Capturing and Understanding the Dynamics and Heterogeneity of Gene Expression in the Living Cell
Abstract
:1. Introduction
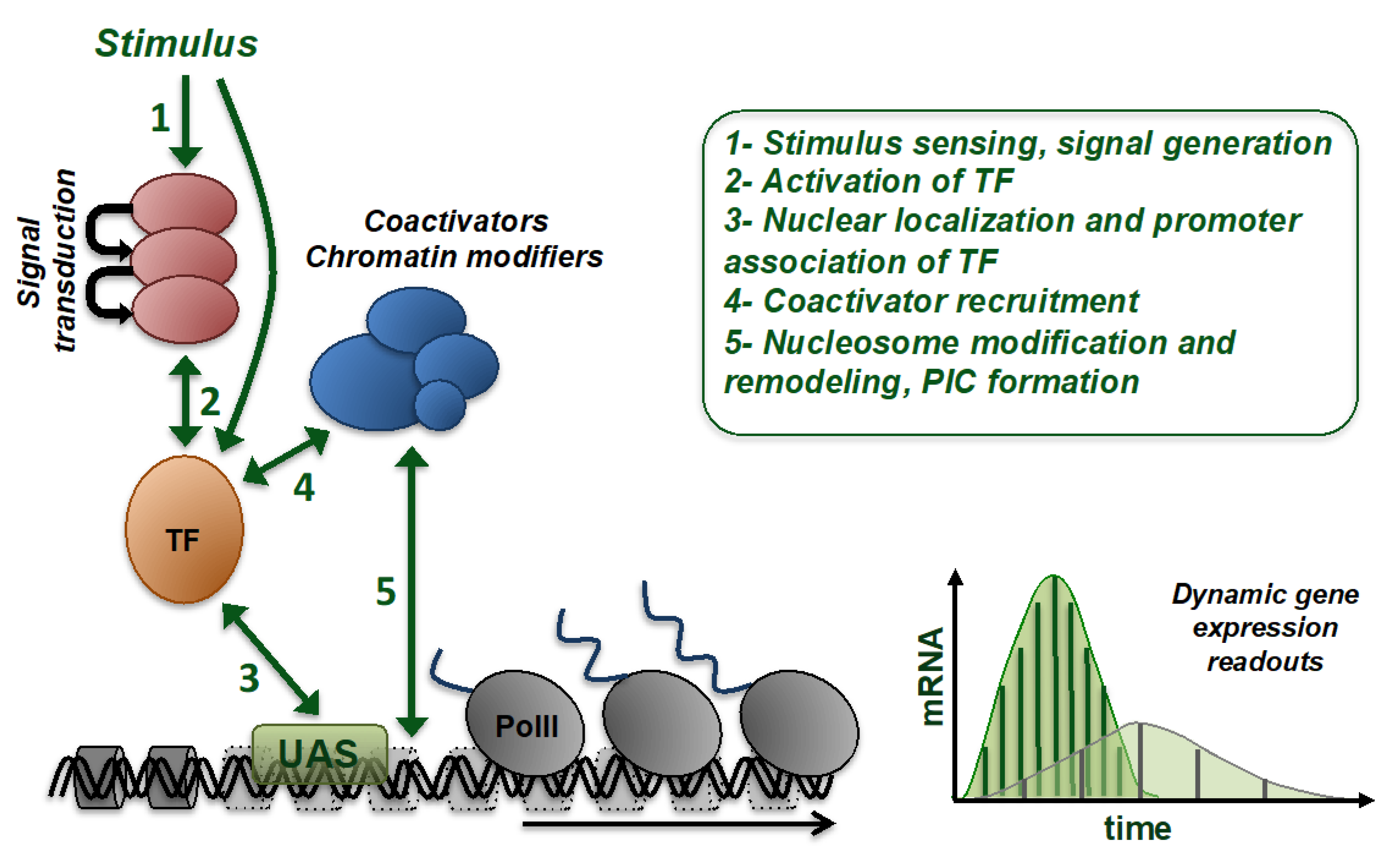
2. Technical Approaches to Capture Gene Expression In Vivo
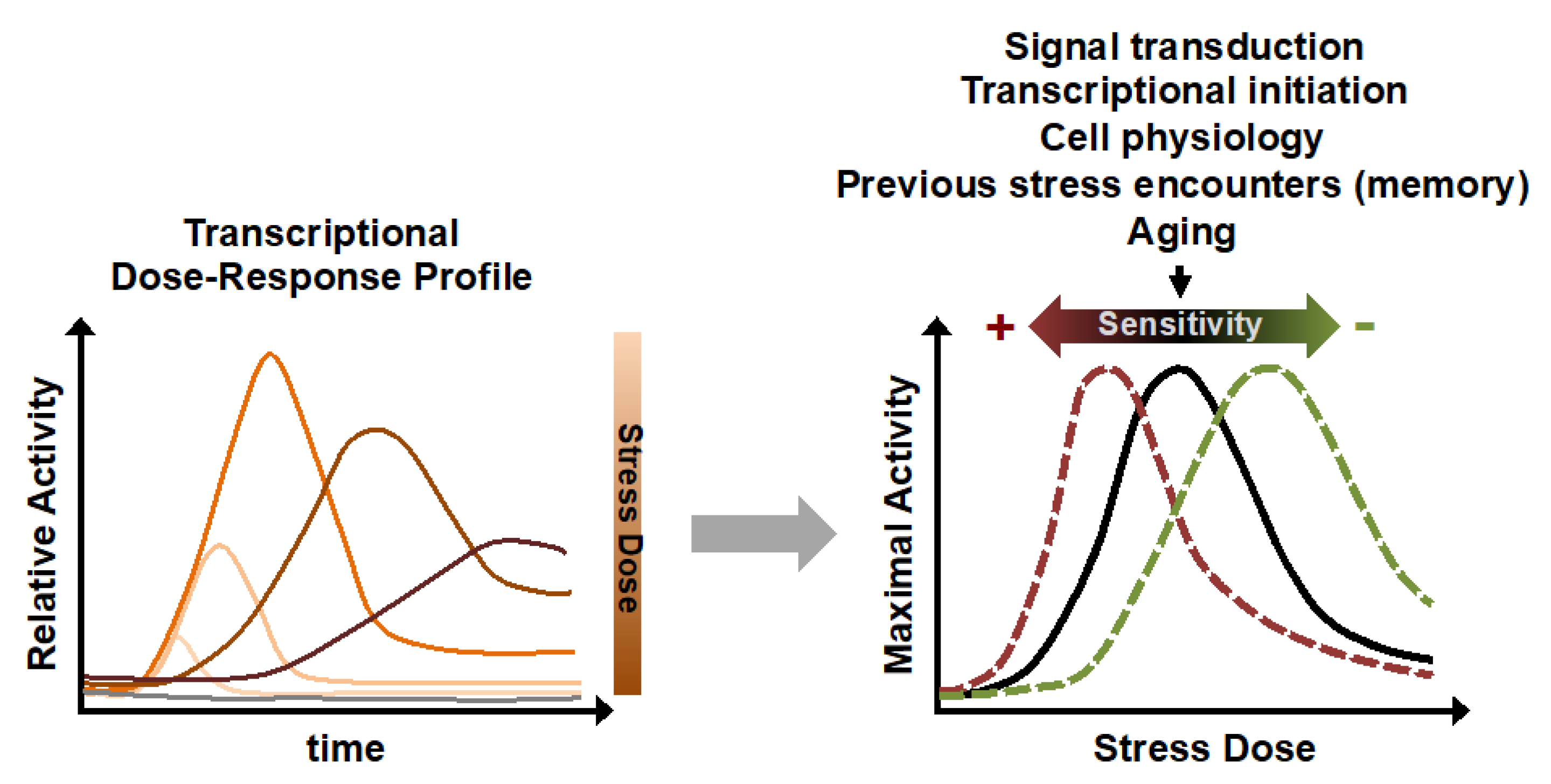
3. Activated Gene Expression: A Highly Dynamic Dose-Dependent Biological Function Sensitive to Many Physiological and Genetic Factors
4. Epigenetic Transcriptional Memory: Modulating Gene Expression Dynamics upon Repeated Stimulation
5. Gene Expression Heterogeneity in Individual Cells
6. Conclusions and Future Perspectives
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| TF | Transcription factor |
| PolII | RNA polymerase II |
| ChIP | Chromatin immunoprecipitation |
| GFP | Green fluorescent protein |
| NPC | Nuclear pore complex |
| PIC | Preinitiation complex |
| MRS | Memory recruitment sequence |
| GRS | Gene recruitment sequence |
| AD | Activation domain |
| DR | Dose–response |
| SAPK | Stress-activated protein kinase |
| CTD | C-terminal domain |
References
- Murray, J.I.; Whitfield, M.L.; Trinklein, N.D.; Myers, R.M.; Brown, P.O.; Botstein, D. Diverse and specific gene expression responses to stresses in cultured human cells. Mol. Biol. Cell 2004, 15, 2361–2374. [Google Scholar] [CrossRef]
- Gasch, A.P.; Spellman, P.T.; Kao, C.M.; Carmel-Harel, O.; Eisen, M.B.; Storz, G.; Botstein, D.; Brown, P.O. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 2000, 11, 4241–4257. [Google Scholar] [CrossRef]
- Ben-Tabou de-Leon, S.; Davidson, E.H. Gene regulation: Gene control network in development. Annu. Rev. Biophys. Biomol. Struct. 2007, 36, 191–212. [Google Scholar] [CrossRef] [Green Version]
- Lenstra, T.L.; Rodriguez, J.; Chen, H.; Larson, D.R. Transcription dynamics in living cells. Annu. Rev. Biophys. 2016, 45, 25–47. [Google Scholar] [CrossRef]
- Coulon, A.; Chow, C.C.; Singer, R.H.; Larson, D.R. Eukaryotic transcriptional dynamics: From single molecules to cell populations. Nat. Rev. Genet. 2013, 14, 572–584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yosef, N.; Regev, A. Impulse control: Temporal dynamics in gene transcription. Cell 2011, 144, 886–896. [Google Scholar] [CrossRef] [Green Version]
- Purvis, J.E.; Lahav, G. Encoding and decoding cellular information through signaling dynamics. Cell 2013, 152, 945–956. [Google Scholar] [CrossRef] [Green Version]
- Weake, V.M.; Workman, J.L. Inducible gene expression: Diverse regulatory mechanisms. Nat. Rev. Genet. 2010, 11, 426–437. [Google Scholar] [CrossRef]
- de Nadal, E.; Ammerer, G.; Posas, F. Controlling gene expression in response to stress. Nat. Rev. Genet. 2011, 12, 833–845. [Google Scholar] [CrossRef]
- Vihervaara, A.; Duarte, F.M.; Lis, J.T. Molecular mechanisms driving transcriptional stress responses. Nat. Rev. Genet. 2018, 19, 385–397. [Google Scholar] [CrossRef]
- Pérez-Ortín, J.E.; Alepuz, P.; Chávez, S.; Choder, M. Eukaryotic mRNA decay: Methodologies, pathways, and links to other stages of gene expression. J. Mol. Biol. 2013, 425, 3750–3775. [Google Scholar] [CrossRef]
- Aparicio, O.; Geisberg, J.V.; Sekinger, E.; Yang, A.; Moqtaderi, Z.; Struhl, K. Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo. Curr. Protoc. Mol. Biol. 2005, 69, 21.3.1–21.3.33. [Google Scholar] [CrossRef]
- wa Maina, C.; Honkela, A.; Matarese, F.; Grote, K.; Stunnenberg, H.G.; Reid, G.; Lawrence, N.D.; Rattray, M. Inference of RNA polymerase II transcription dynamics from chromatin immunoprecipitation time course data. PLoS Comput. Biol. 2014, 10, e1003598. [Google Scholar] [CrossRef] [PubMed]
- Mason, P.B.; Struhl, K. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol. Cell 2005, 17, 831–840. [Google Scholar] [CrossRef]
- Sato, H.; Das, S.; Singer, R.H.; Vera, M. Imaging of DNA and RNA in living eukaryotic cells to reveal spatiotemporal dynamics of gene expression. Annu. Rev. Biochem. 2020, 89, 159–187. [Google Scholar] [CrossRef] [Green Version]
- Janicki, S.M.; Tsukamoto, T.; Salghetti, S.E.; Tansey, W.P.; Sachidanandam, R.; Prasanth, K.V.; Ried, T.; Shav-Tal, Y.; Bertrand, E.; Singer, R.H.; et al. From silencing to gene expression: Real-time analysis in single cells. Cell 2004, 116, 683–698. [Google Scholar] [CrossRef] [Green Version]
- Chao, J.A.; Patskovsky, Y.; Almo, S.C.; Singer, R.H. Structural basis for the coevolution of a viral RNA-protein complex. Nat. Struct. Mol. Biol. 2008, 15, 103–105. [Google Scholar] [CrossRef]
- Bertrand, E.; Chartrand, P.; Schaefer, M.; Shenoy, S.M.; Singer, R.H.; Long, R.M. Localization of ASH1 mRNA particles in living yeast. Mol. Cell 1998, 2, 437–445. [Google Scholar] [CrossRef] [Green Version]
- Campbell, P.D.; Chao, J.A.; Singer, R.H.; Marlow, F.L. Dynamic visualization of transcription and RNA subcellular localization in zebrafish. Development 2015, 142, 1368–1374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Golding, I.; Paulsson, J.; Zawilski, S.M.; Cox, E.C. Real-time kinetics of gene activity in individual bacteria. Cell 2005, 123, 1025–1036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larson, D.R.; Zenklusen, D.; Wu, B.; Chao, J.A.; Singer, R.H. Real-time observation of transcription initiation and elongation on an endogenous yeast gene. Science 2011, 332, 475–478. [Google Scholar] [CrossRef] [Green Version]
