Regulatory RNAs: A Universal Language for Inter-Domain Communication
Abstract
1. Regulatory RNAs in Eukaryotes
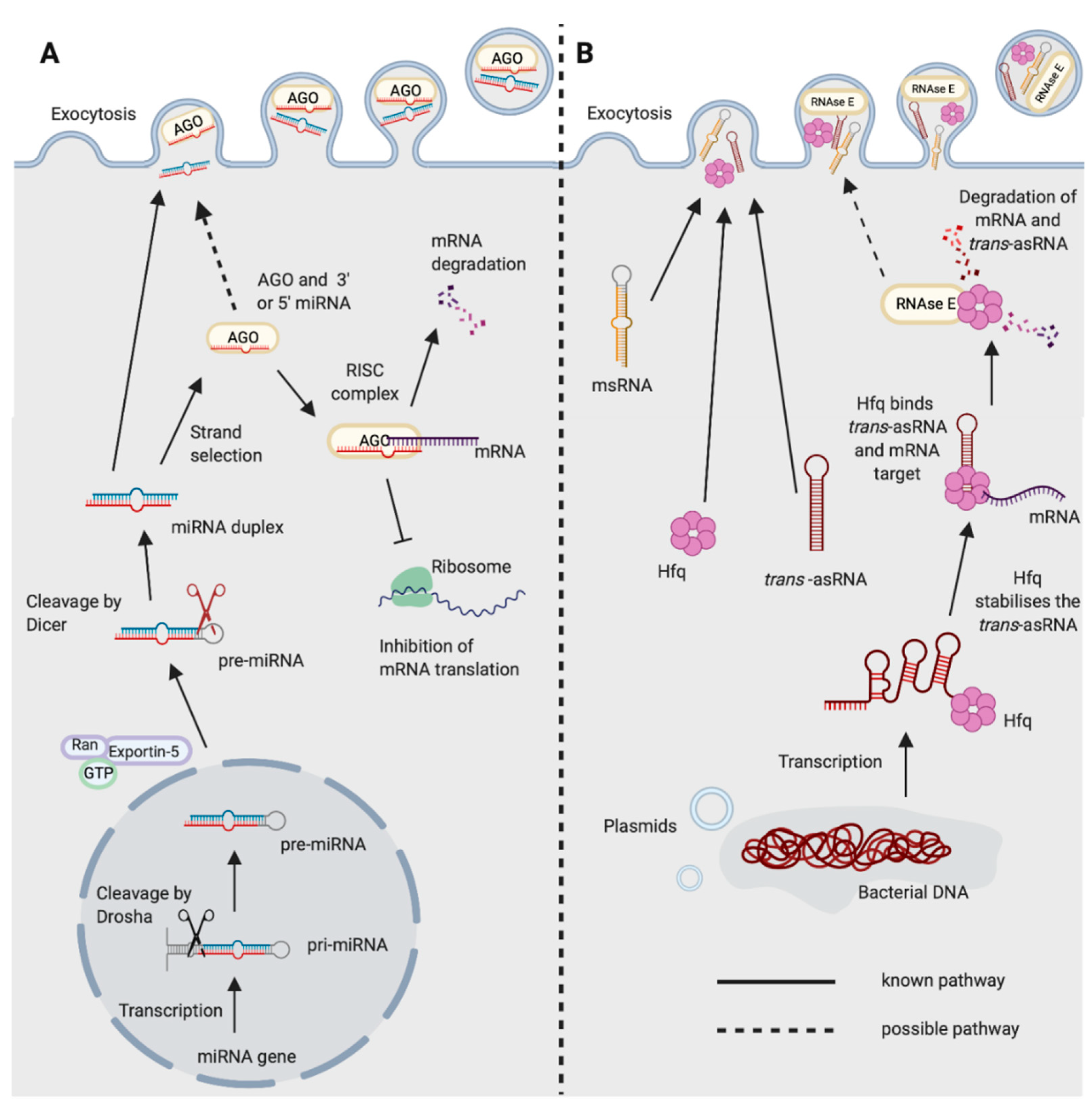
1.1. miRNA Biogenesis
1.2. miRNAs in Disease
1.3. miRNAs and Bacteria
2. Regulatory RNAs in Bacteria
2.1. Bacterial Regulatory RNAs
2.2. Cis-Acting asRNAs
2.3. Trans-Acting asRNAs
2.4. asRNA-Mediated Regulation in Gram-Positive and Gram-Negative Bacteria
2.5. Trans-asRNAs in CRISPR
3. Shared Features of Eukaryotic and Bacterial Regulatory RNA Pathways
3.1. A Conserved Protein Fold between AGO, PIWI and Cas
3.2. CRISPR RNA and piRNA-Guided Immunity
3.3. Shared Features of mRNA Decay in Eukaryotes and Bacteria
3.4. Bacterial Hfq is Reminiscent of Eukaryotic Decapping Proteins
4. miRNAs Facilitate Inter-Kingdom Communications
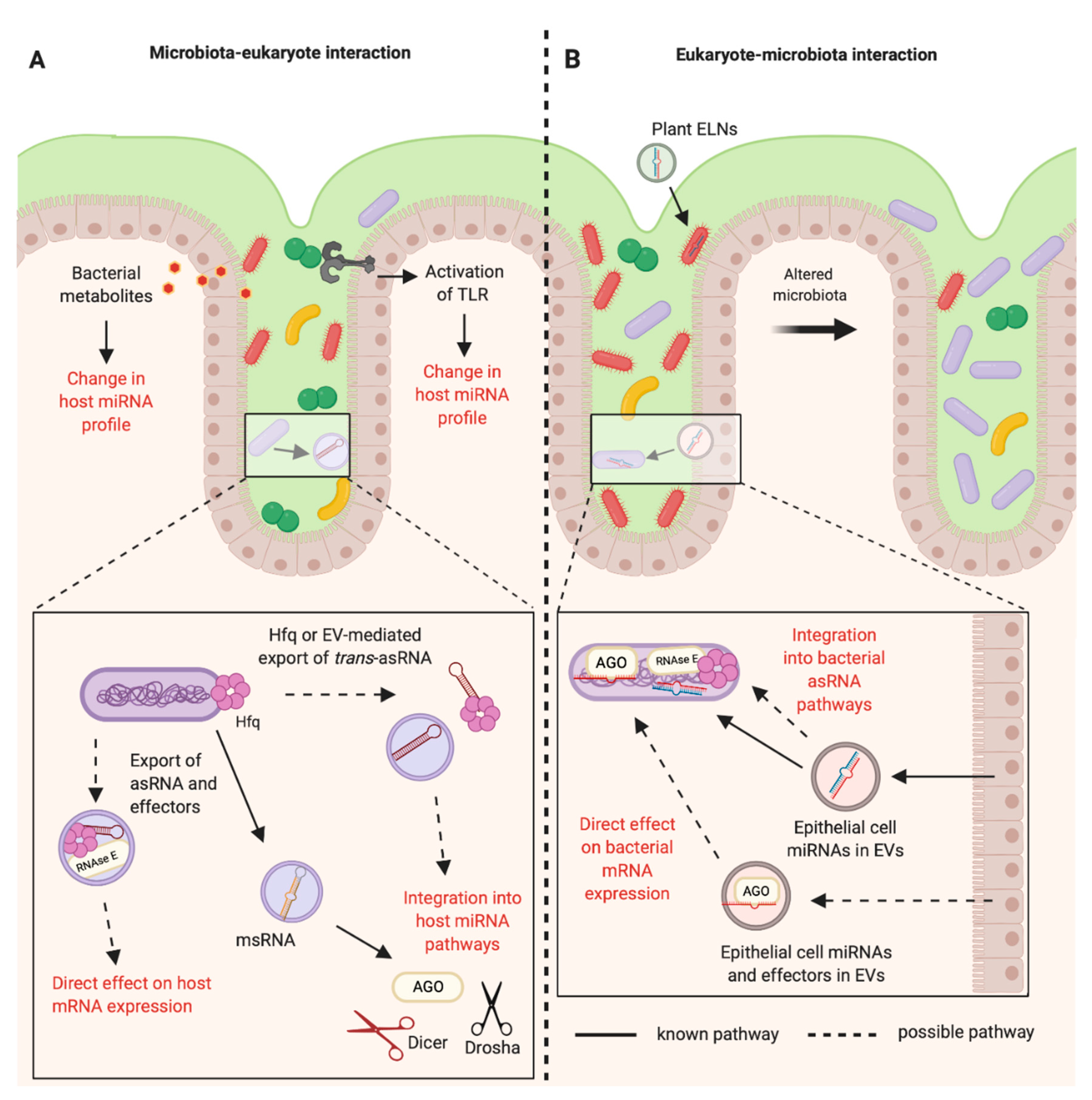
4.1. EVs in Eukaryotes and Bacteria
4.2. Plant miRNAs in Inter-Kingdom Communications
4.3. Human and Plant miRNAs in EVs Can Modulate the Gut Microbiota
4.4. The Microbiota Can Modulate Host miRNA Expression
5. Mechanisms for RNA-based Communication between Eukaryotes and Bacteria
5.1. Bacterial asRNAs Can Hijack Eukaryotic RNAi Pathways
5.2. Proposal of Mechanisms for Regulatory RNAs in Inter-domain Communications
6. Future Directions
7. Concluding Remarks
Authors Contribution
Funding
Conflicts of Interest
References
- Fang, Y.; Fullwood, M. Roles, Functions, and mechanisms of long non-coding RNAs in cancer. Genom. Proteom. Bioinform. 2016, 14, 42–54. [Google Scholar] [CrossRef]
- Maston, G.; Evans, S.; Green, M. Transcriptional regulatory elements in the human genome. Annu. Rev. Genom. Hum. Genet. 2006, 7, 29–59. [Google Scholar] [CrossRef]
- Carthew, R.; Sontheimer, E. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef] [PubMed]
- Lam, J.; Chow, M.; Zhang, Y.; Leung, S. siRNA versus miRNA as therapeutics for gene silencing. Mol. Ther. Nucleic Acids 2015, 4, e252. [Google Scholar] [CrossRef] [PubMed]
- Sijen, T.; Fleenor, J.; Simmer, F.; Thijssen, K.; Parrish, S.; Timmons, L.; Plasterk, R.; Fire, A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 2001, 107, 465–476. [Google Scholar] [CrossRef]
- Zhang, C.; Ruvkun, G. New insights into siRNA amplification and RNAi. RNA Biol. 2012, 9, 1045–1049. [Google Scholar] [CrossRef]
