Abstract
Ctn[15–34], the C-terminal fragment of crotalicidin, an antimicrobial peptide from the South American rattlesnake Crotalus durissus terrificus venom, displays remarkable anti-infective and anti-proliferative activities. Herein, its activity on Candida albicans biofilms and its interaction with the cytoplasmic membrane of the fungal cell and with a biomembrane model in vitro was investigated. A standard C. albicans strain and a fluconazole-resistant clinical isolate were exposed to the peptide at its minimum inhibitory concentration (MIC) (10 µM) and up to 100 × MIC to inhibit biofilm formation and its eradication. A viability test using XTT and fluorescent dyes, confocal laser scanning microscopy, and atomic force microscopy (AFM) were used to observe the antibiofilm effect. To evaluate the importance of membrane composition on Ctn[15–34] activity, C. albicans protoplasts were also tested. Fluorescence assays using di-8-ANEPPS, dynamic light scattering, and zeta potential measurements using liposomes, protoplasts, and C. albicans cells indicated a direct mechanism of action that was dependent on membrane interaction and disruption. Overall, Ctn[15–34] showed to be an effective antifungal peptide, displaying antibiofilm activity and, importantly, interacting with and disrupting fungal plasma membrane.
1. Introduction
After the first report of the presence of cecropins in the hemolymph of Hyalophora cecropia moths [1], in 1981, reports followed of mammalian α-defensins in human neutrophils [2] and magainins in the skin secretions of the amphibian Xenopus laevis four years later [3]. The expression of antimicrobial peptides (AMPs) in organisms of all kingdoms has been recognized as an essential feature of the innate immunity system, and hundreds of AMP sequences belonging to different structural and functional families have been characterized. Dedicated databases maintain information regarding this class of biologically active peptides in either native, synthetic, or de novo-designed versions [4,5,6]. More recently, AMPs from animal venoms have gained attention [7,8,9,10].
The vipericidins are a group of highly conserved cathelicidin-related antimicrobial peptides (CRAMPs) from the venom glands of South American pit vipers, exemplified by lachesicidin, lutzicidin, batroxicidin, and crotalicidin, as disclosed and characterized by some of us [11]. Vipericidins share also a high homology with CRAMPs from Asian elapid snakes, despite the fact that both groups of snakes (pit vipers and elapids) are phylogenetically and geographically distant relatives [12]. One of the best studied vipericidins, crotalicidin (Ctn), a linear 34-residue peptide (KRFKKFFKKVKKSVKKRLKKIFKKPMVIGVTIPF), rich in basic (Lys, Arg) residues and with an α-helical fold, is generated from a precursor made up of a signal peptide and a cathelin domain [1,4]. Its short N- and C-terminal fragments, Ctn[1–14] (KRFKKFFKKVKKSV) and Ctn[15–34] (KKRLKKIFKKPMVIGVTIPF), respectively, result from a careful structural dissection of the full-length Ctn and have been studied for anti-infective (antimicrobial, anti-parasite, and antiviral) and anti-proliferative properties (antitumoral), as reported elsewhere [13,14]. The two fragments adopt distinct folds in solution and display biological activities different from the parental Ctn; thus, Ctn[1–14] preserves helicity but, paradoxically, has no noticeable activities, while Ctn[15–34] adopts a mainly random structure and it is fully active as an AMP against Gram-negative bacteria, being less cytotoxic to healthy cells, highly stable in serum, as well as moderately active against several cancer cell lines in vitro [15,16]. Ctn[15–34] exerts its microbicidal effect by membrane disruption and interaction with microbial DNA, with a rapid onset of action, as observed for bacteria [17]. Recently, the antifungal properties of these peptides were demonstrated against planktonic forms of standard and clinical strains of pathogenic yeasts and dermatophytes, both susceptible and resistant to antifungals used in clinical settings, like polyenes (e.g., amphotericin B) and azoles (e.g., fluconazole) [18,19]. In this first study, it was demonstrated that Ctn[15–34] disrupts the plasma membrane integrity of Candida cells and induces apoptosis-mediated cell death [20]. These findings are interesting, given that systemic fungal infections cause some 1.5 million deaths each year worldwide [21]. The pathogenic fungi mainly responsible for systemic infections belong to the genus Candida and cause candidiasis with higher incidence in immunosuppressed patients [22]. Notably, these opportunistic pathogens are part of the human microbiota, colonizing the oral cavity, the gastrointestinal tract, and the vaginal mucosa [23]. The severity of candidiasis is associated to immune conditions of human host and virulence factors of this eukaryotic pathogen, such as dimorphism, expression of adhesion proteins, production of hydrolytic enzymes, and alteration of cell wall, which invariably exacerbate Candida pathogenicity [24]. Particularly, adhesins on the wall of Candida cells contribute to the formation of biofilms on general surfaces and on medical devices that are in contact with or implanted in patients, and such capacity to form biofilms was associated with invasiveness [25]. Biofilms are complex networks of a single or more species of microorganisms, surrounded by an extracellular polymeric matrix, consisting of carbohydrates, proteins, and nucleic acids. This structured consortium of cells provides some advantages for microorganisms, such as protection against environmental stress, deluding the host immune system, and, importantly, increasing resistance to antimicrobial drugs [25,26]. In fact, resistance to antifungals in biofilms is mainly attributed to the extracellular matrix acting as a shield that delays or prevents drug diffusion to individual cells located deep within a microcolony, therefore reducing the adsorption and/or neutralizing the drug effect [26,27].
Since antimicrobial peptides are poised as potential alternatives to conventional anti-infective chemotherapeutics in fighting infections and may be developed into biopharmaceuticals, in the present study, we evaluated the antifungal effects of Ctn[15–34] on planktonic (free-floating) cells and biofilms of two strains of Candida albicans: a reference (wild-type) drug-sensitive strain and a fluconazole-resistant clinical isolate. To this aim, biophysical methodologies were combined to address the fungicidal efficacy of Ctn[15–34] in C. albicans biofilms, as well as the candidacidal mechanism of action with lipid membranes that simulate the Candida cytoplasmic membrane. By combining fluorescence spectroscopy, dynamic light scattering, zeta potential assays, confocal microscopy, and atomic force microscopy, the membrane selectivity is demonstrated and new insights on the mechanism of action of Ctn[15–34] against C. albicans are revealed.
2. Results
2.1. Ctn[15–34] Activity against C. albicans Biofilms
After the synthesis, purification, and essential biochemical characterization of Ctn[15–34], antifungal activity was confirmed. Ctn[15–34] minimum inhibitory concentrations (MICs) for both C. albicans strains were in agreement with those previously determined. C. albicans breakpoints available from the European Committee on Antimicrobial Susceptibility Testing (EUCAST) and Clinical & Laboratory Standards Institute (CLSI) guidelines were used as parameters to establish yeast resistance [28]. For Ctn[15–34], a MIC of 10 μM (23.7 µg mL−1) was found for both strains tested (Table 1). According to the CLSI and EUCAST breakpoints, C. albicans (LABMIC 0125) was resistant to fluconazole (FCZ), with a MIC of 8 μg mL−1 (25 µM), higher than reported for this antifungal (4 μg mL−1 or 12.48 µM) [29], while for the standard wild-type strain (ATCC 90028), the MIC for FCZ is almost 10-fold lower, i.e., 1 μg mL−1 (3.12 µM).

Table 1.
Minimum inhibitory concentration (MIC) of Ctn[15–34] against Candida albicans.
