E.; Dardis, A. Molecular genetics of Pompe disease: A comprehensive overview. Ann. Transl. Med. 2019, 7, 278. [Google Scholar] [CrossRef] [PubMed]
- Potter, N.T.; Miller, C.A.; Anderson, I.J. Mutation detection in an equivocal case of Friedreich’s ataxia. Pediatric Neurol. 2000, 22, 413–415. [Google Scholar] [CrossRef]
- Bogaert, R.; Tiller, G.E.; Weis, M.A.; Gruber, H.E.; Rimoin, D.L.; Cohn, D.H.; Eyre, D.R. An amino acid substitution (Gly853 → Glu) in the collagen α1(II) chain produces hypochondrogenesis. J. Biol. Chem. 1992, 267, 22522–22526. [Google Scholar]
- Kinghorn, K.J.; Asghari, A.M.; Castillo-Quan, J.I. The emerging role of autophagic-lysosomal dysfunction in Gaucher disease and Parkinson’s disease. Neural Regen. Res. 2017, 12, 380–384. [Google Scholar] [CrossRef] [PubMed]
- Benitez, B.A.; Davis, A.A.; Jin, S.C.; Ibanez, L.; Ortega-Cubero, S.; Pastor, P.; Choi, J.; Cooper, B.; Perlmutter, J.S.; Cruchaga, C. Resequencing analysis of five Mendelian genes and the top genes from genome-wide association studies in Parkinson’s Disease. Mol. Neurodegener. 2016, 11, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Ortiz, A.; Germain, D.P.; Desnick, R.J.; Politei, J.; Mauer, M.; Burlina, A.; Eng, C.; Hopkin, R.J.; Laney, D.; Linhart, A.; et al. Fabry disease revisited: Management and treatment recommendations for adult patients. Mol. Genet. Metab. 2018, 123, 416–427. [Google Scholar] [CrossRef]
- Bräuer, A.U.; Kuhla, A.; Holzmann, C.; Wree, A.; Witt, M. Current challenges in understanding the cellular and molecular mechanisms in niemann-pick disease type C1. Int. J. Mol. Sci. 2019, 20, 4392. [Google Scholar] [CrossRef] [Green Version]
- Wilmer, M.J.; Schoeber, J.P.; Van Den Heuvel, L.P.; Levtchenko, E.N. Cystinosis: Practical tools for diagnosis and treatment. Pediatric Nephrol. 2011, 26, 205–215. [Google Scholar] [CrossRef] [Green Version]
- Mole, S.E.; Cotman, S.L. Genetics of the neuronal ceroid lipofuscinoses (Batten disease). Biochim. Et Biophys. Acta Mol. Basis Dis. 2015, 1852, 2237–2241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caciotti, A.; Catarzi, S.; Tonin, R.; Lugli, L.; Perez, C.R.; Michelakakis, H.; Mavridou, I.; Donati, M.A.; Guerrini, R.; d’Azzo, A.; et al. Galactosialidosis: Review and analysis of CTSA gene mutations. Orphanet J. Rare Dis. 2013, 8, 114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wali, G.; Wali, G.M.; Sue, C.M.; Kumar, K.R. A Novel Homozygous Mutation in the FUCA1 Gene Highlighting Fucosidosis as a Cause of Dystonia: Case Report and Literature Review. Neuropediatrics 2019, 50, 248–252. [Google Scholar] [CrossRef] [PubMed]
- Aula, N.; Salomäki, P.; Timonen, R.; Verheijen, F.; Mancini, G.; Månsson, J.E.; Aula, P.; Peltonen, L. The spectrum of SLC17A5-gene mutations resulting in free sialic acid-storage diseases indicates some genotype-phenotype correlation. Am. J. Hum. Genet. 2000, 67, 832–840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malm, D.; Nilssen, Ø. Alpha-mannosidosis. Orphanet J. Rare Dis. 2008, 3, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Molho-Pessach, V.; Bargal, R.; Abramowitz, Y.; Doviner, V.; Ingber, A.; Raas-Rothschild, A.; Ne’eman, Z.; Zeigler, M.; Zlotogorski, A. Angiokeratoma corporis diffusum in human β-mannosidosis: Report of a new case and a novel mutation. J. Am. Acad. Dermatol. 2007, 57, 407–412. [Google Scholar] [CrossRef]