- Kaul, D.; Khanna, A. Suman Evidence and nature of a novel miRNA encoded by HIV-1. Proc. Indian Acad. Sci. 2006, 72, 91–95. [Google Scholar]
- Zhou, R.; Rana, T. RNA-based mechanisms regulating host-virus interactions. Immunol. Rev. 2013, 253, 97–111. [Google Scholar] [CrossRef]
- Lippman, Z.; Martienssen, R. The role of RNA interference in heterochromatic silencing. Nature 2004, 431, 364–370. [Google Scholar] [CrossRef]
- Shalgi, R.; Pilpel, Y.; Oren, M. Repression of transposable-elements—A microRNA anti-cancer defense mechanism? Trends Genet. 2010, 26, 253–259. [Google Scholar] [CrossRef]
- Castañeda, J.; Genzor, P.; Bortvin, A. piRNAs, transposon silencing, and germline genome integrity. Mutat. Res./Fundam. Mol. Mech. Mutagenesis 2011, 714, 95–104. [Google Scholar]
- Czech, B.; Hannon, G. One loop to rule them all: The Ping-Pong cycle and piRNA-guided silencing. Trends Biochem. Sci. 2016, 41, 324–337. [Google Scholar] [CrossRef] [PubMed]
- Le Thomas, A.; Rogers, A.; Webster, A.; Marinov, G.; Liao, S.; Perkins, E.; Hur, J.; Aravin, A.; Toth, K. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev. 2013, 27, 390–399. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; da Cunha, A.; Rezende, R.; Cialic, R.; Wei, Z.; Bry, L.; Comstock, L.; Gandhi, R.; Weiner, H. The host shapes the gut microbiota via fecal MicroRNA. Cell Host Microbe 2016, 19, 32–43. [Google Scholar] [CrossRef]
- Sundaram, K.; Miller, D.; Kumar, A.; Teng, Y.; Sayed, M.; Mu, J.; Lei, C.; Sriwastva, M.; Zhang, L.; Jun, Y.; et al. Plant-derived exosomal nanoparticles inhibit pathogenicity of Porphyromonas gingivalis. iScience 2019, 21, 308–327. [Google Scholar] [CrossRef]
- Teng, Y.; Ren, Y.; Sayed, M.; Hu, X.; Lei, C.; Kumar, A.; Hutchins, E.; Mu, J.; Deng, Z.; Luo, C.; et al. Plant-derived exosomal MicroRNAs shape the gut microbiota. Cell Host Microbe 2018, 24, 637–652.e8. [Google Scholar] [CrossRef]
- Lee, R.; Feinbaum, R.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Bruscella, P.; Bottini, S.; Baudesson, C.; Pawlotsky, J.; Feray, C.; Trabucchi, M. Viruses and miRNAs: More friends than foes. Front. Microbiol. 2017, 8, 824. [Google Scholar] [CrossRef]
- Lee, H.; Li, L.; Gu, W.; Xue, Z.; Crosthwaite, S.; Pertsemlidis, A.; Lewis, Z.; Freitag, M.; Selker, E.; Mello, C.; et al. Diverse pathways generate MicroRNA-like RNAs and dicer-independent small interfering RNAs in Fungi. Mol. Cell 2010, 38, 803–814. [Google Scholar] [CrossRef]