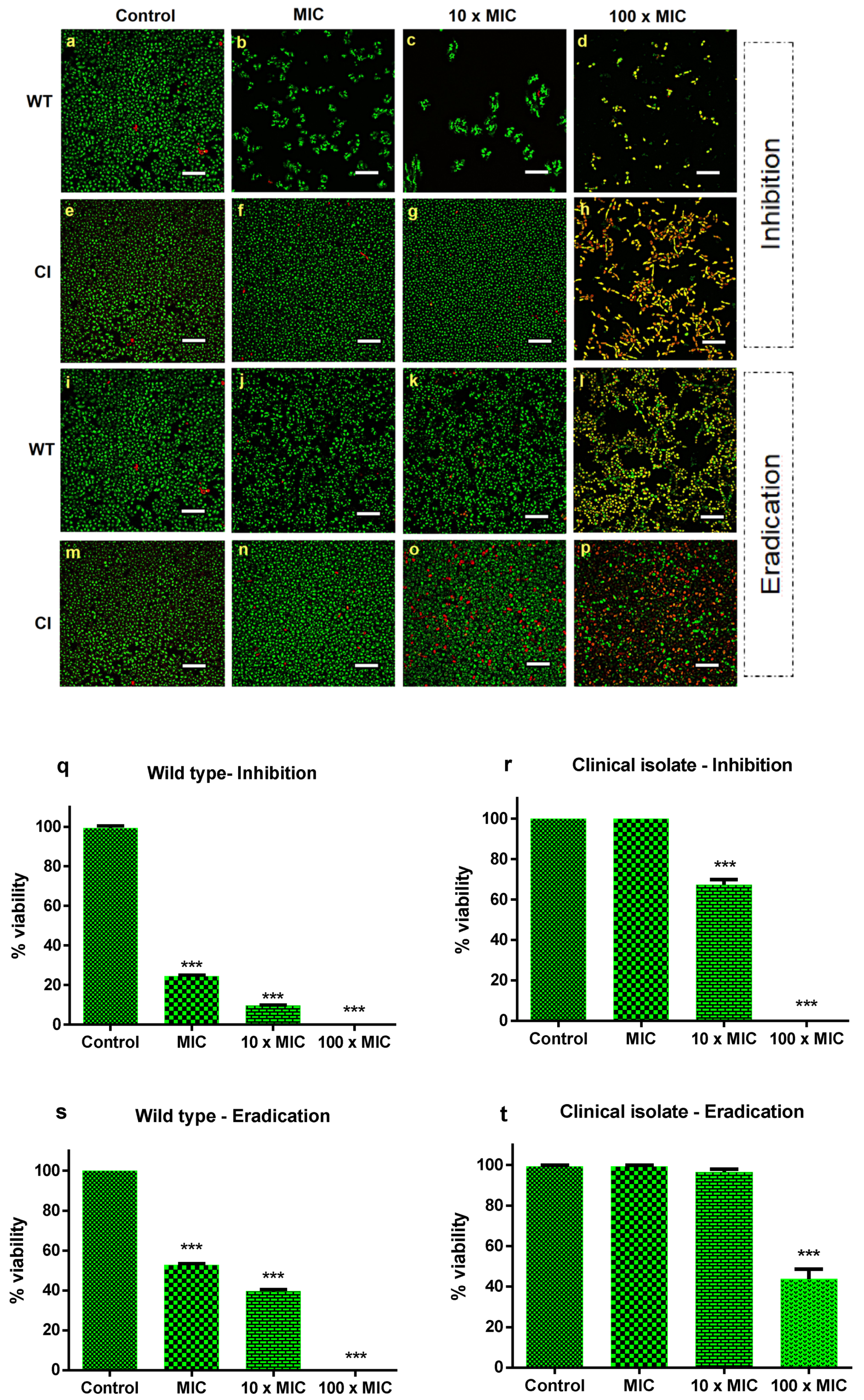
To determine the antibiofilm effect of Ctn[15–34], the best conditions for biofilm formation were established (Supplementary Materials Figure S1). For the drug-resistant Candida isolate, the best time span for biofilm formation corresponds to 72 h, with a concentration of 107 CFU mL−1. The standard strain forms biofilms at the same time interval as the drug-resistant isolate, but with a lower number of cells (104 CFU mL−1). After establishing the best time and cell number to achieve biofilm formation of both strains, the next step was to evaluate the inhibitory activity of Ctn[15–34] on biofilm formation and biofilm eradication, in this later case, once the Candida biofilms were established. For that, a peptide concentration corresponding to the MIC (10 µM), as previously determined for the planktonic cells, was used. The peptide effects on biofilms were visualized by confocal microscopy (Figure 1a–p). After treatment with Ctn[15–34], cell density was clearly lower for the wild-type C. albicans ATCC 90028, when compared to the drug-resistant strain. This behavior was observed both in the assays of biofilm inhibition and eradication (Figure 1a–t). In the inhibition assay, the concentrations of 10 μM (MIC) and 100 μM (10 × MIC) considerably reduced the biofilm of the wild-type strain, whereas for the resistant strain, no change in cell number covering the support surface was observed. At 100 × MIC, there was a complete inhibition of biofilm formation of the susceptible strain. In the eradication assays, at concentrations lower than 100 × MIC, distinct results were observed when comparing the sensitive and the resistant clinical isolate strains. With the wild-type strain, the peptide completely eradicated the biofilm, whereas with the resistant strain, the biofilm was only partially eradicated, with some viable cells remaining (Figure 1). Illustrative xyz slice images of preformed C. albicans biofilm untreated or treated with Ctn[15–34] are presented in Figures S2 and S3. As shown, C. albicans biofilm densities are variable between both strains (Figure S2, control). After treatment with different Ctn[15–34] concentrations, a difference between strains in the action of this peptide was observed (Figures S2 and S3). Overall, treatment with Ctn[15–34] at 100 × MIC showed to be the best concentration to reduce and eradicate C. albicans biofilms.
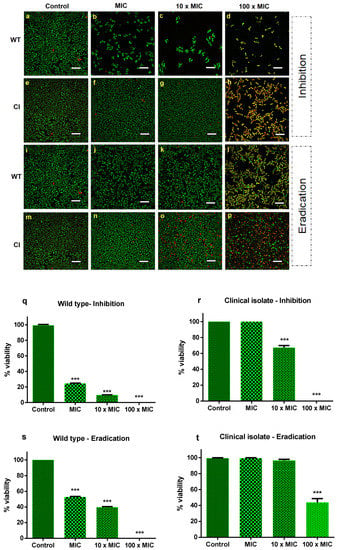
Figure 1.
Effect of Ctn[15–34] on the inhibition and eradication of C. albicans biofilms. Confocal microscopy images of the inhibition of C. albicans biofilms for the sensitive wild-type strain (a–d) and the drug-resistant clinical isolate strain (e–h), and biofilm eradication of wild-type (i–l) and clinical isolate (m–p), in the absence (control) and presence of Ctn[15–34] at different concentrations. Images obtained with live/dead staining (SYTO 9, green; PI, red) using 40-fold magnification. Scale bars: 30 µM. Quantification of viable cells (q–t) obtained using the ImageJ software. Bars represent mean ± SD. *** p < 0.001 relative to control (without peptide).
As shown in Figure 1q–t, on the quantification of peptide-induced yeast cell death, the number of dead cells was low (approximately 2%) in biofilms of both Candida strains not exposed to Ctn[15–34], as expected. In the biofilm inhibition assay, at a peptide concentration equivalent to 100 × the MIC, the inhibition of biofilm formation and eradication were effective only for the drug-susceptible wild-type Candida strain. In contrast, such high concentration was effective in inhibiting biofilm formation with the fluconazole-resistant clinical strain, but eradication was not complete. Despite this, the number of dead cells continued to increase within 24 h of exposure to Ctn[15–34].
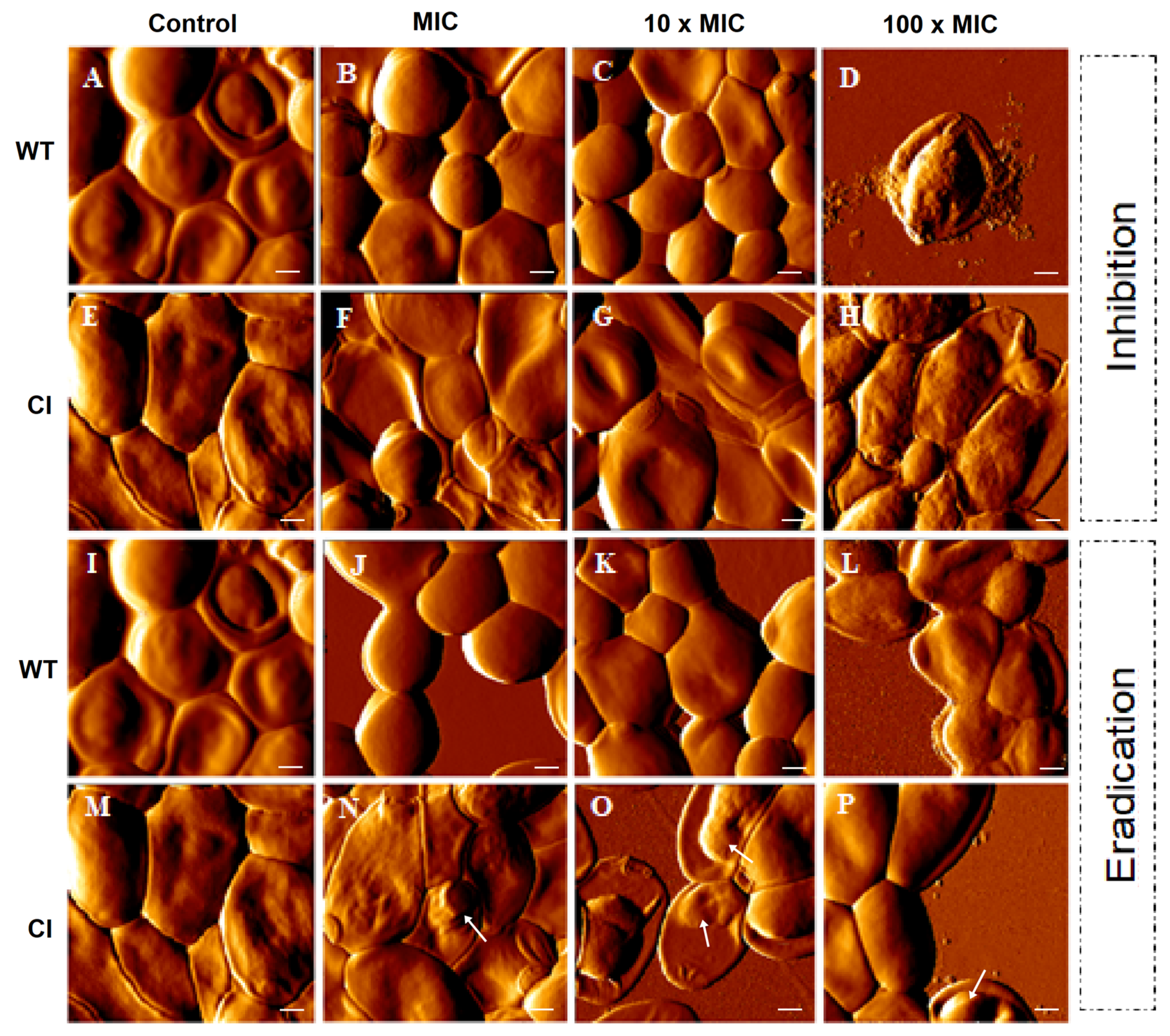
To further evaluate the inhibition and the eradication effects induced by Ctn[15–34] on C. albicans biofilms, atomic force microscopy (AFM) imaging, on intermittent contact mode, was used. Figure 2 highlights the main morphological changes that occurred in drug-resistant and wild-type Candida biofilms after exposure to Ctn[15–34]. As it can be noticed, Ctn[15–34] induced morphological alterations in a peptide concentration-dependent manner, even considering that untreated cells (controls) have some intrinsic distinctions: in the wild-type strain a smooth surface and regular shape were observed (Figure 2A,I), while in the drug-resistant C. albicans a light surface roughness, together with larger size and elongated shape, were normally visible (Figure 2E,M). On AFM images from the biofilm inhibition assay, the concentration-dependent irregularities in the cell surface of wild-type and drug-resistant Candida cells caused at 100 × MIC were evident (Figure 2B–D and Figure 2F–H, respectively). Moreover, the presence of pseudohyphae, which occurs when several elongated cells (blastoconidia) are joined together, was not observed. Additionally, on AFM images of wild-type Candida cells in the biofilm exposed to 100 × MIC, the predominant irregularities on the surface were accompanied by extravasation of intracellular content (Figure 2D). Images from the wild-type Candida cells in the biofilm eradication assay show that an irregular surface becomes evident with the increasing concentration of Ctn[15–34] (Figure 2J–L). Moreover, images from the biofilm along the eradication testing (Figure 2N–P) show apparently small vesicles on the surface of the drug-resistant cells when exposed to Ctn[15–34].