- Saito, H.; Inazawa, J.; Saito, S.; Kasumi, F.; Koi, S.; Sagae, S.; Kudo, R.; Saito, J.; Noda, K.; Nakamura, Y. Detailed Deletion Mapping of Chromosome 17q in Ovarian and Breast Cancers: 2-cM Region on 17q21.3 Often and Commonly Deleted in Tumors. Cancer Res. 1993, 53, 3382–3385. [Google Scholar]
- Gao, X.; Zacharek, A.; Salkowski, A.; Grignon, D.J.; Sakr, W.; Porter, A.T.; Honn, K.V. Loss of heterozygosity of the BRCA1 and other loci on chromosome 17q in human prostate cancer. Cancer Res. 1995, 55, 1002–1005. [Google Scholar]
- Carvill, G.L.; Liu, A.; Mandelstam, S.; Schneider, A.; Lacroix, A.; Zemel, M.; McMahon, J.M.; Bello-Espinosa, L.; Mackay, M.; Wallace, G.; et al. Severe infantile onset developmental and epileptic encephalopathy caused by mutations in autophagy gene WDR45. Epilepsia 2018, 59, e5–e13. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Huang, J.; Pang, S.; Wang, H.; Zhang, A.; Hawley, R.G.; Yan, B. Novel and functional ATG12 gene variants in sporadic Parkinson’s disease. Neurosci. Lett. 2017, 643, 22–26. [Google Scholar] [CrossRef]
- Chen, D.; Pang, S.; Feng, X.; Huang, W.; Hawley, R.G.; Yan, B. Genetic analysis of the ATG7 gene promoter in sporadic Parkinson’s disease. Neurosci. Lett. 2013, 534, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Elisa, R.; Rainero, I.; Chiò, A.; Rogaeva, E.; Galimberti, D.; Fenoglio, P.; Grinberg, Y.; Isaia, G.; Calvo, A.; Gentile, S.; et al. SQSTM1 mutations in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Neurology 2012, 79, 1556–1562. [Google Scholar]
- Bucelli, R.C.; Arhzaouy, K.; Pestronk, A.; Pittman, S.K.; Rojas, L.; Sue, C.M.; Evilä, A.; Hackman, P.; Udd, B.; Harms, M.B.; et al. SQSTM1 splice site mutation in distal myopathy with rimmed vacuoles. Neurology 2015, 85, 665–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nuij, V.J.A.A.; Peppelenbosch, M.P.; Woude, C.J.; Fuhler, G.M. Genetic polymorphism in ATG16L1 gene is associated with adalimumab use in inflammatory bowel disease. J. Transl. Med. 2017, 15, 248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koder, S.; Repnik, K.; Ferkolj, I.; Pernat, C.; Skok, P.; Weersma, R.K.; Potočnik, U. Genetic polymorphism in ATG16L1 gene influences the response to adalimumab in Crohn’s disease patients. Pharmacogenomics 2015, 16, 191–204. [Google Scholar] [CrossRef] [PubMed]

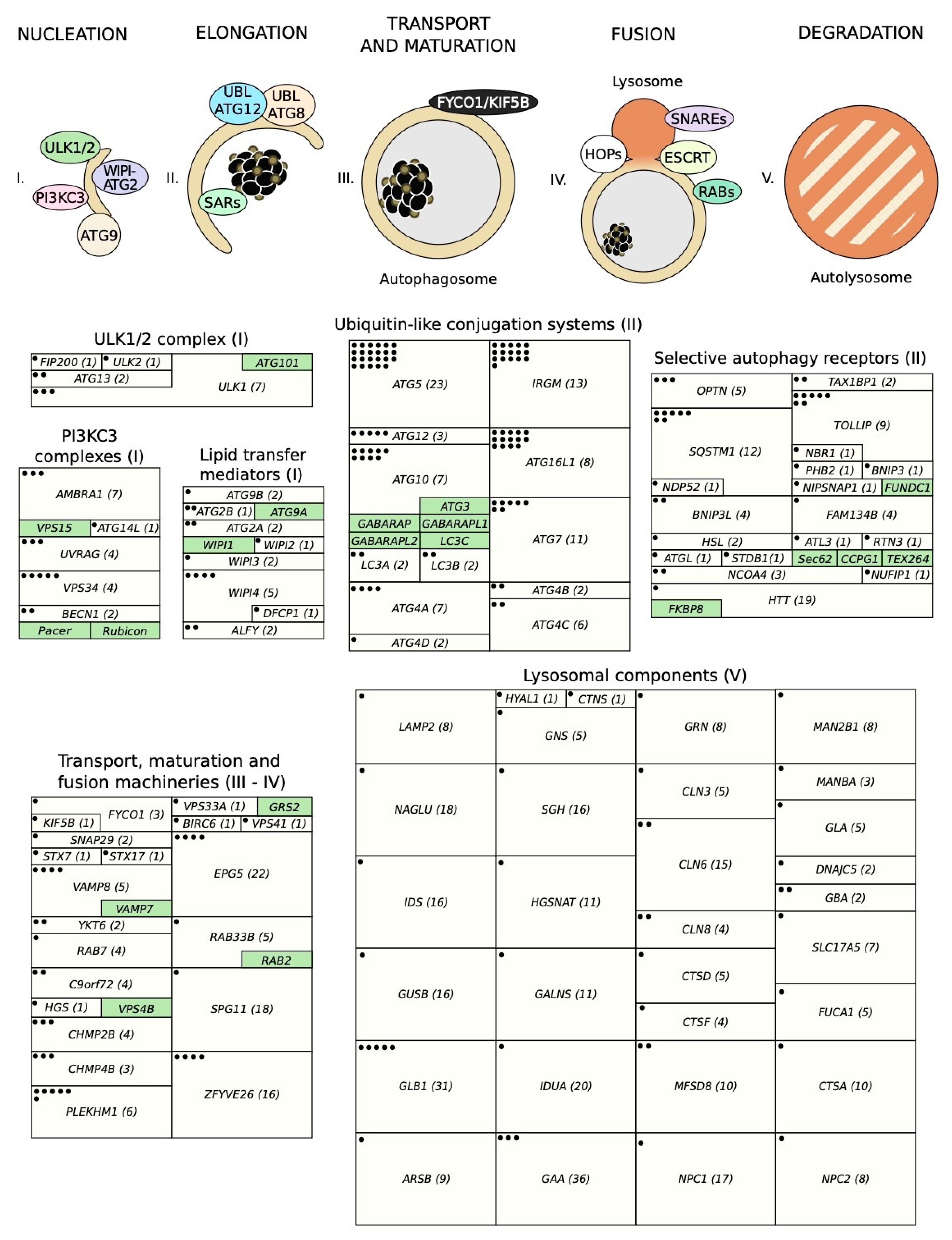
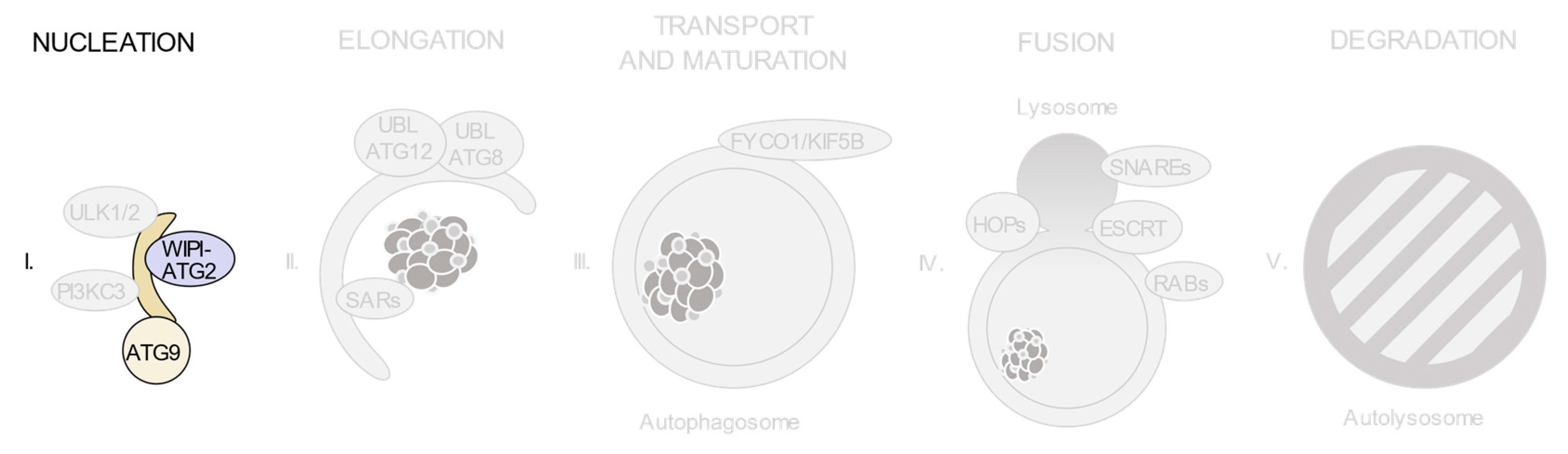

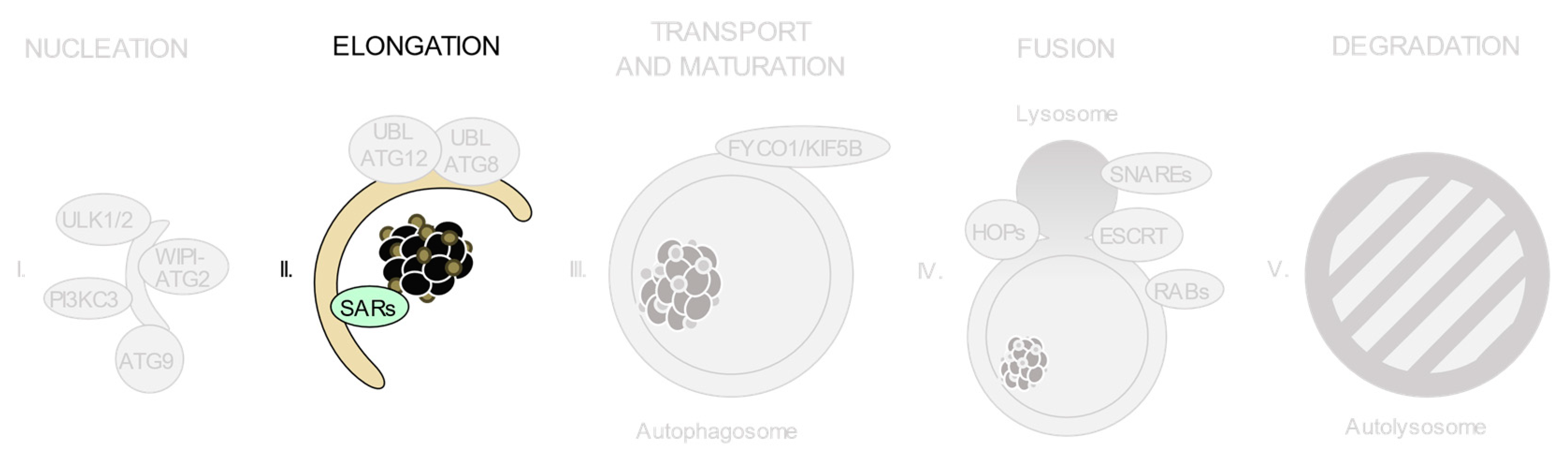

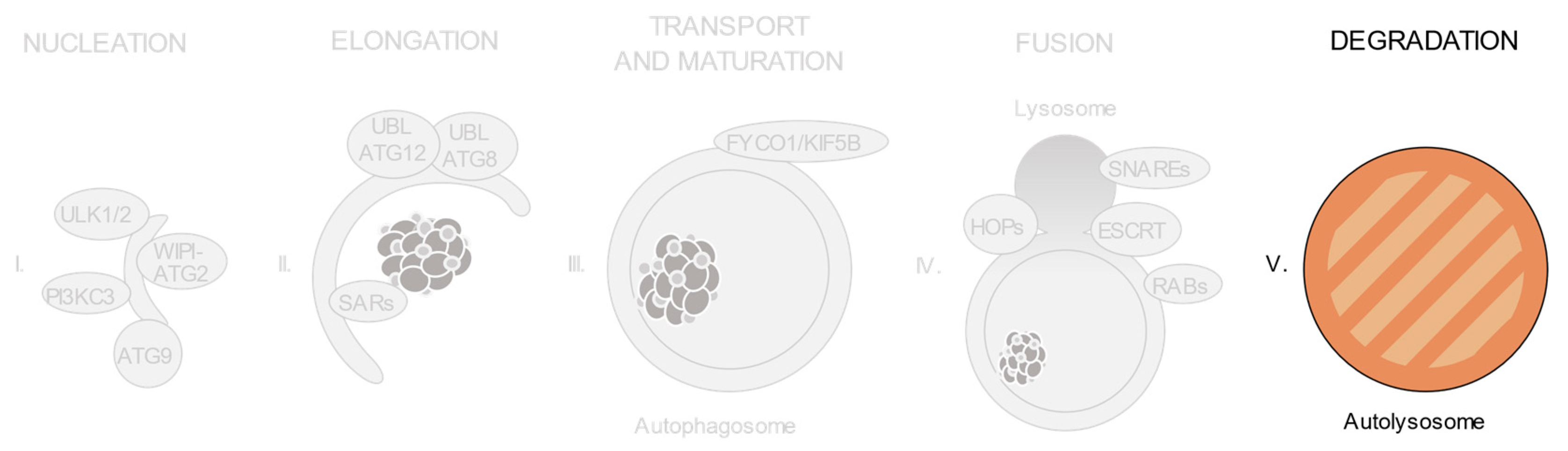
| Gene | Disease | dbSNP rsID |
|---|---|---|
| ATG13 | Selective immunoglobulin A deficiency | rs4565870 |
| ATG13 | Breast cancer | rs10838611 |
| FIP200 | Hypertension | rs1129660 |
| ULK1 | Crohn’s disease | rs12303764; rs10902469; rs7488085 |
| ULK1 | Tuberculosis | rs12297124; rs7138581; rs9481 |