- Wahid, F.; Shehzad, A.; Khan, T.; Kim, Y. MicroRNAs: Synthesis, mechanism, function, and recent clinical trials. Biochim. Et Biophys. Acta (Bba) Mol. Cell Res. 2010, 1803, 1231–1243. [Google Scholar] [CrossRef]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 2002, 21, 4663–4670. [Google Scholar] [CrossRef]
- Yi, R.; Qin, Y.; Macara, I.; Cullen, B. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003, 17, 3011–3016. [Google Scholar] [CrossRef] [PubMed]
- Kuchenbauer, F.; Mah, S.; Heuser, M.; McPherson, A.; Rüschmann, J.; Rouhi, A.; Berg, T.; Bullinger, L.; Argiropoulos, B.; Morin, R.; et al. Comprehensive analysis of mammalian miRNA* species and their role in myeloid cells. Blood 2011, 118, 3350–3358. [Google Scholar] [CrossRef] [PubMed]
- Rivas, F.; Tolia, N.; Song, J.; Aragon, J.; Liu, J.; Hannon, G.; Joshua-Tor, L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 2005, 12, 340–349. [Google Scholar] [CrossRef]
- Zeng, Y.; Yi, R.; Cullen, B. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc. Natl. Acad. Sci. USA 2003, 100, 9779–9784. [Google Scholar] [CrossRef] [PubMed]
- Vasudevan, S. Posttranscriptional upregulation by MicroRNAs. Wiley Interdiscip. Rev. RNA 2011, 3, 311–330. [Google Scholar] [CrossRef]
- Lewis, B.; Shih, I.; Jones-Rhoades, M.; Bartel, D.; Burge, C. Prediction of mammalian MicroRNA targets. Cell 2003, 115, 787–798. [Google Scholar] [CrossRef]
- Baek, Y.; Jang, K.; Lee, H.; Yoon, S.; Baek, A.; Lee, K.; Kim, D. The bacterial endoribonuclease Rnase E can cleave RNA in the absence of the RNA chaperone Hfq. J. Biol. Chem. 2019, 294, 16465–16478. [Google Scholar] [CrossRef]
- Ikeda, Y.; Yagi, M.; Morita, T.; Aiba, H. Hfq binding at RhlB-recognition region of Rnase E is crucial for the rapid degradation of target mRNAs mediated by sRNAs in Escherichia coli. Mol. Microbiol. 2010, 79, 419–432. [Google Scholar] [CrossRef]
- De Lay, N.; Gottesman, S. Rnase E finds some sRNAs Stimulating. Mol. Cell 2012, 47, 825–826. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Khemici, V.; Poljak, L.; Luisi, B.; Carpousis, A. The Rnase E of Escherichia coli is a membrane-binding protein. Mol. Microbiol. 2008, 70, 799–813. [Google Scholar] [PubMed]