- Chubb, J.R.; Trcek, T.; Shenoy, S.M.; Singer, R.H. Transcriptional pulsing of a developmental gene. Curr. Biol. 2006, 16, 1018–1025. [Google Scholar] [CrossRef] [Green Version]
- Garcia, H.G.; Tikhonov, M.; Lin, A.; Gregor, T. Quantitative imaging of transcription in living Drosophila embryos links polymerase activity to patterning. Curr. Biol. 2013, 23, 2140–2145. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Wang, J.; Liang, Y.; Fu, Y.; Li, S.; Huang, J.; Xu, H.; Zou, W.; Chen, B. TriTag: An integrative tool to correlate chromatin dynamics and gene expression in living cells. Nucleic Acids Res. 2020. [Google Scholar] [CrossRef]
- Niedenthal, R.K.; Riles, L.; Johnston, M.; Hegemann, J.H. Green fluorescent protein as a marker for gene expression and subcellular localization in budding yeast. Yeast 1996, 12, 773–786. [Google Scholar] [CrossRef]
- Plautz, J.D.; Day, R.N.; Dailey, G.M.; Welsh, S.B.; Hall, J.C.; Halpain, S.; Kay, S.A. Green fluorescent protein and its derivatives as versatile markers for gene expression in living Drosophila melanogaster, plant and mammalian cells. Gene 1996, 173, 83–87. [Google Scholar] [CrossRef]
- Chalfie, M.; Tu, Y.; Euskirchen, G.; Ward, W.W.; Prasher, D.C. Green fluorescent protein as a marker for gene expression. Science 1994, 263, 802–805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bongaerts, R.J.M.; Hautefort, I.; Sidebotham, J.M.; Hinton, J.C.D. Green fluorescent protein as a marker for conditional gene expression in bacterial cells. In Bacterial Pathogenesis Part C: Identification, Regulation, and Function of Virulence Factors; Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2002; Volume 358, pp. 43–66. ISBN 9780121822613. [Google Scholar]
- Longo, D.; Hasty, J. Dynamics of single-cell gene expression. Mol. Syst. Biol. 2006, 2, 64. [Google Scholar] [CrossRef] [Green Version]
- Zou, F.; Bai, L. Using time-lapse fluorescence microscopy to study gene regulation. Methods 2019, 159–160, 138–145. [Google Scholar] [CrossRef]
- Han, J.; Xia, A.; Huang, Y.; Ni, L.; Chen, W.; Jin, Z.; Yang, S.; Jin, F. Simultaneous visualization of multiple gene expression in single cells using an engineered multicolor reporter toolbox and approach of spectral crosstalk correction. ACS Synth. Biol. 2019, 8, 2536–2546. [Google Scholar] [CrossRef]
- Mateus, C.; Avery, S.V. Destabilized green fluorescent protein for monitoring dynamic changes in yeast gene expression with flow cytometry. Yeast 2000, 16, 1313–1323. [Google Scholar] [CrossRef]
- Li, X.; Zhao, X.; Fang, Y.; Jiang, X.; Duong, T.; Fan, C.; Huang, C.C.; Kain, S.R. Generation of destabilized green fluorescent protein as a transcription reporter. J. Biol. Chem. 1998, 273, 34970–34975. [Google Scholar] [CrossRef] [Green Version]
- Andersen, J.B.; Sternberg, C.; Poulsen, L.K.; Bjorn, S.P.; Givskov, M.; Molin, S. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl. Environ. Microbiol. 1998, 64, 2240–2246. [Google Scholar] [CrossRef] [Green Version]
- He, L.; Binari, R.; Huang, J.; Falo-Sanjuan, J.; Perrimon, N. In vivo study of gene expression with an enhanced dual-color fluorescent transcriptional timer. eLife 2019, 8. [Google Scholar] [CrossRef]
- Allen, M.S.; Wilgus, J.R.; Chewning, C.S.; Sayler, G.S.; Simpson, M.L. A destabilized bacterial luciferase for dynamic gene expression studies. Syst. Synth. Biol. 2007, 1, 3–9. [Google Scholar] [CrossRef] [Green Version]
- Yasunaga, M.; Murotomi, K.; Abe, H.; Yamazaki, T.; Nishii, S.; Ohbayashi, T.; Oshimura, M.; Noguchi, T.; Niwa, K.; Ohmiya, Y.; et al. Highly sensitive luciferase reporter assay using a potent destabilization sequence of calpain 3. J. Biotechnol. 2015, 194, 115–123. [Google Scholar] [CrossRef]
- Leclerc, G.M.; Boockfor, F.R.; Faught, W.J.; Frawley, L.S. Development of a destabilized firefly luciferase enzyme for measurement of gene expression. BioTechniques 2000, 29, 590–591, 594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rienzo, A.; Pascual-Ahuir, A.; Proft, M. The use of a real-time luciferase assay to quantify gene expression dynamics in the living yeast cell. Yeast 2012, 29, 219–231. [Google Scholar] [CrossRef] [Green Version]
- Robertson, J.B.; Stowers, C.C.; Boczko, E.; Johnson, C.H. Real-time luminescence monitoring of cell-cycle and respiratory oscillations in yeast. Proc. Natl. Acad. Sci. USA 2008, 105, 17988–17993. [Google Scholar] [CrossRef] [Green Version]
- Deng, L.; Sugiura, R.; Takeuchi, M.; Suzuki, M.; Ebina, H.; Takami, T.; Koike, A.; Iba, S.; Kuno, T. Real-time monitoring of calcineurin activity in living cells: Evidence for two distinct Ca2+-dependent pathways in fission yeast. Mol. Biol. Cell 2006, 17, 4790–4800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazo-Vargas, A.; Park, H.; Aydin, M.; Buchler, N.E. Measuring fast gene dynamics in single cells with time-lapse luminescence microscopy. Mol. Biol. Cell 2014, 25, 3699–3708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Tjian, R. Visualizing transcription factor dynamics in living cells. J. Cell Biol. 2018, 217, 1181–1191. [Google Scholar] [CrossRef] [Green Version]
- Jin, X.; Hapsari, N.D.; Lee, S.; Jo, K. DNA binding fluorescent proteins as single-molecule probes. Analyst 2020, 145, 4079–4095. [Google Scholar] [CrossRef] [PubMed]