| ULK1 | Ankylosing spondylitis | rs9652059 |
| ULK2 | Asparaginase-associated pancreatitis | rs281366 |
| Gene | Disease | dbSNP rsID |
|---|---|---|
| AMBRA1 | Schizophrenia | rs11819869; rs12574668; rs61882743; rs7112229; rs7130141 |
| AMBRA1 | Autism | rs3802890 |
| AMBRA1 | Selective immunoglobulin A deficiency | rs4565870 |
| ATG14L | Testicular germ cell tumor | rs1009647 |
| BECN1 | Machado–Joseph disease | rs60221525 |
| BECN1 | Diabetes | rs10512488 |
| UVRAG | Multiple sclerosis treatment | rs80191572 |
| UVRAG | Rheumatoid arthritis | rs7111334 |
| UVRAG | Non-segmental vitiligo | rs1458836; rs7933235 |
| VPS34 | Pancreatic cancer | rs76692125 |
| VPS34 | Esophageal squamous cell carcinoma | rs52911 |
| VPS34 | Gastric cancer | rs2162440 |
| VPS34 | Schizophrenia | rs3813065 |
| VPS34 | Systemic lupus erythematosus | rs3813065 |
| Gene | Disease | dbSNP rsID |
|---|---|---|
| ATG2A | Granuloma formation in Crohn’s disease | rs17146441 |
| ATG2A | Hyperuricemia | rs188780113 |
| ATG2B | Non-muscle invasive bladder cancer | rs3759601 |
| ATG2B | Head and neck squamous cell carcinoma | rs3759601 |
| ATG9B | Coronary artery disease | rs2373929; rs7830 |
| ALFY | Microcephaly | rs1553924800 |
| ALFY | Malignant neoplasm of oropharynx | rs6847067 |
| DFCP1 | Tuberculosis | rs2333021 |
| WIPI2 | Osteoporosis | rs4720530 |
| WIPI3 | Neurodevelopmental disorder | rs786205510; rs1555647262 |
| WIPI4 | Rett syndrome | rs886041382; rs886041693 |
| WIPI4 | Neurodegeneration with brain iron accumulation | rs886041382 |
| WIPI4 | Early-onset epileptic encephalopathy | rs1064793294 |
| WIPI4 | β-propeller protein-associated neurodegeneration (BPAN) | rs387907330 |
| Gene | Disease | dbSNP rsID |
|---|---|---|
| ATG10 | Breast cancer | rs10514231; rs1864182; rs7707921 |
| ATG10 | Paget’s disease of the bone | rs1864183 |
| ATG10 | Vogt–Koyanagi–Harada syndrome | rs4703863 |
| ATG10 | Lung cancer | rs10514231; rs1864182; rs1864183; rs10036653 |
| ATG10 | Melanoma | rs1864182 |
| ATG10 | Brain metastasis | rs10036653 |
| ATG10 | Head and neck squamous cell carcinoma | rs10514231; rs1864183; rs4703533 |
| ATG10 | Pneumoconiosis | rs1864182 |
| ATG10 | Hepatocellular carcinoma | rs10514231; rs1864183 |
| ATG12 | Brain metastasis | rs26532 |
| ATG12 | Pneumoconiosis | rs26538 |
| ATG12 | Lung cancer | rs26538 |
| ATG12 | Head and neck squamous cell carcinoma | rs26537 |
| ATG12 | Hepatocellular carcinoma | rs26537 |
| ATG16L1 | Crohn’s disease | rs2241880 |
| ATG16L1 | Palmoplantar pustulosis | rs2241879; rs2241880; rs7587633 |
| ATG16L1 | Psoriasis vulgaris | rs10210302; rs12994971; rs13005285; rs2241879; rs2241880 |
| ATG16L1 | Cell-derived thyroid carcinoma | rs2241880 |
| ATG16L1 | Colorectal cancer | rs2241880 |
| ATG16L1 | Paget’s disease of the bone | rs2241880 |
| ATG16L1 | Prostate cancer | rs78835907 |
| ATG16L1 | Gastric cancer | rs2241880 |
| ATG16L1 | Melanoma | rs2241880 |
| ATG16L1 | Brain metastasis | rs2241880 |
| ATG16L1 | Head and neck squamous cell carcinoma | rs2241880; rs4663402 |
| ATG16L1 | Lung cancer | rs2241880 |