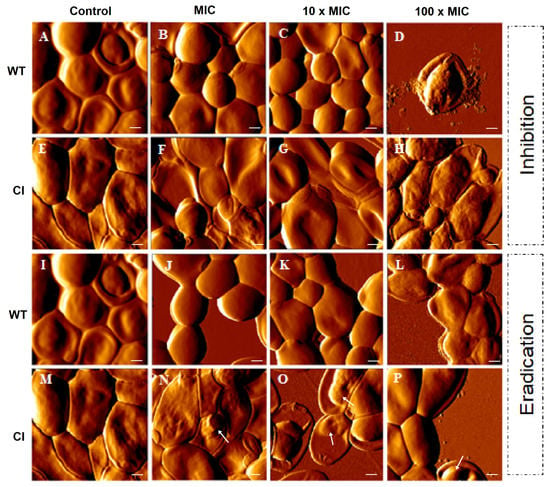
Figure 2.
AFM images of C. albicans biofilms. Formation (control) (A,E,I,M), inhibition, and eradication assays were performed for both strains (wild-type and clinical isolate). Images show the effect on C. albicans of the antimicotic peptide at different concentrations (MIC, 10 × MIC, and 100 × MIC) (B–D, F–H, J–L, N–P). Images of formation of biofilm represent the control without peptide. Total scanning area for each image: 10 × 10 µm2. Vesicles on the cell surface (N–P) were identified using white arrows. Scale bars: 1 µM.
2.2. Evaluation of Ctn[15–34] Interaction with Lipid Membranes
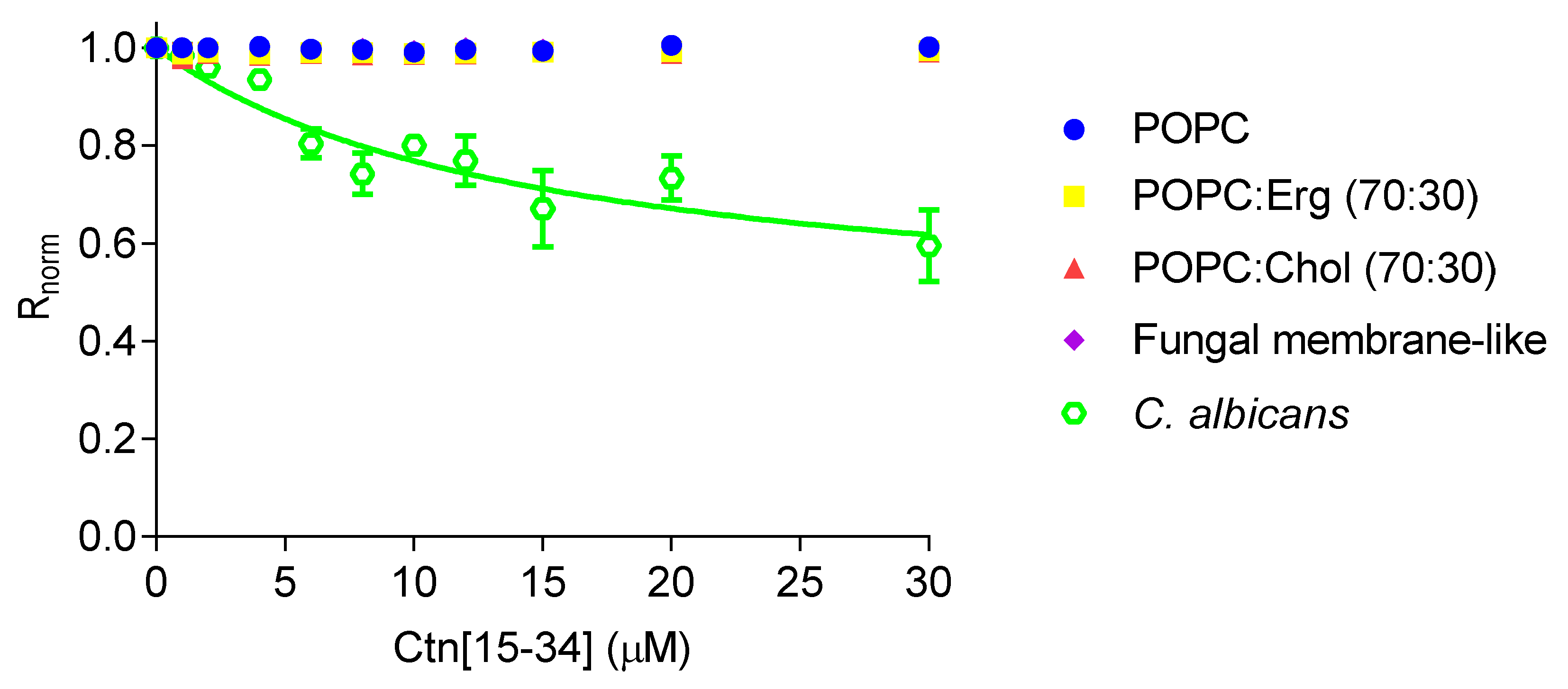
Changes in dipole potential upon peptide-membrane interaction were assessed by labeling lipid vesicles and C. albicans cells with 4-(2-[6-(dioctylamino)-2-naphthalenyl]ethenyl)-1-(3-sulfopropyl)pyridinium inner salt (di-8-ANEPPS; Figure 3). By increasing the concentration of Ctn[15–34], no changes in the dipole potential of the lipid vesicles were observed. In contrast, Ctn[15–34] show measurable affinity with C. albicans cells (ATCC 90028), yielding an apparent dissociation constant (Kd) of 14.5 ± 5.6 µM, indicating a preference (high affinity) of Ctn[15–34] for these cells.
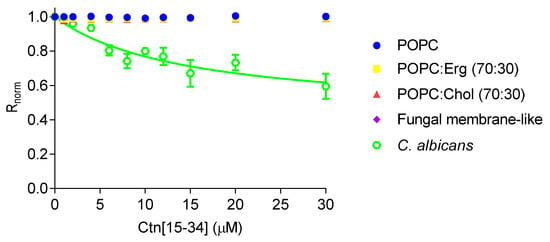
Figure 3.
Interaction of Ctn[15–34] with lipid vesicles and C. albicans protoplasts, assessed with the membrane dipole potential-sensitive probe di-8-ANEPPS. The excitation ratio, R (I455/I525), was normalized by dividing by the R value obtained in the absence of Ctn[15–34]. Equation (1) was used to fit the experimental data. Data are presented as mean ± SD of three independent measurements.