- Dolz-Edo, L.; Rienzo, A.; Poveda-Huertes, D.; Pascual-Ahuir, A.; Proft, M. Deciphering dynamic dose responses of natural promoters and single cis elements upon osmotic and oxidative stress in yeast. Mol. Cell. Biol. 2013, 33, 2228–2240. [Google Scholar] [CrossRef] [Green Version]
- Pascual-Ahuir, A.; González-Cantó, E.; Juyoux, P.; Pable, J.; Poveda-Huertes, D.; Saiz-Balbastre, S.; Squeo, S.; Ureña-Marco, A.; Vanacloig-Pedros, E.; Zaragoza-Infante, L.; et al. Dose dependent gene expression is dynamically modulated by the history, physiology and age of yeast cells. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 1862, 457–471. [Google Scholar] [CrossRef]
- Pelet, S.; Rudolf, F.; Nadal-Ribelles, M.; de Nadal, E.; Posas, F.; Peter, M. Transient activation of the HOG MAPK pathway regulates bimodal gene expression. Science 2011, 332, 732–735. [Google Scholar] [CrossRef] [Green Version]
- Paliwal, S.; Iglesias, P.A.; Campbell, K.; Hilioti, Z.; Groisman, A.; Levchenko, A. MAPK-mediated bimodal gene expression and adaptive gradient sensing in yeast. Nature 2007, 446, 46–51. [Google Scholar] [CrossRef]
- Zhang, Q.; Yoon, Y.; Yu, Y.; Parnell, E.J.; Garay, J.A.R.; Mwangi, M.M.; Cross, F.R.; Stillman, D.J.; Bai, L. Stochastic expression and epigenetic memory at the yeast HO promoter. Proc. Natl. Acad. Sci. USA 2013, 110, 14012–14017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gutin, J.; Joseph-Strauss, D.; Sadeh, A.; Shalom, E.; Friedman, N. Genetic screen of the yeast environmental stress response dynamics uncovers distinct regulatory phases. Mol. Syst. Biol. 2019, 15, e8939. [Google Scholar] [CrossRef]
- Rajkumar, A.S.; Liu, G.; Bergenholm, D.; Arsovska, D.; Kristensen, M.; Nielsen, J.; Jensen, M.K.; Keasling, J.D. Engineering of synthetic, stress-responsive yeast promoters. Nucleic Acids Res. 2016, 44, e136. [Google Scholar] [CrossRef]
- Duveau, F.; Yuan, D.C.; Metzger, B.P.H.; Hodgins-Davis, A.; Wittkopp, P.J. Effects of mutation and selection on plasticity of a promoter activity in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2017, 114, E11218–E11227. [Google Scholar] [CrossRef] [Green Version]
- Redden, H.; Morse, N.; Alper, H.S. The synthetic biology toolbox for tuning gene expression in yeast. FEMS Yeast Res. 2015, 15, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Brouwer, I.; Lenstra, T.L. Visualizing transcription: Key to understanding gene expression dynamics. Curr. Opin. Chem. Biol. 2019, 51, 122–129. [Google Scholar] [CrossRef]
- Rodriguez, J.; Larson, D.R. Transcription in living cells: Molecular mechanisms of bursting. Annu. Rev. Biochem. 2020, 89, 189–212. [Google Scholar] [CrossRef] [Green Version]
- Tunnacliffe, E.; Chubb, J.R. What is a transcriptional burst? Trends Genet. 2020, 36, 288–297. [Google Scholar] [CrossRef] [PubMed]
- Hornung, G.; Bar-Ziv, R.; Rosin, D.; Tokuriki, N.; Tawfik, D.S.; Oren, M.; Barkai, N. Noise-mean relationship in mutated promoters. Genome Res. 2012, 22, 2409–2417. [Google Scholar] [CrossRef] [Green Version]
- Dadiani, M.; van Dijk, D.; Segal, B.; Field, Y.; Ben-Artzi, G.; Raveh-Sadka, T.; Levo, M.; Kaplow, I.; Weinberger, A.; Segal, E. Two DNA-encoded strategies for increasing expression with opposing effects on promoter dynamics and transcriptional noise. Genome Res. 2013, 23, 966–976. [Google Scholar] [CrossRef] [Green Version]
- Raveh-Sadka, T.; Levo, M.; Shabi, U.; Shany, B.; Keren, L.; Lotan-Pompan, M.; Zeevi, D.; Sharon, E.; Weinberger, A.; Segal, E. Manipulating nucleosome disfavoring sequences allows fine-tune regulation of gene expression in yeast. Nat. Genet. 2012, 44, 743–750. [Google Scholar] [CrossRef]
- van Dijk, D.; Sharon, E.; Lotan-Pompan, M.; Weinberger, A.; Segal, E.; Carey, L.B. Large-scale mapping of gene regulatory logic reveals context-dependent repression by transcriptional activators. Genome Res. 2017, 27, 87–94. [Google Scholar] [CrossRef] [Green Version]
- Mehta, G.D.; Ball, D.A.; Eriksson, P.R.; Chereji, R.V.; Clark, D.J.; McNally, J.G.; Karpova, T.S. Single-Molecule Analysis Reveals Linked Cycles of RSC Chromatin Remodeling and Ace1p Transcription Factor Binding in Yeast. Mol. Cell 2018, 72, 875–887.e9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ball, D.A.; Mehta, G.D.; Salomon-Kent, R.; Mazza, D.; Morisaki, T.; Mueller, F.; McNally, J.G.; Karpova, T.S. Single molecule tracking of Ace1p in Saccharomyces cerevisiae defines a characteristic residence time for non-specific interactions of transcription factors with chromatin. Nucleic Acids Res. 2016, 44, e160. [Google Scholar] [CrossRef]
- Karpova, T.S.; Kim, M.J.; Spriet, C.; Nalley, K.; Stasevich, T.J.; Kherrouche, Z.; Heliot, L.; McNally, J.G. Concurrent fast and slow cycling of a transcriptional activator at an endogenous promoter. Science 2008, 319, 466–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donovan, B.T.; Huynh, A.; Ball, D.A.; Patel, H.P.; Poirier, M.G.; Larson, D.R.; Ferguson, M.L.; Lenstra, T.L. Live-cell imaging reveals the interplay between transcription factors, nucleosomes, and bursting. EMBO J. 2019, 38. [Google Scholar] [CrossRef]
- Lenstra, T.L.; Coulon, A.; Chow, C.C.; Larson, D.R. Single-Molecule Imaging Reveals a Switch between Spurious and Functional ncRNA Transcription. Mol. Cell 2015, 60, 597–610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Senecal, A.; Munsky, B.; Proux, F.; Ly, N.; Braye, F.E.; Zimmer, C.; Mueller, F.; Darzacq, X. Transcription factors modulate c-Fos transcriptional bursts. Cell Rep. 2014, 8, 75–83. [Google Scholar] [CrossRef]