| ATG16L1 | Hepatocellular carcinoma | rs4663402 |
| ATG16L1 | Helicobacter pylori infection | rs2241880 |
| ATG4A | Cervical Cancer | rs5973822; rs4036579; rs807181; rs807182; rs807183 |
| ATG4A | Lung cancer | rs807185 |
| ATG4A | Granuloma formation in Crohn’s disease | rs5973822 |
| ATG4A | Clear cell renal cell carcinoma | rs7880351 |
| ATG4B | Obesity | rs7601000 |
| ATG4B | Atherosclerosis | rs139302128 |
| ATG4C | Clear cell renal cell carcinoma | rs6670694; rs6683832 |
| ATG4C | Kashin–Beck disease | rs11208030; rs4409690; rs12097658; rs6587988 |
| ATG4D | Granuloma formation in Crohn’s disease | rs7248036; rs2304165 |
| ATG5 | Systemic lupus erythematosus | rs6937876; rs3827644; rs573775; rs548234 |
| ATG5 | Asthma | rs12212740; rs11751513; rs12201458; rs2299863; rs510432 |
| ATG5 | Parkinson’s disease | rs510432 |
| ATG5 | Systemic sclerosis | rs3827644; rs9373839 |
| ATG5 | Non-medullary thyroid cancer | rs2245214 |
| ATG5 | Neuromyelitis optica | rs548234; rs6937876 |
| ATG5 | Paget’s disease of the bone | rs2245214 |
| ATG5 | Behçet’s disease | rs573775 |
| ATG5 | Spinocerebellar ataxia | rs1131692265 |
| ATG5 | Crohn’s disease | rs510432; rs9373839 |
| ATG5 | Multiple myeloma | rs9372120 |
| ATG5 | Melanoma | rs2245214; rs510432 |
| ATG5 | Sepsis | rs506027; rs510432 |
| ATG5 | Pneumoconiosis | rs510432 |
| ATG5 | Esophageal squamous cell carcinoma | rs1322178; rs3804329; rs671116 |
| ATG5 | Lung cancer | rs510432; rs688810; rs2245214 |
| ATG5 | Cerebral palsy | rs6568431 |
| ATG5 | Breast cancer | rs473543 |
| ATG5 | Clear cell renal cell carcinoma | rs490010 |
| ATG5 | Chronic Q fever | rs2245214 |
| ATG5 | Aplastic anaemia | rs473543; rs510432; rs573775; rs803360 |
| ATG5 | HBV infection | rs510432; rs6568431; rs548234 |
| ATG5 | Hepatocellular carcinoma | rs17067724 |
| ATG7 | Systemic lupus erythematosus | rs11706903; rs2736340 |
| ATG7 | Breast cancer | rs8154 |
| ATG7 | Ischemic stroke | rs2594966; rs2594973; rs4684776 |
| ATG7 | Lung cancer | rs8154 |
| ATG7 | Clear cell renal cell carcinoma | rs2606736; rs6442260 |
| ATG7 | Cerebral palsy | rs1470612; rs2594972 |
| ATG7 | Huntington’s disease | rs36117895 |
| IRGM | Crohn’s disease | rs10065172; rs1000113; rs10065172; rs11747270; rs11749391; rs180802994; rs4958843; rs4958847; rs72553867; rs7714584, rs9637876 |
| IRGM | Systemic lupus erythematosus | rs10065172; rs13361189 |
| IRGM | Ulcerative colitis | rs1000113; rs11747270; rs11749391; rs180802994; rs4958847 |
| IRGM | Tuberculosis | rs10051924; rs12654043; rs4958843; rs72553867 |
| IRGM | Celiac disease | rs10065172 |
| IRGM | Inflammatory bowel diseases | rs10065172; rs4958847 |
| IRGM | Ankylosing spondylitis | rs10065172; rs11749391 |
| IRGM | Arthritis | rs11747270; rs4958847 |
| IRGM | Chronic periodontitis | rs11747270 |
| IRGM | Asthma | rs11747270 |
| IRGM | Multiple sclerosis | rs11747270 |
| IRGM | Cholangitis | rs11749391 |
| IRGM | Psoriasis vulgaris | rs11749391 |
| IRGM | Pathologic fistula | rs4958847 |
| IRGM | Malignant neoplasm of stomach | rs4958847 |