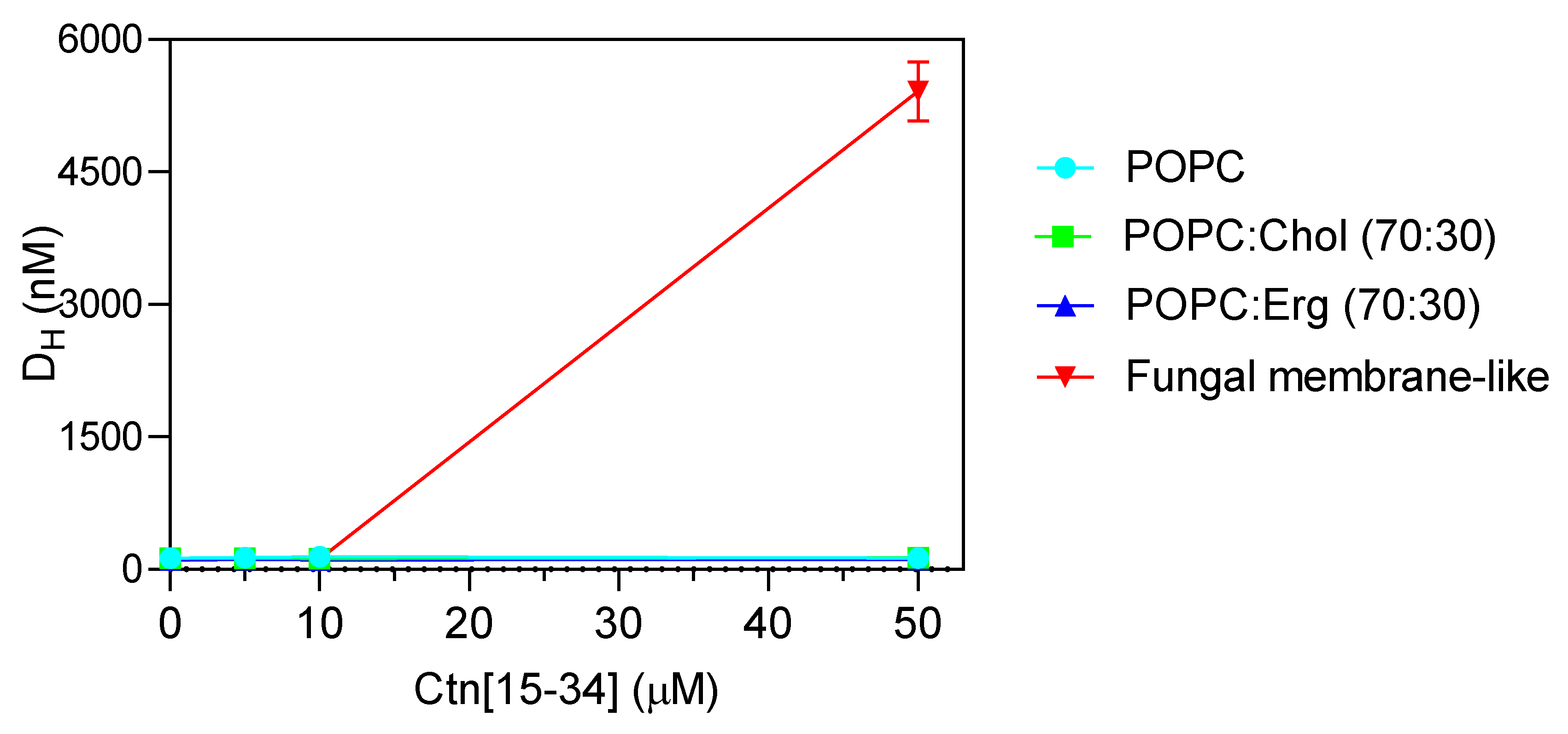
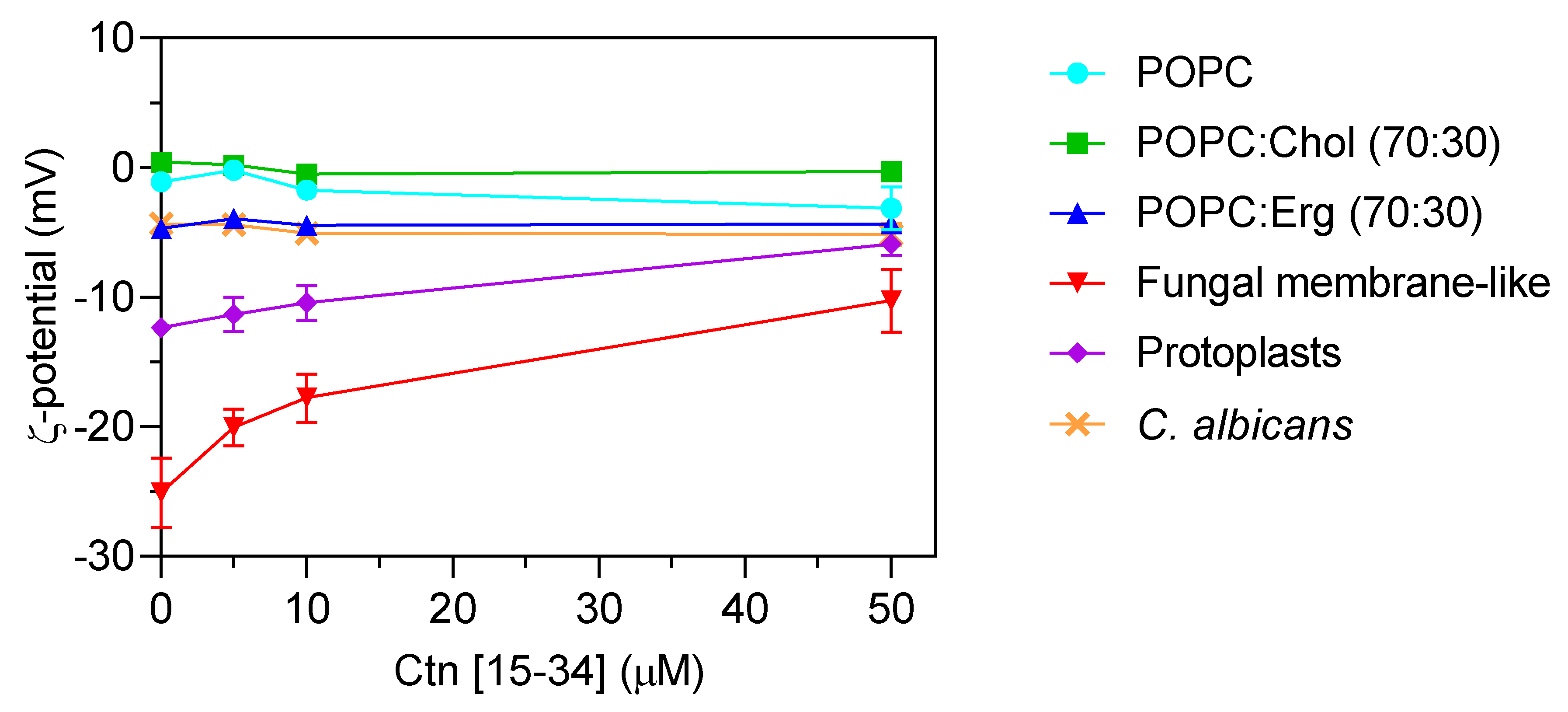
To evaluate whether membranes properties are altered upon the interaction with Ctn[15–34], two biophysical parameters were evaluated by dynamic light scattering, namely, lipid vesicle hydrodynamic diameter (DH; Figure 4) and zeta potential (Figure 5). Ctn[15–34] did not promote any significant change in the DH of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), POPC:Chol, and POPC:Erg (Figure 4). In contrast, using lipid vesicles with a composition that mimics the fungal cell membrane, Ctn[15–34] induced their aggregation at a peptide concentration of 50 μM (5 × MIC; Figure 4). Zeta potential results showed important differences between vesicles and cells. Despite the fact that the initial charge of the lipid vesicles of POPC, POPC:Chol, and POPC:Erg were slightly different, the increasing peptide concentrations did not promote additional changes in the zeta potential of these vesicles (Figure 5). Furthermore, no alteration in the zeta potential of C. albicans cells exposed to Ctn[15–34] (−4.32 mV) was observed.
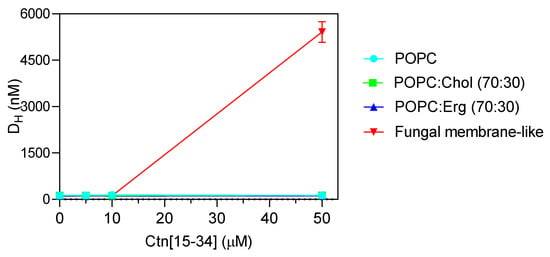
Figure 4.
Hydrodynamic diameter (DH) of the different lipid vesicles (200 µM total lipid concentration) exposed to increasing concentrations of Ctn[15–34]. Extensive aggregation is clear at the concentration of 50 µM for the fungal membrane-like lipid composition. Data are presented as mean ± SD of three independent replicates.
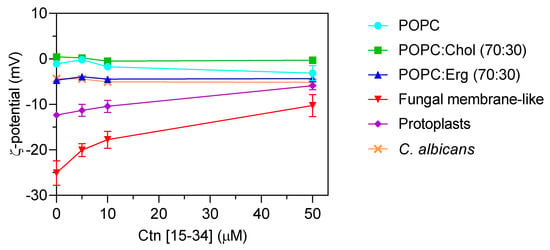
Figure 5.
Zeta potential (ζ) of lipid vesicles, C. albicans protoplasts, and C. albicans cells. Lipid vesicles were tested at a total lipid concentration of 200 µM. C. abicans cells and protoplasts were tested at the concentration of 105 cells mL−1. The results showed the importance of some membrane components of the cell membrane for Ctn[15–34] interaction. C. albicans and POPC:Erg vesicles show similar zeta potential behavior upon increasing Ctn[15–34] concentration. The initial values are −12 mV for protoplasts and –25 mV for fungal membrane-like vesicles. Data are presented as mean ± SD of three independent replicates.
Comparing zeta potential values for C. albicans cells with those for POPC:Erg, similarities can be found. Regardless of the inability to neutralize the surface charge, importantly, Ctn[15–34] induced a change in the zeta potential of the fungal membrane-like lipid vesicles from −25.1 ± 2.4 to −10.28 ± 2.6 mV (Figure 5). Antifungal agents may act on different cell components, but lipid membranes are commonly the first target to overcome. Thus, in addition to liposomes, the effect of Ctn[15–34] interaction with membranes and the neutralization of the membrane charge in C. albicans protoplasts is a robust evidence of the plasma membrane as the main initial target for antifungal activity. These cell-driven structures had an initial zeta potential of −12.37 ± 0.22 mV, which, after exposure to Ctn[15–34] (5 × MIC), increased to −5.9 ± 0.87 mV (Figure 5).
3. Discussion
The development of new antimicrobial drugs remains a major challenge to overcome the spread of drug resistance of clinically-relevant microbes, as strains are increasingly becoming less sensitive to conventional antibiotics. Previous studies have demonstrated that the crotalicidin fragment Ctn[15–34] is an effective antimicrobial peptide, with distinct anti-infective and anti-proliferative activities [13,14]. Its physicochemical properties, including small size, positive net charge, high hydrophobicity, and intermediate hydrophobic moment, endow Ctn[15–34] with a well-balanced structure-activity profile that makes it biologically active against bacteria and certain tumor cells, as well as stable in biological fluids [15,16]. In previous studies, we reported the antifungal properties of Ctn[15–34] against standard and clinical strains of susceptible and drug-resistant Candida species and dermatophytes, like Cryptococcus [18,19,20]. Here, we examined the antibiofilm potential of Ctn[15–34] on two C. albicans strains, including a drug-resistant clinical isolate, that are able to form biofilms. Moreover, the ability of Ctn[15–34] to interact with fungal membranes and biomembrane model systems was investigated as the initial mechanism of the peptide antifungal action. Firstly, the antifungal activity was confirmed, quantified as their minimum inhibitory concentrations against planktonic (free-floating) C. albicans cells of both a fluconazole-resistant clinical isolate and a standard wild-type ATCC strain. Both strains were shown to be susceptible to the fungicide activity of Ctn[15–34], with a MIC of 10 µM for both of them (Table 1).