- Stavreva, D.A.; Garcia, D.A.; Fettweis, G.; Gudla, P.R.; Zaki, G.F.; Soni, V.; McGowan, A.; Williams, G.; Huynh, A.; Palangat, M.; et al. Transcriptional Bursting and Co-bursting Regulation by Steroid Hormone Release Pattern and Transcription Factor Mobility. Mol. Cell 2019, 75, 1161–1177.e11. [Google Scholar] [CrossRef]
- Nelson, D.E.; Ihekwaba, A.E.C.; Elliott, M.; Johnson, J.R.; Gibney, C.A.; Foreman, B.E.; Nelson, G.; See, V.; Horton, C.A.; Spiller, D.G.; et al. Oscillations in NF-kappaB signaling control the dynamics of gene expression. Science 2004, 306, 704–708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lahav, G.; Rosenfeld, N.; Sigal, A.; Geva-Zatorsky, N.; Levine, A.J.; Elowitz, M.B.; Alon, U. Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat. Genet. 2004, 36, 147–150. [Google Scholar] [CrossRef]
- Izeddin, I.; Récamier, V.; Bosanac, L.; Cissé, I.I.; Boudarene, L.; Dugast-Darzacq, C.; Proux, F.; Bénichou, O.; Voituriez, R.; Bensaude, O.; et al. Single-molecule tracking in live cells reveals distinct target-search strategies of transcription factors in the nucleus. ELife 2014, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suter, D.M.; Molina, N.; Gatfield, D.; Schneider, K.; Schibler, U.; Naef, F. Mammalian genes are transcribed with widely different bursting kinetics. Science 2011, 332, 472–474. [Google Scholar] [CrossRef] [Green Version]
- Keller, S.H.; Jena, S.G.; Yamazaki, Y.; Lim, B. Regulation of spatiotemporal limits of developmental gene expression via enhancer grammar. Proc. Natl. Acad. Sci. USA 2020, 117, 15096–15103. [Google Scholar] [CrossRef] [PubMed]
- Ochiai, H.; Hayashi, T.; Umeda, M.; Yoshimura, M.; Harada, A.; Shimizu, Y.; Nakano, K.; Saitoh, N.; Liu, Z.; Yamamoto, T.; et al. Genome-wide kinetic properties of transcriptional bursting in mouse embryonic stem cells. Sci. Adv. 2020, 6, eaaz6699. [Google Scholar] [CrossRef] [PubMed]
- Hoppe, C.; Bowles, J.R.; Minchington, T.G.; Sutcliffe, C.; Upadhyai, P.; Rattray, M.; Ashe, H.L. Modulation of the promoter activation rate dictates the transcriptional response to graded BMP signaling levels in the drosophila embryo. Dev. Cell 2020, 54, 727–741.e7. [Google Scholar] [CrossRef]
- Bakker, R.; Mani, M.; Carthew, R.W. The Wg and Dpp morphogens regulate gene expression by modulating the frequency of transcriptional bursts. Elife 2020, 9. [Google Scholar] [CrossRef]
- Klemm, S.L.; Shipony, Z.; Greenleaf, W.J. Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet. 2019, 20, 207–220. [Google Scholar] [CrossRef]
- Nocetti, N.; Whitehouse, I. Nucleosome repositioning underlies dynamic gene expression. Genes Dev. 2016, 30, 660–672. [Google Scholar] [CrossRef] [Green Version]
- Cosma, M.P.; Tanaka, T.; Nasmyth, K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell 1999, 97, 299–311. [Google Scholar] [CrossRef] [Green Version]
- Govind, C.K.; Yoon, S.; Qiu, H.; Govind, S.; Hinnebusch, A.G. Simultaneous recruitment of coactivators by Gcn4p stimulates multiple steps of transcription in vivo. Mol. Cell. Biol. 2005, 25, 5626–5638. [Google Scholar] [CrossRef] [Green Version]
- Biggar, S.R.; Crabtree, G.R. Continuous and widespread roles for the Swi-Snf complex in transcription. EMBO J. 1999, 18, 2254–2264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rando, O.J.; Winston, F. Chromatin and transcription in yeast. Genetics 2012, 190, 351–387. [Google Scholar] [CrossRef] [Green Version]
- Shen, C.H.; Leblanc, B.P.; Alfieri, J.A.; Clark, D.J. Remodeling of yeast CUP1 chromatin involves activator-dependent repositioning of nucleosomes over the entire gene and flanking sequences. Mol. Cell. Biol. 2001, 21, 534–547. [Google Scholar] [CrossRef] [Green Version]
- Shen, C.H.; Clark, D.J. DNA sequence plays a major role in determining nucleosome positions in yeast CUP1 chromatin. J. Biol. Chem. 2001, 276, 35209–35216. [Google Scholar] [CrossRef] [Green Version]
- Erkina, T.Y.; Zou, Y.; Freeling, S.; Vorobyev, V.I.; Erkine, A.M. Functional interplay between chromatin remodeling complexes RSC, SWI/SNF and ISWI in regulation of yeast heat shock genes. Nucleic Acids Res. 2010, 38, 1441–1449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitra, D.; Parnell, E.J.; Landon, J.W.; Yu, Y.; Stillman, D.J. SWI/SNF binding to the HO promoter requires histone acetylation and stimulates TATA-binding protein recruitment. Mol. Cell. Biol. 2006, 26, 4095–4110. [Google Scholar] [CrossRef] [Green Version]
- Sudarsanam, P.; Cao, Y.; Wu, L.; Laurent, B.C.; Winston, F. The nucleosome remodeling complex, Snf/Swi, is required for the maintenance of transcription in vivo and is partially redundant with the histone acetyltransferase, Gcn5. EMBO J. 1999, 18, 3101–3106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbaric, S.; Luckenbach, T.; Schmid, A.; Blaschke, D.; Hörz, W.; Korber, P. Redundancy of chromatin remodeling pathways for the induction of the yeast PHO5 promoter in vivo. J. Biol. Chem. 2007, 282, 27610–27621. [Google Scholar] [CrossRef] [Green Version]
- Proft, M.; Struhl, K. Hog1 kinase converts the Sko1-Cyc8-Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol. Cell 2002, 9, 1307–1317. [Google Scholar] [CrossRef]
- Lemieux, K.; Gaudreau, L. Targeting of Swi/Snf to the yeast GAL1 UAS G requires the Mediator, TAF IIs, and RNA polymerase II. EMBO J. 2004, 23, 4040–4050. [Google Scholar] [CrossRef] [Green Version]