| IRGM | Non-alcoholic fatty liver disease | rs4958847 |
| MAP1LC3A | Chronic Q fever | rs1040747 |
| MAP1LC3A | Coronary artery disease | rs2424994 |
| MAP1LC3B | Myopia | rs1054521 |
| MAP1LC3B | Systemic lupus erythematosus | rs933717 |
| Gene | Disease | dbSNP rsID |
|---|---|---|
| ATGL | Neutral lipid storage disease with myopathy | rs121918259 |
| ATL3 | Hereditary sensory autonomic neuropathy | rs587777108 |
| BNIP3 | Major depressive disorder | rs9419139 |
| BNIP3L | Schizophrenia | rs1042992; rs73219805; rs73219806 |
| BNIP3L | Cognitive decline | rs77609452 |
| HSL | Lipodystrophy | rs766817317; rs587777699 |
| HTT | Huntington’s disease | rs1210554604; rs10015979; rs110501; rs11731237; rs2071655; rs2269499; rs2285086; rs2298969; rs2471347; rs362272; rs363066; rs363092; rs363096; rs3856973; rs6855981; rs82333; rs916171; rs118005095; rs13102260 |
| NBR1 | Brooke–Spiegler syndrome | rs202122812 |
| NCOA4 | Prostate cancer | rs10740051; rs10761581 |
| NCOA4 | Papillary thyroid carcinoma | rs782237788 |
| NDP52 | Crohn’s disease | rs2303015 |
| NIPSNAP1 | Breast cancer | rs183421746 |
| NUFIP1 | Asthma | rs114280567 |
| OPTN | Amyotrophic lateral sclerosis | rs267606928; rs267606929 |
| OPTN | Primary open-angle glaucoma | rs28939688 |
| OPTN | Paget’s disease of the bone | rs1561570; rs2234968 |
| PHB2 | Bisphosphonate-associated osteonecrosis of the jaw | rs11064477 |
| FAM134B | Hereditary sensory autonomic neuropathy | rs137852737; rs137852738; rs137852739; rs886037748 |
| RTN3 | Malaria | rs542998 |
| SQSTM1 | Frontotemporal dementia | rs776749939; rs772889843; rs1355424687 |
| SQSTM1 | Paget’s disease of the bone | rs796051869; rs104893941 |
| SQSTM1 | Amyotrophic lateral sclerosis | rs796052214; rs796051870; rs796051870 |
| SQSTM1 | Neurodegeneration | rs886039780 |
| SQSTM1 | Parkinson’s disease | rs200396166 |
| SQSTM1 | Atypical apraxia of speech | rs796052214 |
| SQSTM1 | Sporadic inclusion body myositis | rs11548633 |
| STBD1 | Parkinson’s disease | rs6812193 |
| TAX1BP1 | Head and neck carcinoma | rs11540483 |
| TAX1BP1 | Hypospadias | rs10214930 |
| TOLLIP | Leishmaniasis | rs3750920; rs5743899 |
| TOLLIP | Leprosy | rs3793964; rs3750920 |
| TOLLIP | Malaria | rs3750920 |
| TOLLIP | Tuberculosis | rs3750920; rs5743867 |
| TOLLIP | Sepsis | rs5743867 |
| TOLLIP | Idiopathic pulmonary fibrosis | rs5743890; rs111521887; rs3750920 |
| TOLLIP | Fibrotic idiopathic interstitial pneumonia | rs3168046; rs3750920; rs3793964; rs3829223; rs5744034 |
| Gene | Disease | dbSNP rsID |
|---|---|---|
| BIRC6 | Glaucoma | rs2754511 |
| C9orf72 | Amyotrophic lateral sclerosis | rs3849943; rs774359; rs3849942 |
| C9orf72 | Familial frontotemporal dementia with amyotrophic lateral sclerosis | rs71492753 |
| CHMP2B | Neuroblastoma | rs63750355; rs63750653 |
| CHMP2B | Frontotemporal dementia | rs78268395 |
| CHMP2B | Amyotrophic lateral sclerosis | rs281864934 |
| CHMP4B | Bilateral cataracts | rs118203966 |
| CHMP4B | Diabetes mellitus, non-insulin-dependent | rs7274168 |
| CHMP4B | Dysphagia | rs2747539 |
| EPG5 | Alzheimer’s disease | rs9963463; rs11082498 |