- Dauros-Singorenko, P.; Blenkiron, C.; Phillips, A.; Swift, S. The functional RNA cargo of bacterial membrane vesicles. FEMS Microbiol. Lett. 2018, 365. [Google Scholar] [CrossRef]
- Ghosal, A.; Upadhyaya, B.; Fritz, J.; Heintz-Buschart, A.; Desai, M.; Yusuf, D.; Huang, D.; Baumuratov, A.; Wang, K.; Galas, D.; et al. The extracellular RNA complement of Escherichia coli. MicrobiologyOpen 2015, 4, 252–266. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, X.; Wang, H.; Wu, J.; Ren, J.; Meng, L.; Wu, Q.; Dong, H.; Wu, J.; Kao, T.; et al. Escherichia coli noncoding RNAs can affect gene expression and physiology of Caenorhabditis elegans. Nat. Commun. 2012, 3, 1073. [Google Scholar] [CrossRef] [PubMed]
- Eddy, J.; Gielda, L.; Caulfield, A.; Rangel, S.; Lathem, W. Production of outer membrane vesicles by the plague pathogen Yersinia pestis. PloS ONE 2014, 9, e107002. [Google Scholar] [CrossRef]
- Friedman, R.; Farh, K.; Burge, C.; Bartel, D. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2008, 19, 92–105. [Google Scholar] [CrossRef]
- Mirbase.org. Mirbase. 2020. Available online: http://www.mirbase.org/summary.shtml?org=hsa (accessed on 10 April 2020).
- Colamatteo, A.; Micillo, T.; Bruzzaniti, S.; Fusco, C.; Garavelli, S.; De Rosa, V.; Galgani, M.; Spagnuolo, M.; Di Rella, F.; Puca, A.; et al. Metabolism and autoimmune responses: The microRNA connection. Front. Immunol. 2019, 10, 1969. [Google Scholar] [CrossRef]
- Hwang, H.; Mendell, J. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br. J. Cancer 2006, 94, 776–780. [Google Scholar] [CrossRef]
- Ivey, K.; Srivastava, D. microRNAs as Developmental Regulators. Cold Spring Harb. Perspect. Biol. 2015, 7, a008144. [Google Scholar] [CrossRef]
- Slattery, M.; Mullany, L.; Sakoda, L.; Wolff, R.; Samowitz, W.; Herrick, J. Dysregulated genes and miRNAs in the apoptosis pathway in colorectal cancer patients. Apoptosis 2018, 23, 237–250. [Google Scholar] [CrossRef] [PubMed]