- Rienzo, A.; Poveda-Huertes, D.; Aydin, S.; Buchler, N.E.; Pascual-Ahuir, A.; Proft, M. Different Mechanisms Confer Gradual Control and Memory at Nutrient- and Stress-Regulated Genes in Yeast. Mol. Cell. Biol. 2015, 35, 3669–3683. [Google Scholar] [CrossRef] [Green Version]
- Kundu, S.; Horn, P.J.; Peterson, C.L. SWI/SNF is required for transcriptional memory at the yeast GAL gene cluster. Genes Dev. 2007, 21, 997–1004. [Google Scholar] [CrossRef] [Green Version]
- Dhasarathy, A.; Kladde, M.P. Promoter occupancy is a major determinant of chromatin remodeling enzyme requirements. Mol. Cell. Biol. 2005, 25, 2698–2707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Acar, M.; Becskei, A.; van Oudenaarden, A. Enhancement of cellular memory by reducing stochastic transitions. Nature 2005, 435, 228–232. [Google Scholar] [CrossRef]
- Vanacloig-Pedros, E.; Lozano-Pérez, C.; Alarcón, B.; Pascual-Ahuir, A.; Proft, M. Live-cell assays reveal selectivity and sensitivity of the multidrug response in budding yeast. J. Biol. Chem. 2019, 294, 12933–12946. [Google Scholar] [CrossRef] [Green Version]
- Thakur, J.K.; Arthanari, H.; Yang, F.; Pan, S.-J.; Fan, X.; Breger, J.; Frueh, D.P.; Gulshan, K.; Li, D.K.; Mylonakis, E.; et al. A nuclear receptor-like pathway regulating multidrug resistance in fungi. Nature 2008, 452, 604–609. [Google Scholar] [CrossRef] [PubMed]
- Hao, N.; Budnik, B.A.; Gunawardena, J.; O’Shea, E.K. Tunable signal processing through modular control of transcription factor translocation. Science 2013, 339, 460–464. [Google Scholar] [CrossRef] [Green Version]
- Hansen, A.S.; O’Shea, E.K. Encoding four gene expression programs in the activation dynamics of a single transcription factor. Curr. Biol. 2016, 26, R269–R271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hao, N.; O’Shea, E.K. Signal-dependent dynamics of transcription factor translocation controls gene expression. Nat. Struct. Mol. Biol. 2011, 19, 31–39. [Google Scholar] [CrossRef]
- Babazadeh, R.; Lahtvee, P.-J.; Adiels, C.B.; Goksör, M.; Nielsen, J.B.; Hohmann, S. The yeast osmostress response is carbon source dependent. Sci. Rep. 2017, 7, 990. [Google Scholar] [CrossRef] [Green Version]
- Vanacloig-Pedros, E.; Bets-Plasencia, C.; Pascual-Ahuir, A.; Proft, M. Coordinated gene regulation in the initial phase of salt stress adaptation. J. Biol. Chem. 2015, 290, 10163–10175. [Google Scholar] [CrossRef] [Green Version]
- Nikopoulou, C.; Parekh, S.; Tessarz, P. Ageing and sources of transcriptional heterogeneity. Biol. Chem. 2019, 400, 867–878. [Google Scholar] [CrossRef] [Green Version]
- Feser, J.; Truong, D.; Das, C.; Carson, J.J.; Kieft, J.; Harkness, T.; Tyler, J.K. Elevated histone expression promotes life span extension. Mol. Cell 2010, 39, 724–735. [Google Scholar] [CrossRef] [Green Version]
- Hu, Z.; Chen, K.; Xia, Z.; Chavez, M.; Pal, S.; Seol, J.-H.; Chen, C.-C.; Li, W.; Tyler, J.K. Nucleosome loss leads to global transcriptional up-regulation and genomic instability during yeast aging. Genes Dev. 2014, 28, 396–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sen, P.; Dang, W.; Donahue, G.; Dai, J.; Dorsey, J.; Cao, X.; Liu, W.; Cao, K.; Perry, R.; Lee, J.Y.; et al. H3K36 methylation promotes longevity by enhancing transcriptional fidelity. Genes Dev. 2015, 29, 1362–1376. [Google Scholar] [CrossRef] [Green Version]
- Feser, J.; Tyler, J. Chromatin structure as a mediator of aging. FEBS Lett. 2011, 585, 2041–2048. [Google Scholar] [CrossRef] [Green Version]
- Liu, P.; Song, R.; Elison, G.L.; Peng, W.; Acar, M. Noise reduction as an emergent property of single-cell aging. Nat. Commun. 2017, 8, 680. [Google Scholar] [CrossRef]
- Işıldak, U.; Somel, M.; Thornton, J.M.; Dönertaş, H.M. Temporal changes in the gene expression heterogeneity during brain development and aging. Sci. Rep. 2020, 10, 4080. [Google Scholar] [CrossRef] [Green Version]
- Wiley, C.D.; Flynn, J.M.; Morrissey, C.; Lebofsky, R.; Shuga, J.; Dong, X.; Unger, M.A.; Vijg, J.; Melov, S.; Campisi, J. Analysis of individual cells identifies cell-to-cell variability following induction of cellular senescence. Aging Cell 2017, 16, 1043–1050. [Google Scholar] [CrossRef] [PubMed]
- Enge, M.; Arda, H.E.; Mignardi, M.; Beausang, J.; Bottino, R.; Kim, S.K.; Quake, S.R. Single-Cell Analysis of Human Pancreas Reveals Transcriptional Signatures of Aging and Somatic Mutation Patterns. Cell 2017, 171, 321–330.e14. [Google Scholar] [CrossRef] [Green Version]
- Bahar, R.; Hartmann, C.H.; Rodriguez, K.A.; Denny, A.D.; Busuttil, R.A.; Dollé, M.E.T.; Calder, R.B.; Chisholm, G.B.; Pollock, B.H.; Klein, C.A.; et al. Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature 2006, 441, 1011–1014. [Google Scholar] [CrossRef] [PubMed]
- Angelidis, I.; Simon, L.M.; Fernandez, I.E.; Strunz, M.; Mayr, C.H.; Greiffo, F.R.; Tsitsiridis, G.; Ansari, M.; Graf, E.; Strom, T.-M.; et al. An atlas of the aging lung mapped by single cell transcriptomics and deep tissue proteomics. Nat. Commun. 2019, 10, 963. [Google Scholar] [CrossRef] [Green Version]
- Koohy, H.; Bolland, D.J.; Matheson, L.S.; Schoenfelder, S.; Stellato, C.; Dimond, A.; Várnai, C.; Chovanec, P.; Chessa, T.; Denizot, J.; et al. Genome organization and chromatin analysis identify transcriptional downregulation of insulin-like growth factor signaling as a hallmark of aging in developing B cells. Genome Biol. 2018, 19, 126. [Google Scholar] [CrossRef] [PubMed]