Fungal biofilms differ from planktonic cells by their higher virulence, better adherence to mammalian tissue, and greater resistance to antifungals, especially azoles [30]. Although all Candida species are potentially prone to form biofilms, a well-recognized fact in clinical settings, the formation of biofilms depends on several factors, involving both the microorganisms themselves and the host [25]. Surfaces can play a determinant role on the formation of Candida biofilms [25,30]. Here, the biofilms of wild-type and fluconazole-resistant C. albicans were grown on polystyrene surfaces and differences in the amount of cells in the initial inoculum necessary to establish the biofilm were evidenced (Figure S1). As observed, despite the cell number- and time-dependence for formation, both strains formed biofilms (Figure 1 and Figure 2).
Most antimycotic drugs that disrupt yeast biofilms act in the initial stage of biofilm formation, preventing cell adhesion and subsequent biofilm consolidation [31]. In tune with this, Ctn[15–34] was able to inhibit biofilm formation and to eradicate the formed biofilms at concentrations 100 × the Ctn[15–34] MIC for the wild-type Candida strain. Drug diffusion and action are known to be restrained by biofilms [26,31] and, at this high peptide concentration (100 × MIC), the peptide caused only the inhibition of biofilm formation in the drug-resistant clinical isolate, without its eradication, despite the increased number of dead cells observed in the biofilm milieu (Figure 2 and Figure 3). As visible by AFM, these two Candida strains showed different phenotypes (Figure 2). Treatment with Ctn[15–34]-induced lethal morphological changes, as vesicle shedding, surface protruding, and bleb formation were evident in cells from the Candida biofilms exposed to 100 × MIC. Similar effects have been reported for Candida biofilms exposed to Psd1, an antimicrobial peptide (defensin) isolated from Pisum sativum seeds [32].
The mechanisms by which Ctn[15–34] kills pathogenic fungi was, up to now, not fully understood, although membrane perturbation with disruption and leakage of intracellular content (e.g., lactate dehydrogenease) and apoptosis were shown to be involved in the activity against Candida [19,20]. Furthermore, detailed studies in bacteria indicated that the cytoplasmic membrane was the first molecular target for the microbicidal effect; Ctn[15–34] interacts with the plasma membrane of target cells by electrostatic forces, accumulates on the cell surface, and ultimately causes membrane permeabilization, resulting in rapid cell lysis. Secondary mechanisms, such as DNA binding, have also been invoked to reinforce the action of the Ctn[15–34] on bacterial cells [17].
In the absence of intrinsic Ctn[15–34] fluorescence, given the lack of tryptophan or tyrosine residues, a biophysical assay to verify the interaction of Ctn[15–34] and lipid membranes was done with the membrane probe di-8-ANEPPS. This probe is sensitive to changes in the membrane dipole potential, with its fluorescence spectra shifting in response to changes in the surrounding electric field, enabling the quantification of eventual binding of the peptide to the lipid membranes [33]. While no Ctn[15–34] interaction with lipid vesicles was detectable, the peptide-membrane interaction was indeed quantifiably evident when C. albicans cells were tested.
Dynamic light scattering was used to measure vesicle size before and after exposure to Ctn[15–34] (Figure 4). This technique relies on the principle of the Brownian motion, so that scattered light follows a pattern that depends on vesicle size [34]. Ctn[15–34], at 50 μM, induced a significant aggregation of fungal membrane-like lipid vesicles (Figure 4). To evaluate the influence of the fungal cell wall on the interaction of Ctn[15–34] and fungal cells, protoplasts of C. albicans were prepared and exposed to different peptide concentrations. Changes of zeta potential of C. albicans cells and protoplasts, as well as lipid vesicles exposed to Ctn[15–34], indicated that it interacts selectively with protoplasts and vesicles that mimic the fungal membrane (Figure 5). Therefore, it can be concluded that the Candida cell wall is not directly involved in the action of Ctn[15–34], but, importantly, the plasma membrane is the main target, playing a critical role on antifungal activity.
It is known that AMPs can cause changes in the topology of lipid membranes by interacting through distinct orientations (e.g., planar alignment parallel or perpendicular to the membrane surface, transmembrane orientation) [35]. For Ctn[15–34], the less negative zeta potentials observed with fungal membrane-like lipid vesicles and protoplasts suggest that the peptide was embedded in the lipid bilayer, contributing consequently to a less anionic surface charge and subsequent destabilization of cytoplasmic membrane with further membrane disruption and cell lysis. Moreover, the reduction in the surface charge of membrane-like lipid vesicles and protoplasts showed that Ctn[15–34] is capable of inducing membrane depolarization by a membrane composition-dependent process.
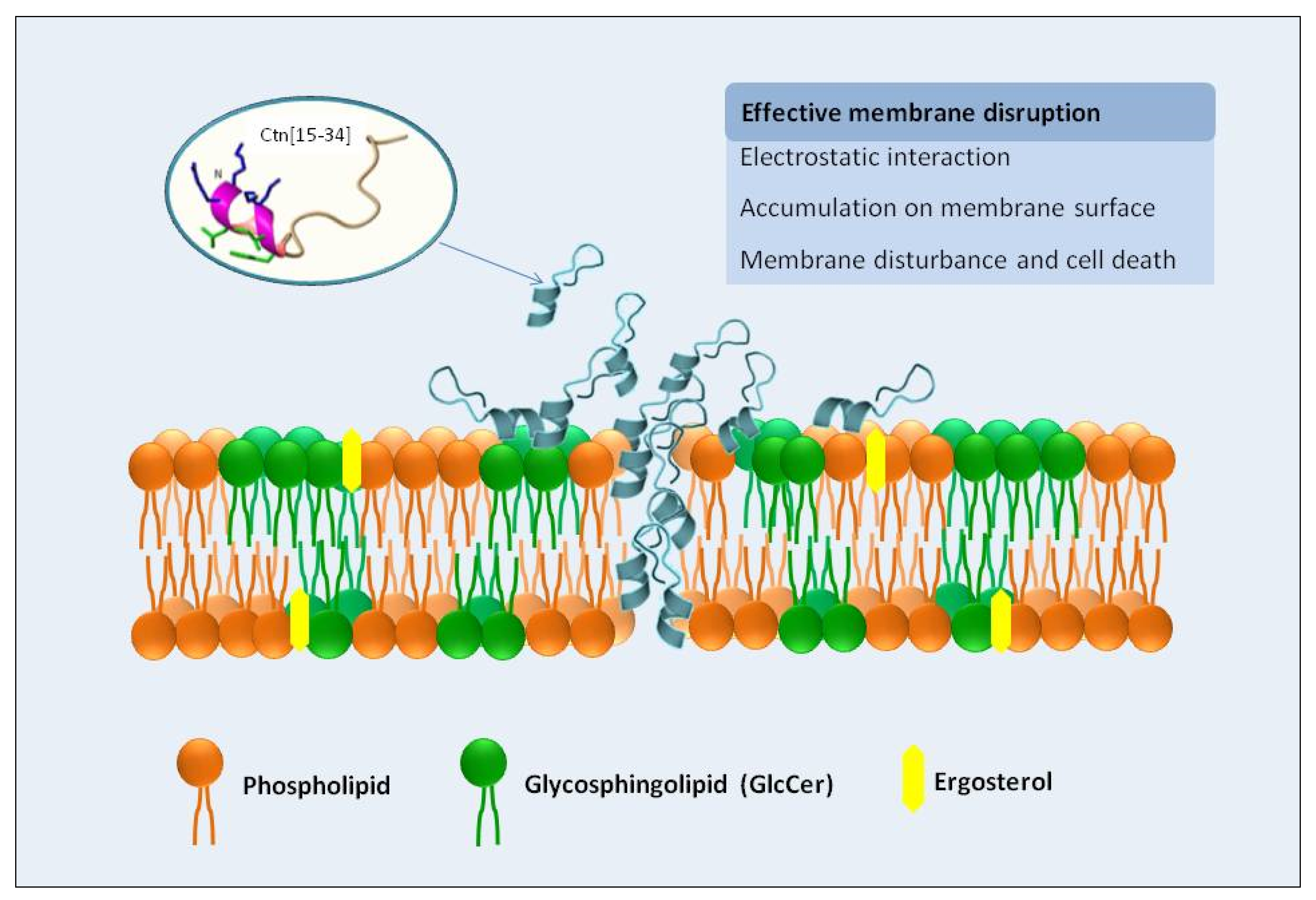
Overall, Ctn[15–34] is a multi-effector antimicrobial peptide with antifungal properties, with the fungal plasma membrane as first and main target, as demonstrated here by different biophysical and biochemical techniques. A scheme of this general behavior is presented in Figure 6. Ctn[15–34] can insert into the lipid bilayer and cause changes in membrane dipole potential. Moreover, Ctn[15–34] induces morphological changes in Candida cells within biofilms, causing their destabilization, although higher peptide concentrations are required, presumably to diffuse into the biofilm. The number of dead cells in the biofilm microenvironments increases along the time of exposure to Ctn[15–34]. Phenotypic differences between standard strain and fluconazole-resistant clinical isolate are indicative of the strain sensitivity and the biofilm stability of Candida exposed to the peptide action.