- Smith-Vikos, T.; Slack, F. MicroRNAs and their roles in aging. J. Cell Sci. 2012, 125, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Nicot, C. miR-28-3p Is a cellular restriction factor that inhibits human T cell leukemia virus, type 1 (HTLV-1) replication and virus infection. J. Biol. Chem. 2015, 290, 5381–5390. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Ji, Y.; Li, A.; Zhang, Q.; Song, W.; Li, Y.; Huang, H.; Qian, J.; Zhai, A.; Yu, X.; et al. MiR-122 directly inhibits human papillomavirus E6 gene and enhances interferon signaling through blocking suppressor of cytokine signaling 1 in SiHa cells. PloS ONE 2014, 9, e108410. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Li, X.; Wu, M. miRNAs reshape immunity and inflammatory responses in bacterial infection. Signal Transduct. Target. Ther. 2018, 3, 1–13. [Google Scholar] [CrossRef]
- Gu, H.; Zhao, C.; Zhang, T.; Liang, H.; Wang, X.; Pan, Y.; Chen, X.; Zhao, Q.; Li, D.; Liu, F.; et al. Salmonella produce microRNA-like RNA fragment Sal-1 in the infected cells to facilitate intracellular survival. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef]
- Hariharan, M.; Scaria, V.; Pillai, B.; Brahmachari, S. Targets for human encoded microRNAs in HIV genes. Biochem. Biophys. Res. Commun. 2005, 337, 1214–1218. [Google Scholar] [CrossRef]
- Kaul, D.; Ahlawat, A.; Gupta, S. HIV-1 genome-encoded hiv1-mir-H1 impairs cellular responses to infection. Mol. Cell. Biochem. 2008, 323, 143–148. [Google Scholar] [CrossRef]
- Kogan, M.; Rappaport, J. HIV-1 accessory protein Vpr: Relevance in the pathogenesis of HIV and potential for therapeutic intervention. Retrovirology 2011, 8, 25. [Google Scholar] [CrossRef]
- Piedade, D.; Azevedo-Pereira, J. The role of microRNAs in the pathogenesis of herpes virus infection. Viruses 2016, 8, 156. [Google Scholar] [CrossRef]
- Hanna, J.; Hossain, G.; Kocerha, J. The potential for microRNA therapeutics and clinical research. Front. Genet. 2019, 10, 478. [Google Scholar] [CrossRef] [PubMed]
- De Palma, G.; Lynch, M.; Lu, J.; Dang, V.; Deng, Y.; Jury, J.; Umeh, G.; Miranda, P.; Pigrau Pastor, M.; Sidani, S.; et al. Transplantation of fecal microbiota from patients with irritable bowel syndrome alters gut function and behavior in recipient mice. Sci. Transl. Med. 2017, 9, eaaf6397. [Google Scholar] [CrossRef] [PubMed]