- Bochkis, I.M.; Przybylski, D.; Chen, J.; Regev, A. Changes in nucleosome occupancy associated with metabolic alterations in aged mammalian liver. Cell Rep. 2014, 9, 996–1006. [Google Scholar] [CrossRef] [Green Version]
- Ucar, D.; Márquez, E.J.; Chung, C.-H.; Marches, R.; Rossi, R.J.; Uyar, A.; Wu, T.-C.; George, J.; Stitzel, M.L.; Palucka, A.K.; et al. The chromatin accessibility signature of human immune aging stems from CD8+ T cells. J. Exp. Med. 2017, 214, 3123–3144. [Google Scholar] [CrossRef] [PubMed]
- Cheung, P.; Vallania, F.; Warsinske, H.C.; Donato, M.; Schaffert, S.; Chang, S.E.; Dvorak, M.; Dekker, C.L.; Davis, M.M.; Utz, P.J.; et al. Single-Cell Chromatin Modification Profiling Reveals Increased Epigenetic Variations with Aging. Cell 2018, 173, 1385–1397.e14. [Google Scholar] [CrossRef] [Green Version]
- Booth, L.N.; Brunet, A. The Aging Epigenome. Mol. Cell 2016, 62, 728–744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez-Jimenez, C.P.; Eling, N.; Chen, H.-C.; Vallejos, C.A.; Kolodziejczyk, A.A.; Connor, F.; Stojic, L.; Rayner, T.F.; Stubbington, M.J.T.; Teichmann, S.A.; et al. Aging increases cell-to-cell transcriptional variability upon immune stimulation. Science 2017, 355, 1433–1436. [Google Scholar] [CrossRef] [Green Version]
- Frenk, S.; Houseley, J. Gene expression hallmarks of cellular ageing. Biogerontology 2018, 19, 547–566. [Google Scholar] [CrossRef] [Green Version]
- Riera, C.E.; Merkwirth, C.; De Magalhaes Filho, C.D.; Dillin, A. Signaling networks determining life span. Annu. Rev. Biochem. 2016, 85, 35–64. [Google Scholar] [CrossRef] [Green Version]
- Guan, Q.; Haroon, S.; Bravo, D.G.; Will, J.L.; Gasch, A.P. Cellular memory of acquired stress resistance in Saccharomyces cerevisiae. Genetics 2012, 192, 495–505. [Google Scholar] [CrossRef] [Green Version]
- Meriem, Z.B.; Khalil, Y.; Hersen, P.; Fabre, E. Hyperosmotic stress response memory is modulated by gene positioning in yeast. Cells 2019, 8, 582. [Google Scholar] [CrossRef] [Green Version]
- D’Urso, A.; Brickner, J.H. Epigenetic transcriptional memory. Curr. Genet. 2017, 63, 435–439. [Google Scholar] [CrossRef]
- Avramova, Z. Transcriptional “memory” of a stress: Transient chromatin and memory (epigenetic) marks at stress-response genes. Plant J. 2015, 83, 149–159. [Google Scholar] [CrossRef]
- Gialitakis, M.; Arampatzi, P.; Makatounakis, T.; Papamatheakis, J. Gamma interferon-dependent transcriptional memory via relocalization of a gene locus to PML nuclear bodies. Mol. Cell. Biol. 2010, 30, 2046–2056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, Y.; Liu, N.; Virlouvet, L.; Riethoven, J.-J.; Fromm, M.; Avramova, Z. Four distinct types of dehydration stress memory genes in Arabidopsis thaliana. BMC Plant Biol. 2013, 13, 229. [Google Scholar] [CrossRef] [Green Version]
- Liu, N.; Ding, Y.; Fromm, M.; Avramova, Z. Different gene-specific mechanisms determine the “revised-response” memory transcription patterns of a subset of A. thaliana dehydration stress responding genes. Nucleic Acids Res. 2014, 42, 5556–5566. [Google Scholar] [CrossRef]
- Ding, Y.; Fromm, M.; Avramova, Z. Multiple exposures to drought “train” transcriptional responses in Arabidopsis. Nat. Commun. 2012, 3, 740. [Google Scholar] [CrossRef]
- Brickner, D.G.; Cajigas, I.; Fondufe-Mittendorf, Y.; Ahmed, S.; Lee, P.-C.; Widom, J.; Brickner, J.H. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 2007, 5, e81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sood, V.; Cajigas, I.; D’Urso, A.; Light, W.H.; Brickner, J.H. Epigenetic Transcriptional Memory of GAL Genes Depends on Growth in Glucose and the Tup1 Transcription Factor in Saccharomyces cerevisiae. Genetics 2017, 206, 1895–1907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kundu, S.; Peterson, C.L. Dominant role for signal transduction in the transcriptional memory of yeast GAL genes. Mol. Cell. Biol. 2010, 30, 2330–2340. [Google Scholar] [CrossRef] [Green Version]
- Zacharioudakis, I.; Gligoris, T.; Tzamarias, D. A yeast catabolic enzyme controls transcriptional memory. Curr. Biol. 2007, 17, 2041–2046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lavy, T.; Yanagida, H.; Tawfik, D.S. Gal3 Binds Gal80 Tighter than Gal1 Indicating Adaptive Protein Changes Following Duplication. Mol. Biol. Evol. 2016, 33, 472–477. [Google Scholar] [CrossRef] [PubMed]
- Sood, V.; Brickner, J.H. Genetic and epigenetic strategies potentiate gal4 activation to enhance fitness in recently diverged yeast species. Curr. Biol. 2017, 27, 3591–3602.e3. [Google Scholar] [CrossRef] [Green Version]
- D’Urso, A.; Takahashi, Y.-H.; Xiong, B.; Marone, J.; Coukos, R.; Randise-Hinchliff, C.; Wang, J.-P.; Shilatifard, A.; Brickner, J.H. Set1/COMPASS and Mediator are repurposed to promote epigenetic transcriptional memory. Elife 2016, 5. [Google Scholar] [CrossRef]
- Light, W.H.; Freaney, J.; Sood, V.; Thompson, A.; D’Urso, A.; Horvath, C.M.; Brickner, J.H. A conserved role for human Nup98 in altering chromatin structure and promoting epigenetic transcriptional memory. PLoS Biol. 2013, 11, e1001524. [Google Scholar] [CrossRef] [Green Version]