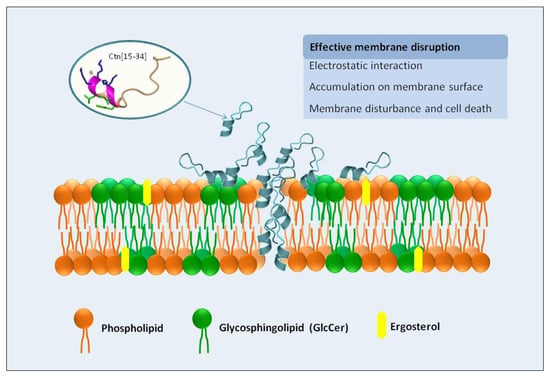
Figure 6.
Depicted mechanism of action of Ctn[15–34] on the interaction and disruption of plasma membrane of the pathogenic yeast Candida albicans. The plasma membrane of yeast cells is the primary target of Ctn[15–34] actions, as demonstrated experimentally by biophysical methods. Ctn[15–34] is recruited to the plasma membrane, through a process facilitated by the electrostatic attraction between the cationic residues of the peptide and the anionic lipids present in the plasma membrane of planktonic yeast or yeast cells in biofilms. After accumulation on the cell surface, peptide can efficiently insert into the lipid bilayer, causing membrane destabilization, as observed by changes in membrane dipole potential, with consequent death of the C. albicans cells.
4. Material and Methods
4.1. Peptides
Ctn (KRFKKFFKKVKKSVKKRLKKIFKKPMVIGVTIPF), Ctn[1–14] (KRFKKFFKKVKKSV), and Ctn[15–34] (KKRLKKIFKKPMVIGVTIPF) were synthesized by solid phase methods, purified by HPLC to >95% homogeneity, and characterized by ESI-mass spectrometry, as previously described [13]. Peptide stock solutions (1 mM) were prepared in sterile Milli-Q water and stored in aliquots at −20 °C, until use.
4.2. Microorganisms
Two C. albicans strains were used: a wild-type standard strain (ATCC 90028) and a fluconazole (FCZ)-resistant clinical isolate (LABMIC 0125), provided by the Santa Casa de Misericordia Hospital in Sobral (Sobral, Brazil). The identification of the clinical isolate was performed using a CHROMagar-Candida (CHROMagar, Paris, France) and VITEK 2 automated identification system (BioMérieux, Lyon, France) with an YST card, and by PCR-AGE using the transcribed internal spacer (ITS) region.
4.3. Susceptibility of Planktonic C. albicans
C. albicans was cultured on Sabouraud-dextrose agar (SDA) and incubated at 35 °C for 24 h. Yeast colonies were transferred to tubes containing sterile phosphate buffer saline (PBS; 0.01 M Na2HPO4, 0.0018 M KH2PO4, 0.137 M NaCl, 0.0027 M KCl, pH 7.4) to obtain suspensions with a turbidity equivalent to Standard McFarland 0.5 (106 CFU mL−1). These suspensions were then diluted 1:2000 in RPMI 1640 (with glutamine) medium/morpholinepropanesulfonic acid (MOPS), pH 7.4 [36]. All reagents were from Sigma (St. Louis, MO, USA).
The minimum inhibitory concentration (MIC) and minimum fungicidal concentration (MFC) of the peptides were determined by the broth microdilution method, using 96-well plates, according to the procedures of the Clinical and Laboratory Standards Institute (CLSI) M27-A3 standard [36]. Candida cells were exposed to peptide concentration ranging from 0.03 to 71.9 μg mL−1 (0.02 to 40 μM), in culture medium, in a microplate, incubated at 35 °C and fungal growth and/or inhibition was observed after 24 h. The MIC was defined as the lowest concentration where no visual growth (no turbidity) was observed, corresponding to 90% inhibition of fungal growth. The MFC was determined after the transference of 100 μL Candida suspension in which no turbidity was observed to capped test tubes containing SDA and incubated at 35 °C, for 48 h. The MFC was determined in accordance with fungal growth in the culture medium. Each experiment was performed in duplicate.
4.4. C. albicans Culture Preparation for Antibiofilm Assays
Stock cells were inoculated on 4% glucose SAD plates and incubated for 24 h at 35 °C. One colony was grown overnight in SDB and incubated at 35 °C with shaking at 180 rpm. Then, 100 µL of the overnight culture were diluted in 5 mL of SDB and incubated for 3 h with shaking at 35 °C. The cell concentration was measured by optical density at 660 nm.
4.5. Biofilm Development and XTT/Menadione Assay
For biofilm formation, RPMI 1640/MOPS supplemented with 2% glucose was used as culture medium. In this assay, wild-type C. albicans (ATCC 90028) and the clinical isolate LABMIC 0125 inocula were prepared as described above, but at cell suspensions of 104, 105, 106, and 107 CFU mL−1. Then, 100 µL of these cell suspensions were individually transferred to wells in a 96-well round bottom polypropylene plate. The plate was incubated for 12, 24, 48, 72, and 96 h, at 37 °C, with shaking at 180 rpm. At the end of the incubation period, the biofilm was washed three times with PBS to remove planktonic cells. Then, 100 µL of 200 μg of XTT/mL with 25 μM menadione was added to the wells of each plate and incubated for 2 h, at 37 °C, protected from light. The supernatants were transferred to a flat bottom plate and the optical density was measured at 490 nm [37].
4.6. Biofilm Inhibition and Eradication Evaluation by Confocal Microscopy
For biofilm inhibition and eradication, 8-wells µ-Slide assays plates (ibidi, Gräfelfing, Germany) were used. Cell concentrations and incubation times were determined by the biofilm formation assay described in 4.5, but with 104 CFU/mL for C. albicans ATCC 90028 and 107 CFU/mL for the clinical isolate, and an incubation time of 72 h. To perform the inhibition assays, 300 µL of each cell diluted in RPMI with 2% glucose was treated with Ctn[15–34] at the MIC, 10 × MIC, and 100 × MIC. For the eradication test, once the biofilm was formed, peptide was also added at the MIC, 10 × MIC, and 100 × MIC. After treatment, microplates were incubated for 24 h, at 37 °C. All plates were washed 10 × with sterile 10 mM HEPES, 150 mM NaCl, pH 7.4. To quantify peptide action on cells, the Live/Dead FungaLight Yeast Viability Kit (Life Technologies, Carlsbad, CA, USA) was used. Optical microscopy measurements with an inverted Zeiss LSM 710 confocal point-scanning microscope (Jena, Germany) were carried out in order to examine the architecture and the viability of Candida cells before and after exposure to antifungal agents. Argon (488 nm; 45 mW) and diode-pumped solid-state (561 nm; 15 mW) lasers were used with a 40 × dry-objective. After incubation, biofilms were stained with 2 µL SYTO 9 and propidium iodide (PI) dye probes (1:1) and incubated for 15 min. Images were acquired and analyzed with ImageJ 1.47v (rsbweb.nih.gov/ij/).
4.7. Atomic Force Microscopy Imaging
C. albicans biofilms were prepared as described for confocal microscopy assays (Section 4.6). After biofilm treatment with and without peptide, the cellular mass was carefully washed with Milli-Q water and air-dried at 25 °C. Images of untreated and treated yeast cells were conducted using a JPK NanoWizard IV atomic force microscope (Berlin, Germany) mounted on a Zeiss Axiovert 200 inverted microscope (Jena, Germany). Measurements were carried out in intermittent contact mode (air) using ACL silicon cantilevers (AppNano, Huntingdon, UK). Height and error images were recorded with similar AFM parameters (setpoint, scan rate, and gain). Scan rate was set between 0.3 and 0.6 Hz and setpoint close to 0.3 V. Images were analyzed with the JPK image processing software v. 6.1.149.
4.8. Fungal Protoplast Preparation
Protoplasts were prepared by removing the cell wall of C. albicans, which consist in a microfibrillar network composed by glucans and chitin embedded in an amorphous material composed mainly of mannoproteins that together provide rigidity to the cell wall [38,39]. Protoplasts were isolated with a yeast lytic enzyme (Zymolyase-20T, Grisp Research Solutions, Porto, Portugal), as described elsewhere [40]. Briefly, C. albicans (ATCC 90028) cultured in SDA was inoculated into yeast peptone glucose (YPG) broth and incubated overnight at 37 °C. After this period, cells were harvested by centrifugation at 3000× g, for 10 min, at room temperature. The pellet was washed with sterile Milli-Q water, resuspended, and incubated in pre-incubation medium (50 mM EDTA, pH 9.0, with 35 mM β-mercaptoethanol), for 30 min, at 37 °C, with shaking at 180 rpm. Cells was centrifuged and washed once with washing solution I (1.2 M sorbitol, 50 mM EDTA, pH 7.5). A second wash was carried out with washing solution II (1.2 M sorbitol, Tris 50 mM, pH 7.5). Cell concentration was determined by optical density at 660 nm. A suspension of 5 × 106 CFU mL−1 was washed with enzyme buffer (1.2 M sorbitol, 50 mM Tris, 0.1 mM calcium acetate, 0.5 mM magnesium acetate). After washing, cells were resuspended in enzyme solution (enzyme buffer and 3 mg mL−1 Zymolyase-20T) and incubated for 2 h at 37 °C, with shaking at 180 rpm. Protoplasts were imaged with an optical microscope, by Gram staining, and maintained in HEPES buffer, at 4 °C, for up to 24 h after isolation.