- Evans, S.; Bassis, C.; Hein, R.; Assari, S.; Flowers, S.; Kelly, M.; Young, V.; Ellingrod, V.; McInnis, M. The gut microbiome composition associates with bipolar disorder and illness severity. J. Psychiatr. Res. 2017, 87, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Pinto-Sanchez, M.; Hall, G.; Ghajar, K.; Nardelli, A.; Bolino, C.; Lau, J.; Martin, F.; Cominetti, O.; Welsh, C.; Rieder, A.; et al. Probiotic Bifidobacterium longum NCC3001 reduces depression scores and alters brain activity: A pilot study in patients with irritable bowel syndrome. Gastroenterology 2017, 153, 448–459.e8. [Google Scholar] [CrossRef]
- Zheng, P.; Zeng, B.; Zhou, C.; Liu, M.; Fang, Z.; Xu, X.; Zeng, L.; Chen, J.; Fan, S.; Du, X.; et al. Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host’s metabolism. Mol. Psychiatry 2016, 21, 786–796. [Google Scholar] [CrossRef]
- Saberi, F.; Kamali, M.; Najafi, A.; Yazdanparast, A.; Moghaddam, M. Natural antisense RNAs as mRNA regulatory elements in bacteria: A review on function and applications. Cell. Mol. Biol. Lett. 2016, 21, 6. [Google Scholar] [CrossRef]
- Ellis, M.; Haniford, D. Riboregulation of bacterial and archaeal transposition. Wiley Interdiscip. Rev. RNA 2016, 7, 382–398. [Google Scholar] [CrossRef]
- Holmqvist, E.; Wagner, E. Impact of bacterial sRNAs in stress responses. Biochem. Soc. Trans. 2017, 45, 1203–1212. [Google Scholar] [CrossRef]
- Olaya-Abril, A.; Luque-Almagro, V.; Pérez, M.; López, C.; Amil, F.; Cabello, P.; Sáez, L.; Moreno-Vivián, C.; Roldán, M. Putative small RNAs controlling detoxification of industrial cyanide-containing wastewaters by Pseudomonas pseudoalcaligenes CECT5344. PLoS ONE 2019, 14, e0212032. [Google Scholar] [CrossRef]
- Saramago, M.; Bárria, C.; Arraiano, C.; Domingues, S. Ribonucleases, antisense RNAs and the control of bacterial plasmids. Plasmid 2015, 78, 26–36. [Google Scholar] [CrossRef]
- Shao, Y.; Bassler, B. Quorum-sensing non-coding small RNAs use unique pairing regions to differentially control mRNA targets. Mol. Microbiol. 2012, 83, 599–611. [Google Scholar] [CrossRef] [PubMed]
- Kawai, G. Translational control by antisense RNA, bacteria. Encycl. Syst. Biol. 2013, 2282–2285. [Google Scholar] [CrossRef]
- Gottesman, S.; Storz, G. Bacterial small RNA regulators: Versatile roles and rapidly evolving variations. Cold Spring Harb. Perspect. Biol. 2010, 3, a003798. [Google Scholar] [CrossRef] [PubMed]
- Brantl, S. Regulatory mechanisms employed by cis-encoded antisense RNAs. Curr. Opin. Microbiol. 2007, 10, 102–109. [Google Scholar] [CrossRef]