- Light, W.H.; Brickner, D.G.; Brand, V.R.; Brickner, J.H. Interaction of a DNA zip code with the nuclear pore complex promotes H2A.Z incorporation and INO1 transcriptional memory. Mol. Cell 2010, 40, 112–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fabrizio, P.; Garvis, S.; Palladino, F. Histone methylation and memory of environmental stress. Cells 2019, 8, 339. [Google Scholar] [CrossRef] [Green Version]
- Lämke, J.; Brzezinka, K.; Altmann, S.; Bäurle, I. A hit-and-run heat shock factor governs sustained histone methylation and transcriptional stress memory. EMBO J. 2016, 35, 162–175. [Google Scholar] [CrossRef] [Green Version]
- Bevington, S.L.; Cauchy, P.; Piper, J.; Bertrand, E.; Lalli, N.; Jarvis, R.C.; Gilding, L.N.; Ott, S.; Bonifer, C.; Cockerill, P.N. Inducible chromatin priming is associated with the establishment of immunological memory in T cells. EMBO J. 2016, 35, 515–535. [Google Scholar] [CrossRef]
- To, T.K.; Kim, J.M. Epigenetic regulation of gene responsiveness in Arabidopsis. Front. Plant Sci. 2014, 4, 548. [Google Scholar] [CrossRef] [Green Version]
- Maxwell, C.S.; Kruesi, W.S.; Core, L.J.; Kurhanewicz, N.; Waters, C.T.; Lewarch, C.L.; Antoshechkin, I.; Lis, J.T.; Meyer, B.J.; Baugh, L.R. Pol II docking and pausing at growth and stress genes in C. elegans. Cell Rep. 2014, 6, 455–466. [Google Scholar] [CrossRef] [Green Version]
- Elowitz, M.B.; Levine, A.J.; Siggia, E.D.; Swain, P.S. Stochastic gene expression in a single cell. Science 2002, 297, 1183–1186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rogers, K.W.; Schier, A.F. Morphogen gradients: From generation to interpretation. Annu. Rev. Cell Dev. Biol. 2011, 27, 377–407. [Google Scholar] [CrossRef] [Green Version]
- Losick, R.; Desplan, C. Stochasticity and cell fate. Science 2008, 320, 65–68. [Google Scholar] [CrossRef] [Green Version]
- Natoli, G.; Saccani, S.; Bosisio, D.; Marazzi, I. Interactions of NF-kappaB with chromatin: The art of being at the right place at the right time. Nat. Immunol. 2005, 6, 439–445. [Google Scholar] [CrossRef]
- Kellogg, R.A.; Tay, S. Noise facilitates transcriptional control under dynamic inputs. Cell 2015, 160, 381–392. [Google Scholar] [CrossRef] [Green Version]
- Wheat, J.C.; Sella, Y.; Willcockson, M.; Skoultchi, A.I.; Bergman, A.; Singer, R.H.; Steidl, U. Single-molecule imaging of transcription dynamics in somatic stem cells. Nature 2020, 583, 431–436. [Google Scholar] [CrossRef]
- Swain, P.S.; Elowitz, M.B.; Siggia, E.D. Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc. Natl. Acad. Sci. USA 2002, 99, 12795–12800. [Google Scholar] [CrossRef] [Green Version]
- Kaern, M.; Elston, T.C.; Blake, W.J.; Collins, J.J. Stochasticity in gene expression: From theories to phenotypes. Nat. Rev. Genet. 2005, 6, 451–464. [Google Scholar] [CrossRef]
- Acar, M.; Mettetal, J.T.; van Oudenaarden, A. Stochastic switching as a survival strategy in fluctuating environments. Nat. Genet. 2008, 40, 471–475. [Google Scholar] [CrossRef]
- Balaban, N.Q.; Merrin, J.; Chait, R.; Kowalik, L.; Leibler, S. Bacterial persistence as a phenotypic switch. Science 2004, 305, 1622–1625. [Google Scholar] [CrossRef] [Green Version]
- Schmutzer, M.; Wagner, A. Gene expression noise can promote the fixation of beneficial mutations in fluctuating environments. PLoS Comput. Biol. 2020, 16, e1007727. [Google Scholar] [CrossRef]
- Levy, S.F.; Ziv, N.; Siegal, M.L. Bet hedging in yeast by heterogeneous, age-correlated expression of a stress protectant. PLoS Biol. 2012, 10, e1001325. [Google Scholar] [CrossRef] [Green Version]
- Levy, S.F. Cellular Heterogeneity: Benefits besides Bet-Hedging. Curr. Biol. 2016, 26, R355–R357. [Google Scholar] [CrossRef] [Green Version]
- Gefen, O.; Balaban, N.Q. The importance of being persistent: Heterogeneity of bacterial populations under antibiotic stress. FEMS Microbiol. Rev. 2009, 33, 704–717. [Google Scholar] [CrossRef]
- Sharma, S.V.; Lee, D.Y.; Li, B.; Quinlan, M.P.; Takahashi, F.; Maheswaran, S.; McDermott, U.; Azizian, N.; Zou, L.; Fischbach, M.A.; et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 2010, 141, 69–80. [Google Scholar] [CrossRef] [Green Version]
- Roesch, A.; Fukunaga-Kalabis, M.; Schmidt, E.C.; Zabierowski, S.E.; Brafford, P.A.; Vultur, A.; Basu, D.; Gimotty, P.; Vogt, T.; Herlyn, M. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell 2010, 141, 583–594. [Google Scholar] [CrossRef] [Green Version]
- Shaffer, S.M.; Dunagin, M.C.; Torborg, S.R.; Torre, E.A.; Emert, B.; Krepler, C.; Beqiri, M.; Sproesser, K.; Brafford, P.A.; Xiao, M.; et al. Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature 2017, 546, 431–435. [Google Scholar] [CrossRef] [Green Version]
- Raser, J.M.; O’Shea, E.K. Control of stochasticity in eukaryotic gene expression. Science 2004, 304, 1811–1814. [Google Scholar] [CrossRef] [Green Version]
- Lidstrom, M.E.; Konopka, M.C. The role of physiological heterogeneity in microbial population behavior. Nat. Chem. Biol. 2010, 6, 705–712. [Google Scholar] [CrossRef]
- Brown, R.; Curry, E.; Magnani, L.; Wilhelm-Benartzi, C.S.; Borley, J. Poised epigenetic states and acquired drug resistance in cancer. Nat. Rev. Cancer 2014, 14, 747–753. [Google Scholar] [CrossRef]
- Bar-Even, A.; Paulsson, J.; Maheshri, N.; Carmi, M.; O’Shea, E.; Pilpel, Y.; Barkai, N. Noise in protein expression scales with natural protein abundance. Nat. Genet. 2006, 38, 636–643. [Google Scholar] [CrossRef]