4.9. Liposome Preparation
Large unilamellar vesicles (LUVs), with a diameter of approximately 100 nm, were obtained by extrusion of multilamellar vesicles (MLVs) as described elsewhere [41]. 1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoinositol (POPI), and 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPS) were from Avanti Polar Lipids (Alabaster, AL, USA), while cholesterol (Chol) and ergosterol (Erg) were from Sigma. The LUVs studied were mostly zwitterionic (pure POPC, POPC:Chol 70:30, POPC:Erg 70:30, and C. albicans membrane-like mixture, i.e., POPC:POPE:POPS:POPI:Erg 59:21:3:4:13 [42]), mimicking membrane systems relevant for the study [32,41]. Liposomes were extruded in the day of the measurement. HEPES buffer was used in all the measurements, except when otherwise indicated.
4.10. C. albicans Strains and Culture Conditions
C. albicans strains were maintained and grown as previously described [32]. From stock cultures kept at −80 °C until use, 10 µL of each strain were plated in SDA and incubated overnight, at 37 °C. An isolated colony was inoculated into 5 mL of SDB and allowed to grow overnight, at 37 °C, with shaking at 180 rpm. On the day of the measurement, 100 µL of the cell suspension were diluted in 5 mL of SDB and left at 37 °C until reaching log-phase, with a final cell counting of 10 × CFU mL−1. Then, cells were washed 3 × with SDB and centrifuged at 4000× g, at 10 °C, for 25 min. Finally, cells were suspended in HEPES or SDB, and reserved until use.
4.11. Fluorescence Spectroscopy Measurements Using di-8-ANEPPS
C. albicans at 105 CFU mL−1 were incubated with 100 μM di-8-ANEPPS (4-(2-[6-(dioctylamino)-2-naphthalenyl]ethenyl)-1-(3-sulfopropyl)pyridinium inner salt) in HEPES buffer with 0.1% (w/v) Pluronic F-127 (Sigma, St. Louis, MO, USA), for 1 h, at 25 °C, with gentle agitation and protection from light. From this suspension, 104 CFU mL−1 of cells and 10 μM of di-8-ANEPPS in a final volume of 500 µL were incubated for 1 h under stirring and protected from light. A suspension with the same concentration of unlabeled cells was used as control. Prior to the measurement, the suspensions were incubated with peptide (0–30 µM) for 1 h.
For lipid vesicle assays, LUVs were diluted in HEPES buffer to a final lipid concentration of 500 µM and labeled with 100 µM di-8-ANEPPS, for 1 h, under stirring in the dark. The same concentration of unlabeled vesicles was used as control. Labeled LUVs were incubated with peptide (0–30 µM) for 1 h.
Excitation spectra and the ratio of intensities were obtained at the excitation wavelengths of 455 and 525 nm (R = I455/I525), with emission at 670 nm. Excitation and emission slits were set to 5 and 10 nm, respectively. The variation of the ratio with the peptide concentrations was analyzed by a single binding site model using the expression
where R is divided by R0, the normalized ratio of intensities obtained without treatment with peptide, Rmin is the minimum asymptotic value of R and Kd is the apparent dissociation constant [43]. Fitting of the equation to the experimental data was performed by non-linear regression with GraphPad Prism 6.
4.12. Dynamic Light Scattering and Zeta Potential Measurements
Dynamic light scattering (DLS) and zeta potential measurements were carried out on a Malvern Zetasizer Nano ZS device (Malvern, UK) and data was processed using the Malvern’s DTS software [44]. DLS was performed to assess possible changes induced by Ctn[15–34] in the size of LUVs. LUVs were diluted in 200 µM HEPES buffer, filtered through a 0.45 µM filter, and treated with 5, 10, or 50 µM peptide. An untreated control was also used. All measurements were conducted at 25 °C, using 1 cm optical path disposable cells, with 15 measurements (100 runs each) for each sample, after 15 min of equilibration time. Viscosity and refractive index were set at 0.8872 cP and 1.330, respectively. The results presented correspond to the average of three independent samples. Zeta potential measurements were performed with LUVs, and with C. albicans cells and protoplasts. LUVs were diluted in HEPES buffer at a lipid concentration of 200 µM and filtered through a 0.45 µM pore size filter. Vesicles were incubated for 1 h with 5, 10, or 50 µM of peptide prior to measurements. C. albicans and protoplasts were tested at 5 × 105 CFU mL−1, in HEPES buffer. Applied current was set to 40 mV, with 60 s waiting between measurements.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/21/21/8339/s1. Figure S1: Colorimetric readings from XTT/menadione assays of biofilms formed by the two C. albicans strains. Each column represents the mean ± standard deviation (SD) from eight independent biofilm readings. Figure S2: Effect of Ctn[15–34] on the inhibition of C. albicans biofilms. Z-tack images of the inhibition of C. albicans biofilms for wild type (WT) and clinical isolate (CI), in the absence (control) and presence of Ctn[15–34] at different concentrations. Images obtained with live/dead staining (SYTO 9 and PI, respectively) using 40-fold magnification. Images obtained using ImageJ software. Figure S3: Effect of Ctn[15–34] on the eradication of C. albicans biofilms. Z-tack images of the eradication of C. albicans biofilms for wild type (WT) and clinical isolate (CI), in the absence (control) and presence of Ctn[15–34] at different concentrations. Images obtained with live/dead staining (SYTO 9 and PI, respectively) using 40-fold magnification. Images obtained using ImageJ software.
Author Contributions
Conceptualization, N.C.S. and G.R.B.; methodology, N.C.S. and S.G.; validation, N.C.S., S.G., D.A., and G.R.B.; formal analysis, F.L.L.d.A., C.S.d.P.C., and S.G.; investigation, F.L.L.d.A. and S.G.; resources, N.C.S., G.R.B., and D.A.; data curation, C.S.d.P.C.; writing—original draft preparation, F.L.L.d.A. and G.R.B.; writing—review and editing, N.C.S., S.G., D.A., and G.R.B.; visualization, F.L.L.d.A. and S.G.; supervision, N.C.S., S.G., and G.R.B.; project administration, N.C.S., D.A., and G.R.B.; funding acquisition, N.C.S., D.A., and G.R.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Coordination for the Improvement of Higher Education Personnel (CAPES), Finance Code 001, from the Ministry of Education and Culture (MEC), Federal Government of Brazil (F.L.L.d.A.), and by Fundação para a Ciência e a Tecnologia—Ministério da Ciência, Tecnologia e Ensino Superior (FCT-MCTES, Portugal). Partial research funding was from the National Council of Research and Development, (CNPq), the Ministry of Science, Technology and Innovation (MCTI), grant numbers 307733/2016-5 and 431077/2016-9 (G.R.B.).
Acknowledgments
Authors acknowledge the technical help of iMM Bioimaging Unit (Lisbon, Portugal).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Steiner, H.; Hultmark, D.; Engström, Å.; Bennich, H.; Boman, H.G. Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature 1981, 292, 246–248. [Google Scholar] [CrossRef] [PubMed]
- Ganz, T.; Selsted, M.E.; Szklarek, D.; Harwig, S.S.; Daher, K.; Bainton, D.F.; Lehrer, R.I. Defensins. Natural peptide antibiotics of human neutrophils. J. Clin. Investig. 1985, 76, 1427–1435. [Google Scholar] [CrossRef] [PubMed]
- Zasloff, M. Magainins, a class of antimicrobial peptides from Xenopus skin: Isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc. Natl. Acad. Sci. USA 1987, 84, 5449–5453. [Google Scholar] [CrossRef] [PubMed]
- Kang, X.; Dong, F.; Shi, C.; Liu, S.; Sun, J.; Chen, J.; Li, H.; Xu, H.; Lao, X.; Zheng, H. DRAMP 2.0, an updated data repository of antimicrobial peptides. Sci. Data 2019, 6, 148. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Li, X.; Wang, Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef]
- Das, D.; Jaiswal, M.; Khan, F.N.; Ahamad, S.; Kumar, S. PlantPepDB: A manually curated plant peptide database. Sci. Rep. 2020, 10, 2194. [Google Scholar] [CrossRef] [PubMed]
- Yacoub, T.; Rima, M.; Karam, M.; Fajloun, J.-M.S. Antimicrobials from Venomous Animals: An Overview. Molecules 2020, 25, 2402. [Google Scholar] [CrossRef]
- Harrison, P.L.; Abdel-Rahman, M.A.; Miller, K.; Strong, P.N. Antimicrobial peptides from scorpion venoms. Toxicon Off. J. Int. Soc. Toxinology 2014, 88, 115–137. [Google Scholar] [CrossRef]
- Van Hoek, M.L. Antimicrobial peptides in reptiles. Pharmaceuticals 2014, 7, 723–753. [Google Scholar] [CrossRef]