- Georg, J.; Hess, W. cis-antisense RNA, another level of gene regulation in bacteria. Microbiol. Mol. Biol. Rev. 2011, 75, 286–300. [Google Scholar] [CrossRef]
- Rath, D.; Amlinger, L.; Rath, A.; Lundgren, M. The CRISPR-Cas immune system: Biology, mechanisms and applications. Biochimie 2015, 117, 119–128. [Google Scholar] [CrossRef]
- Dugar, G.; Leenay, R.; Eisenbart, S.; Bischler, T.; Aul, B.; Beisel, C.; Sharma, C. CRISPR RNA-dependent binding and cleavage of endogenous RNAs by the Campylobacter jejuni Cas9. Mol. Cell 2018, 69, 893–905.e7. [Google Scholar] [CrossRef]
- Lee, E.; Groisman, E. An antisense RNA that governs the expression kinetics of a multifunctional virulence gene. Mol. Microbiol. 2010, 76, 1020–1033. [Google Scholar] [CrossRef]
- Lioliou, E.; Sharma, C.; Caldelari, I.; Helfer, A.; Fechter, P.; Vandenesch, F.; Vogel, J.; Romby, P. Global regulatory functions of the Staphylococcus aureus endoribonuclease III in gene expression. PLoS Genet. 2012, 8, e1002782. [Google Scholar] [CrossRef]
- Sayed, N.; Jousselin, A.; Felden, B. A cis-antisense RNA acts in trans in Staphylococcus aureus to control translation of a human cytolytic peptide. Nat. Struct. Mol. Biol. 2011, 19, 105–112. [Google Scholar] [CrossRef]
- Šetinová, D.; Šmídová, K.; Pohl, P.; Musić, I.; Bobek, J. RNase III-Binding-mRNAs revealed novel complementary transcripts in streptomyces. Front. Microbiol. 2018, 8, 2693. [Google Scholar] [CrossRef] [PubMed]
- Stazic, D.; Lindell, D.; Steglich, C. Antisense RNA protects mRNA from RNase E degradation by RNA–RNA duplex formation during phage infection. Nucleic Acids Res. 2011, 39, 4890–4899. [Google Scholar] [CrossRef] [PubMed]
- Cho, K. The structure and function of the gram-positive bacterial RNA degradosome. Front. Microbiol. 2017, 8, 154. [Google Scholar] [CrossRef] [PubMed]
- Durand, S.; Gilet, L.; Bessières, P.; Nicolas, P.; Condon, C. Three essential ribonucleases—Rnase Y, J1, and III—Control the abundance of a majority of Bacillus subtilis mRNAs. PloS Genet. 2012, 8, e1002520. [Google Scholar] [CrossRef]
- Viegas, S.; Silva, I.; Saramago, M.; Domingues, S.; Arraiano, C. Regulation of the small regulatory RNA MicA by ribonuclease III: A target-dependent pathway. Nucleic Acids Res. 2010, 39, 2918–2930. [Google Scholar] [CrossRef]