- Barroso, G.V.; Puzovic, N.; Dutheil, J.Y. The Evolution of Gene-Specific Transcriptional Noise Is Driven by Selection at the Pathway Level. Genetics 2018, 208, 173–189. [Google Scholar] [CrossRef] [Green Version]
- Newman, J.R.S.; Ghaemmaghami, S.; Ihmels, J.; Breslow, D.K.; Noble, M.; DeRisi, J.L.; Weissman, J.S. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature 2006, 441, 840–846. [Google Scholar] [CrossRef]
- Gasch, A.P.; Yu, F.B.; Hose, J.; Escalante, L.E.; Place, M.; Bacher, R.; Kanbar, J.; Ciobanu, D.; Sandor, L.; Grigoriev, I.V.; et al. Single-cell RNA sequencing reveals intrinsic and extrinsic regulatory heterogeneity in yeast responding to stress. PLoS Biol. 2017, 15, e2004050. [Google Scholar] [CrossRef] [Green Version]
- Charlebois, D.A.; Abdennur, N.; Kaern, M. Gene expression noise facilitates adaptation and drug resistance independently of mutation. Phys. Rev. Lett. 2011, 107, 218101. [Google Scholar] [CrossRef]
- Charlebois, D.A. Effect and evolution of gene expression noise on the fitness landscape. Phys. Rev. E Stat. Nonlinear Soft Matter Phys. 2015, 92, 022713. [Google Scholar] [CrossRef] [Green Version]
- Jones, D.L.; Brewster, R.C.; Phillips, R. Promoter architecture dictates cell-to-cell variability in gene expression. Science 2014, 346, 1533–1536. [Google Scholar] [CrossRef] [Green Version]
- Sanchez, A.; Golding, I. Genetic determinants and cellular constraints in noisy gene expression. Science 2013, 342, 1188–1193. [Google Scholar] [CrossRef] [Green Version]
- Sanchez, A.; Choubey, S.; Kondev, J. Regulation of noise in gene expression. Annu. Rev. Biophys. 2013, 42, 469–491. [Google Scholar] [CrossRef]
- Sánchez, A.; Kondev, J. Transcriptional control of noise in gene expression. Proc. Natl. Acad. Sci. USA 2008, 105, 5081–5086. [Google Scholar] [CrossRef] [Green Version]
- Das, D.; Dey, S.; Brewster, R.C.; Choubey, S. Effect of transcription factor resource sharing on gene expression noise. PLoS Comput. Biol. 2017, 13, e1005491. [Google Scholar] [CrossRef]
- Engl, C.; Jovanovic, G.; Brackston, R.D.; Kotta-Loizou, I.; Buck, M. The route to transcription initiation determines the mode of transcriptional bursting in E. coli. Nat. Commun. 2020, 11, 2422. [Google Scholar] [CrossRef]
- Brown, C.R.; Boeger, H. Nucleosomal promoter variation generates gene expression noise. Proc. Natl. Acad. Sci. USA 2014, 111, 17893–17898. [Google Scholar] [CrossRef] [Green Version]
- Brown, C.R.; Mao, C.; Falkovskaia, E.; Jurica, M.S.; Boeger, H. Linking stochastic fluctuations in chromatin structure and gene expression. PLoS Biol. 2013, 11, e1001621. [Google Scholar] [CrossRef] [Green Version]
- Buenrostro, J.D.; Wu, B.; Litzenburger, U.M.; Ruff, D.; Gonzales, M.L.; Snyder, M.P.; Chang, H.Y.; Greenleaf, W.J. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 2015, 523, 486–490. [Google Scholar] [CrossRef]
- Wu, S.; Li, K.; Li, Y.; Zhao, T.; Li, T.; Yang, Y.-F.; Qian, W. Independent regulation of gene expression level and noise by histone modifications. PLoS Comput. Biol. 2017, 13, e1005585. [Google Scholar] [CrossRef] [Green Version]
- Lagha, M.; Bothma, J.P.; Esposito, E.; Ng, S.; Stefanik, L.; Tsui, C.; Johnston, J.; Chen, K.; Gilmour, D.S.; Zeitlinger, J.; et al. Paused Pol II coordinates tissue morphogenesis in the Drosophila embryo. Cell 2013, 153, 976–987. [Google Scholar] [CrossRef] [Green Version]
- Buettner, F.; Natarajan, K.N.; Casale, F.P.; Proserpio, V.; Scialdone, A.; Theis, F.J.; Teichmann, S.A.; Marioni, J.C.; Stegle, O. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat. Biotechnol. 2015, 33, 155–160. [Google Scholar] [CrossRef]
- Battich, N.; Stoeger, T.; Pelkmans, L. Control of transcript variability in single mammalian cells. Cell 2015, 163, 1596–1610. [Google Scholar] [CrossRef] [Green Version]
- Ansel, J.; Bottin, H.; Rodriguez-Beltran, C.; Damon, C.; Nagarajan, M.; Fehrmann, S.; François, J.; Yvert, G. Cell-to-cell stochastic variation in gene expression is a complex genetic trait. PLoS Genet. 2008, 4, e1000049. [Google Scholar] [CrossRef] [Green Version]
- You, S.-T.; Jhou, Y.-T.; Kao, C.-F.; Leu, J.-Y. Experimental evolution reveals a general role for the methyltransferase Hmt1 in noise buffering. PLoS Biol. 2019, 17, e3000433. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pascual-Ahuir, A.; Fita-Torró, J.; Proft, M. Capturing and Understanding the Dynamics and Heterogeneity of Gene Expression in the Living Cell. Int. J. Mol. Sci. 2020, 21, 8278. https://doi.org/10.3390/ijms21218278
Pascual-Ahuir A, Fita-Torró J, Proft M. Capturing and Understanding the Dynamics and Heterogeneity of Gene Expression in the Living Cell. International Journal of Molecular Sciences. 2020; 21(21):8278. https://doi.org/10.3390/ijms21218278
Chicago/Turabian StylePascual-Ahuir, Amparo, Josep Fita-Torró, and Markus Proft. 2020. "Capturing and Understanding the Dynamics and Heterogeneity of Gene Expression in the Living Cell" International Journal of Molecular Sciences 21, no. 21: 8278. https://doi.org/10.3390/ijms21218278
APA StylePascual-Ahuir, A., Fita-Torró, J., & Proft, M. (2020). Capturing and Understanding the Dynamics and Heterogeneity of Gene Expression in the Living Cell. International Journal of Molecular Sciences, 21(21), 8278. https://doi.org/10.3390/ijms21218278