- Dodou Lima, H.V.; de Paula Cavalcante, C.S.; Rádis-Baptista, G. Antifungal In Vitro Activity of Pilosulin- and Ponericin-Like Peptides from the Giant Ant Dinoponera quadriceps and Synergistic Effects with Antimycotic Drugs. Antibiotics 2020, 9, 354. [Google Scholar] [CrossRef]
- Falcao, C.B.; de La Torre, B.G.; Perez-Peinado, C.; Barron, A.E.; Andreu, D.; Radis-Baptista, G. Vipericidins: A novel family of cathelicidin-related peptides from the venom gland of South American pit vipers. Amino Acids 2014, 46, 2561–2571. [Google Scholar] [CrossRef] [PubMed]
- Rádis-Baptista, G. Vipericidins, Snake Venom Cathelicidin-Related Peptides, in the Milieu of Reptilian Antimicrobial Polypeptides. In Snake Venoms; Inagaki, H., Vogel, C.-W., Mukherjee, A.K., Rahmy, T.R., Eds.; Springer: Dordrecht, The Netherlands, 2017; pp. 297–325. [Google Scholar] [CrossRef]
- Falcao, C.B.; Radis-Baptista, G. Crotamine and crotalicidin, membrane active peptides from Crotalus durissus terrificus rattlesnake venom, and their structurally-minimized fragments for applications in medicine and biotechnology. Peptides 2020, 126, 170234. [Google Scholar] [CrossRef]
- Pérez-Peinado, C.; Defaus, S.; Andreu, D. Hitchhiking with Nature: Snake Venom Peptides to Fight Cancer and Superbugs. Toxins 2020, 12, 255. [Google Scholar] [CrossRef] [PubMed]
- Falcao, C.B.; Pérez-Peinado, C.; de la Torre, B.G.; Mayol, X.; Zamora-Carreras, H.; Jiménez, M.; Rádis-Baptista, G.; Andreu, D. Structural Dissection of Crotalicidin, a Rattlesnake Venom Cathelicidin, Retrieves a Fragment with Antimicrobial and Antitumor Activity. J. Med. Chem. 2015, 58, 8553–8563. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Peinado, C.; Dias, S.A.; Mendonça, D.A.; Castanho, M.; Veiga, A.S.; Andreu, D. Structural determinants conferring unusual long life in human serum to rattlesnake-derived antimicrobial peptide Ctn[15–34]. J. Pept. Sci. Off. Publ. Eur. Pept. Soc. 2019, 25, e3195. [Google Scholar] [CrossRef] [PubMed]
- Perez-Peinado, C.; Dias, S.A.; Domingues, M.M.; Benfield, A.H.; Freire, J.M.; Radis-Baptista, G.; Gaspar, D.; Castanho, M.; Craik, D.J.; Henriques, S.T.; et al. Mechanisms of bacterial membrane permeabilization by crotalicidin (Ctn) and its fragment Ctn[15–34], antimicrobial peptides from rattlesnake venom. J. Biol. Chem. 2018, 293, 1536–1549. [Google Scholar] [CrossRef]
- Cavalcante, C.S.; Falcao, C.B.; Fontenelle, R.O.; Andreu, D.; Radis-Baptista, G. Anti-fungal activity of Ctn[15–34], the C-terminal peptide fragment of crotalicidin, a rattlesnake venom gland cathelicidin. J. Antibiot. 2017, 70, 231–237. [Google Scholar] [CrossRef]
- De Aguiar, F.L.L.; Cavalcante, C.; Dos Santos Fontenelle, R.O.; Falcão, C.B.; Andreu, D.; Rádis-Baptista, G. The antiproliferative peptide Ctn[15–34] is active against multidrug-resistant yeasts Candida albicans and Cryptococcus neoformans. J. Appl. Microbiol. 2020, 128, 414–425. [Google Scholar] [CrossRef] [PubMed]
- Cavalcante, C.S.P.; de Aguiar, F.L.L.; Fontenelle, R.O.S.; de Menezes, R.; Martins, A.M.C.; Falcao, C.B.; Andreu, D.; Radis-Baptista, G. Insights into the candidacidal mechanism of Ctn[15–34]—A carboxyl-terminal, crotalicidin-derived peptide related to cathelicidins. J. Med. Microbiol. 2018, 67, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Bongomin, F.; Gago, S.; Oladele, R.O.; Denning, D.W. Global and Multi-National Prevalence of Fungal Diseases-Estimate Precision. J. Fungi 2017, 3, 57. [Google Scholar] [CrossRef]
- Yapar, N. Epidemiology and risk factors for invasive candidiasis. Ther. Clin. Risk Manag. 2014, 10, 95–105. [Google Scholar] [CrossRef]
- Sardi, J.C.O.; Scorzoni, L.; Bernardi, T.; Fusco-Almeida, A.M.; Mendes Giannini, M.J.S. Candida species: Current epidemiology, pathogenicity, biofilm formation, natural antifungal products and new therapeutic options. J. Med. Microbiol. 2013, 62, 10–24. [Google Scholar] [CrossRef]
- Staniszewska, M. Virulence Factors in Candida species. Curr. Protein Pept. Sci. 2020, 21, 313–323. [Google Scholar] [CrossRef]
- Ramage, G.; Saville, S.P.; Thomas, D.P.; López-Ribot, J.L. Candida biofilms: An update. Eukaryot. Cell 2005, 4, 633–638. [Google Scholar] [CrossRef] [PubMed]
- Al-Fattani, M.A.; Douglas, L.J. Biofilm matrix of Candida albicans and Candida tropicalis: Chemical composition and role in drug resistance. J. Med. Microbiol. 2006, 55, 999–1008. [Google Scholar] [CrossRef] [PubMed]
- Silva, S.; Rodrigues, C.F.; Araujo, D.; Rodrigues, M.E.; Henriques, M. Candida Species Biofilms’ Antifungal Resistance. J. Fungi 2017, 3, 8. [Google Scholar] [CrossRef]
- EUCAST. Determination of minimum inhibitory concentrations (MICs) of antibacterial agents by broth dilution. Clin. Microbiol. Infect. 2003, 9, ix. [Google Scholar] [CrossRef]
- Alastruey-Izquierdo, A.; Melhem, M.S.; Bonfietti, L.X.; Rodriguez-Tudela, J.L. Susceptibility test for fungi: Clinical and laboratorial correlations in medical mycology. Rev. Inst. Med. Trop. Sao Paulo 2015, 57 (Suppl. S19), 57–64. [Google Scholar] [CrossRef]
- Uppuluri, P.; Acosta Zaldivar, M.; Anderson, M.Z.; Dunn, M.J.; Berman, J.; Lopez Ribot, J.L.; Kohler, J.R. Candida albicans Dispersed Cells Are Developmentally Distinct from Biofilm and Planktonic Cells. mBio 2018, 9. [Google Scholar] [CrossRef]
- Sherry, L.; Rajendran, R.; Lappin, D.F.; Borghi, E.; Perdoni, F.; Falleni, M.; Tosi, D.; Smith, K.; Williams, C.; Jones, B.; et al. Biofilms formed by Candida albicans bloodstream isolates display phenotypic and transcriptional heterogeneity that are associated with resistance and pathogenicity. BMC Microbiol. 2014, 14, 182. [Google Scholar] [CrossRef]
- Gonçalves, S.; Silva, P.M.; Felício, M.R.; de Medeiros, L.N.; Kurtenbach, E.; Santos, N.C. Psd1 Effects on Candida albicans Planktonic Cells and Biofilms. Front. Cell Infect. Microbiol. 2017, 7, 249. [Google Scholar] [CrossRef]
- Matos, P.M.; Gonçalves, S.; Santos, N.C. Interaction of peptides with biomembranes assessed by potential-sensitive fluorescent probes. J. Pept. Sci. 2009, 14, 407–415. [Google Scholar] [CrossRef]
- Domingues, M.M.; Santiago, P.S.; Castanho, M.A.R.B.; Santos, N.C. What can light scattering spectroscopy do for membrane-active peptide studies? J. Pept. Sci. 2008, 14, 394–400. [Google Scholar] [CrossRef]
- Harmouche, N.; Bechinger, B. Lipid-Mediated Interactions between the Antimicrobial Peptides Magainin 2 and PGLa in Bilayers. Biophys. J. 2018, 115, 1033–1044. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts, 3rd ed.; CLSI Document M27-A3; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2008. [Google Scholar]
- Pierce, C.G.; Uppuluri, P.; Tristan, A.R.; Wormley, F.L., Jr.; Mowat, E.; Ramage, G.; Lopez-Ribot, J.L. A simple and reproducible 96-well plate-based method for the formation of fungal biofilms and its application to antifungal susceptibility testing. Nat. Protoc. 2008, 3, 1494–1500. [Google Scholar] [CrossRef]
- Rico, H.; Carrillo, C.; Aguado, C.; Mormeneo, S.; Sentandreu, R. Initial steps of wall protoplast regeneration in Candida albicans. Res. Microbiol. 1997, 148, 593–603. [Google Scholar] [CrossRef]
- Shepherd, M.G.; Poulter, R.T.; Sullivan, P.A. Candida albicans: Biology, genetics, and pathogenicity. Annu. Rev. Microbiol. 1985, 39, 579–614. [Google Scholar] [CrossRef]
- Terpitz, U.; Raimunda, D.; Westhoff, M.; Sukhorukov, V.L.; Beaugé, L.; Bamberg, E.; Zimmermann, D. Electrofused giant protoplasts of Saccharomyces cerevisiae as a novel system for electrophysiological studies on membrane proteins. Biochim. Biophys. Acta 2008, 1778, 1493–1500. [Google Scholar] [CrossRef]
- Mayer, L.D.; Hope, M.J.; Cullis, P.R. Vesicles of variable sizes produced by a rapid extrusion procedure. Biochim. Biophys. Acta 1986, 858, 161–168. [Google Scholar] [CrossRef]
- Warschawski, D.E.; Arnold, A.A.; Beaugrand, M.; Gravel, A.; Chartrand, É.; Marcotte, I. Choosing membrane mimetics for NMR structural studies of transmembrane proteins. Biochim. Biophys. Acta 2011, 1808, 1957–1974. [Google Scholar] [CrossRef]
- Cladera, J.; O’Shea, P. Intramembrane molecular dipoles affect the membrane insertion and folding of a model amphiphilic peptide. Biophys. J. 1998, 74, 2434–2442. [Google Scholar] [CrossRef]
- Domingues, M.M.; Castanho, M.A.; Santos, N.C. rBPI(21) promotes lipopolysaccharide aggregation and exerts its antimicrobial effects by (hemi)fusion of PG-containing membranes. PLoS ONE 2009, 4, e8385. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).