| EPG5 | Depressive disorders | rs58682566 |
| EPG5 | Vici syndrome | rs1470797555; rs1555673917; rs1568107449; rs1568112516; rs1568112543; rs1568118775; rs1568133724; rs1568133760; rs201757275; rs587776940; rs587776941; rs587776942; rs762639913; rs767638289; rs780889226; rs863225064; rs866435487; rs961245497; rs863225064 |
| EPG5 | Cataract | rs201757275 |
| FYCO1 | Cataract | rs387906963; rs387906964; rs387906965 |
| HGS | Age-related macular degeneration | rs8070488 |
| KIF5B | Bipolar disorder | rs1775715 |
| PLEKHM1 | Osteopetrosis | rs786205055 |
| PLEKHM1 | Parkinson’s disease | rs11012 |
| PLEKHM1 | Alopecia | rs144733372 |
| PLEKHM1 | Unipolar depression | rs144733372 |
| PLEKHM1 | Major depressive disorder | rs144733372 |
| PLEKHM1 | Ovarian cancer | rs1879586; rs2077606; rs17631303 |
| RAB33B | Smith–McCort dysplasia | rs1085307129; rs886044716; rs1085307131; rs1085307128; rs587776958 |
| RAB7 | Charcot–Marie–Tooth disease type 2B | rs121909080; rs121909078; rs121909079; rs121909081 |
| SNAP29 | Cednik syndrome | rs387907363; rs869312906 |
| SPG11 | Spastic paraplegia | rs1085307097; rs118203963; rs140385286; rs1555447432; rs141848292; rs312262720; rs312262721; rs312262722; rs312262737; rs312262749; rs312262752; rs312262764; rs312262779; rs371334506; rs747220413; rs764647588; rs765477482; rs767798272 |
| STX17 | Alopecia | rs10760706 |
| STX7 | Neuronal heterotopia | rs864309676 |
| VAMP8 | Cerebrovascular accident | rs1010 |
| VAMP8 | Tuberculosis | rs1010 |
| VAMP8 | Coronary artery disease | rs1010 |
| VAMP8 | Prostate cancer | rs10187424; rs3731827 |
| VPS33A | MPS-like disorder | rs767748011 |
| VPS41 | Major depressive disorder | rs10274968 |
| YKT6 | Diabetes | rs2908282 |
| YKT6 | Birth weight and subsequent risk factors | rs138715366 |
| ZFYVE26 | Spastic paraplegia | rs1049504575; rs1057518016; rs1214483973; rs1555394376; rs200832994; rs558285072; rs767164213; rs768176054; rs769329153; rs774809466; rs941230062; rs981804211 |
| ZFYVE26 | Amyotrophic lateral sclerosis | rs12891047 |
| ZFYVE26 | Breast cancer | rs200595749 |
| ZFYVE26 | Movement disorders | rs752283089; rs869312914 |
| Gene | Disease | dbSNP rsID |
|---|---|---|
| ARSB | Mucopolysaccharidosis type VI (Maroteaux–Lamy syndrome) | rs118203938; rs118203939; rs118203940; rs431905493; rs431905495; rs431905496; rs118203942; rs118203944; rs118203943 |
| CLN3 | Neuronal ceroid lipofuscinosis type 3 | rs121434286; rs267606737; rs386833720; rs786201028; rs121434286 |
| CLN6 | Neuronal ceroid lipofuscinosis type 6 | rs104894483; rs104894486; rs121908079; rs121908080; rs397515352; rs774543080; rs786205065; rs786205066; rs786205067; rs104894484 |
| CLN6 | Adult neuronal ceroid lipofuscinosis | rs154774633; rs154774634; rs154774635; rs154774636 |
| CLN8 | Neuronal ceroid lipofuscinosis type 8 | rs104894060; rs137852883; rs28940569 |
| CLN8 | Northern epilepsy syndrome | rs104894064 |
| CTNS | Cystinosis | rs375952052 |
| CTSA | Galactosialidosis | rs137854540; rs137854544; rs137854546; rs137854547; rs137854548; rs137854549; rs786200859; rs875989777; rs137854544; rs137854543 |
| CTSD | Neuronal ceroid lipofuscinosis type 10 | rs786205105; rs797045137; rs797045138; rs121912789; rs121912790 |
| CTSF | Neuronal ceroid lipofuscinosis type 13 | rs753084727; rs797045136; rs143889283; rs397514731 |
| DNAJC5 | Neuronal ceroid lipofuscinosis, Parry type, | rs587776892; rs387907043 |
| FUCA1 | Fucosidosis | rs118204450; rs80358195; rs80358196; rs80358197; rs80358198 |
| GAA | Glycogen storage disease type II (Pompe disease) | rs1057516581; rs12450199; rs140826989; rs121907940; rs121907941; rs1393386120; rs1414146587; rs121907942; rs1344266804; rs121907943; rs121907944; rs1221948995; rs1245412108; rs121907938; rs121907945; rs121907936; rs1800309; rs121907937; rs1800307; rs147804176; rs1555600061; rs1555601773; rs1800312; rs200856561; rs1555601773; rs1800312; rs200856561; rs369531647; rs1057516277; rs886043343; rs892129065; rs28940868;rs1057516215; rs1055945806; |
| GAA | Friedreich ataxia | rs1245992455 |
| GAA | Hypochondrogenesis | rs1289257741 |
| GALNS | Mucopolysaccharidosis IVA (Morchio A syndrome) | rs1028668536; rs118204438; rs118204449; rs786205899; rs118204435; rs118204441; rs118204442; rs118204446; rs118204447; rs118204448; rs267606838 |
| GBA | Parkinson’s disease | rs75548401 |
| GBA | Gaucher disease | rs421016 |
| GLA | Fabry disease | rs104894828; rs104894834; rs104894845; rs28935197; rs869312142 |
| GLB1 | GM1 gangliosidosis | rs192732174; rs376663785; rs587776524; rs794727165; rs794729217; rs781658798; rs778423653; rs778700089; rs879050821; rs72555361; rs72555364; rs72555368; rs72555370; rs72555390; rs72555393; rs794729217; rs72555392; rs72555362; rs1214295886; rs1553606128; rs1553610382; rs1553610553; rs1553612189; rs1559401428; rs192732174; rs189115557; |
| GLB1 | Mucopolysaccharidosis IVB (Morchio B syndrome) | rs72555363; rs1553606128; rs1553610382; rs1553610553; rs1553612220; rs189115557; rs192732174; rs794729217; rs794727165; rs778700089; rs778423653 |
| GLB1 | Neuraminidase 1 deficiency | rs1356418704 |
| GLB1 | Respiratory tract diseases | rs9828592 |
| GLB1 | Asthma | rs79337446 |
| GNS | Mucopolysaccharidosis type IIID (Sanfilippo syndrome) | rs119461974; rs119461975; rs483352898; rs483352899; rs483352900 |
| GRN | Presenile dementia | rs373885474 |
| GRN | Frontotemporal lobar degeneration | rs606231220; rs63749801; rs63750077; rs63751006; rs63750331; rs63751294; rs63751243 |
| GUSB | Mucopolysaccharidosis type VII (Sly syndrome) | rs121918179; rs121918181; rs121918185; rs377519272; rs786200863; rs121918180; rs121918173; rs121918174; rs121918175; rs121918176; rs121918177; rs121918178; rs121918182; rs121918183; rs121918184; rs121918172 |
| HGSNAT | Mucopolysaccharidosis type IIIC (Sanfilippo syndrome type C) | rs121908282; rs121908283; rs121908284; rs121908285; rs121908286; rs193066451; rs483352896; rs753355844; rs754875934; rs764206492; rs797045120 |
| HYAL1 | Mucopolysaccharidosis IX | rs104893743 |
| IDS | Mucopolysaccharidosis type II (Hunter syndrome) | rs113993946; rs113993947; rs199422230; rs483352904; rs483352905; rs797044671; rs869025304; rs869025305; rs869025306; rs869025307; rs869025308; rs104894856; rs104894861; rs199422228; rs199422229; rs199422231 |
| IDUA | Mucopolysaccharidosis type I (Hurler and Scheie syndrome) | rs121965025; rs121965033; rs199801029; rs387906504; rs398123258; rs762411583; rs786200915; rs869025584; rs121965021; rs121965026; rs121965027; rs121965031; rs121965023; rs121965019; rs121965021; rs121965030; rs764196171; rs121965019; rs121965033; rs121965024; |
| LAMP2 | Danon disease | rs104894857; rs104894858; rs1060502302; rs137852527; rs727503118; rs727503119; rs727503120; rs727504742 |
| MAN2B1 | Alpha-mannosidosis | rs121434331; rs121434332; rs775200333; rs80338677; rs80338678; rs80338679; rs80338680; rs80338681 |
| MANBA | Beta-mannosidosis | rs121434334; rs121434335; rs121434336 |
| MFSD8 | Neuronal ceroid lipofuscinosis type 7 | rs11820397; rs587778809; rs724159971; rs727502801; rs118203975; rs118203976; rs140948465; rs267607235; rs749704755 |
| MFSD8 | Late-infantile neuronal ceroid lipofuscinosis | rs200319160 |
| NAGLU | Mucopolysaccharidosis type IIIB (Sanfilippo syndrome type B) | rs104894591; rs104894592; rs104894597; rs104894598; rs118204025; rs746006696; rs886039894; rs886039895; rs118204024; rs104894590; rs104894593; rs104894594; rs104894595; rs104894597; rs104894598; rs753520553; rs796052122; rs104894601 |
| NPC1 | Niemann–Pick disease, type C | rs1055204017; rs1057518711; rs1474434210; rs753768576; rs139751448; rs143124972; rs28942104; rs756815030; rs758231839; rs886042270; rs80358257; rs80358254; rs80358259; rs150334966; rs1555634422; rs768999208; rs80358259 |
| NPC2 | Niemann–Pick disease type C | rs80358262; rs80358263; rs80358266; rs80358268; rs11694; rs80358261; rs80358264; rs104894458 |
| SGSH | Mucopolysaccharidosis type IIIA (Sanfilippo syndrome type A) | rs104894635; rs104894637; rs138504221; rs1057521801; rs374621913; rs770947426; rs777956287; rs778700037; rs104894638; rs104894640; rs104894642; rs104894643; rs104894636; rs104894641; rs138504221; rs104894635 |
| SLC17A5 | Sialic acid storage diseases (SASDs) | rs386833987; rs386833994; rs727504156; rs119491109; rs119491110; rs80338795; rs80338794 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tamargo-Gómez, I.; Fernández, Á.F.; Mariño, G. Pathogenic Single Nucleotide Polymorphisms on Autophagy-Related Genes. Int. J. Mol. Sci. 2020, 21, 8196. https://doi.org/10.3390/ijms21218196
Tamargo-Gómez I, Fernández ÁF, Mariño G. Pathogenic Single Nucleotide Polymorphisms on Autophagy-Related Genes. International Journal of Molecular Sciences. 2020; 21(21):8196. https://doi.org/10.3390/ijms21218196
Chicago/Turabian StyleTamargo-Gómez, Isaac, Álvaro F. Fernández, and Guillermo Mariño. 2020. "Pathogenic Single Nucleotide Polymorphisms on Autophagy-Related Genes" International Journal of Molecular Sciences 21, no. 21: 8196. https://doi.org/10.3390/ijms21218196
APA StyleTamargo-Gómez, I., Fernández, Á. F., & Mariño, G. (2020). Pathogenic Single Nucleotide Polymorphisms on Autophagy-Related Genes. International Journal of Molecular Sciences, 21(21), 8196. https://doi.org/10.3390/ijms21218196