- Holmqvist, E.; Reimegård, J.; Sterk, M.; Grantcharova, N.; Römling, U.; Wagner, E. Two antisense RNAs target the transcriptional regulator CsgD to inhibit curli synthesis. EMBO J. 2010, 29, 1840–1850. [Google Scholar] [CrossRef]
- Moll, I.; Afonyushkin, T.; Vytvytska, O.; Kaberdin, V.; BLÄSI, U. Coincident Hfq binding and RNase E cleavage sites on mRNA and small regulatory RNAs. RNA 2003, 9, 1308–1314. [Google Scholar] [CrossRef]
- Malabirade, A.; Morgado-Brajones, J.; Trépout, S.; Wien, F.; Marquez, I.; Seguin, J.; Marco, S.; Velez, M.; Arluison, V. Membrane association of the bacterial riboregulator Hfq and functional perspectives. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef]
- Faner, M.; Feig, A. Identifying and characterizing Hfq–RNA interactions. Methods 2013, 63, 144–159. [Google Scholar] [CrossRef]
- Nielsen, J.; Lei, L.; Ebersbach, T.; Olsen, A.; Klitgaard, J.; Valentin-Hansen, P.; Kallipolitis, B. Defining a role for Hfq in Gram-positive bacteria: Evidence for Hfq-dependent antisense regulation in Listeria monocytogenes. Nucleic Acids Res. 2009, 38, 907–919. [Google Scholar] [CrossRef]
- Chao, Y.; Vogel, J. The role of Hfq in bacterial pathogens. Curr. Opin. Microbiol. 2010, 13, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Davis, B.; Waldor, M. Hfq is essential for Vibrio cholerae virulence and downregulates σE expression. Mol. Microbiol. 2004, 53, 345–354. [Google Scholar] [CrossRef] [PubMed]
- Sittka, A.; Pfeiffer, V.; Tedin, K.; Vogel, J. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol. Microbiol. 2007, 63, 193–217. [Google Scholar] [CrossRef] [PubMed]
- Condon, C.; Putzer, H. The phylogenetic distribution of bacterial ribonucleases. Nucleic Acids Res. 2002, 30, 5339–5346. [Google Scholar] [CrossRef]
- Deltcheva, E.; Chylinski, K.; Sharma, C.; Gonzales, K.; Chao, Y.; Pirzada, Z.; Eckert, M.; Vogel, J.; Charpentier, E. CRISPR RNA maturation by trans-encoded small RNA and host factor Rnase III. Nature 2011, 471, 602–607. [Google Scholar] [CrossRef]
- Karvelis, T.; Gasiunas, G.; Miksys, A.; Barrangou, R.; Horvath, P.; Siksnys, V. crRNA and tracrRNA guide Cas9-mediated DNA interference in Streptococcus thermophilus. RNA Biol. 2013, 10, 841–851. [Google Scholar] [CrossRef]
- Moelling, K.; Broecker, F. The reverse transcriptase-RNase H: From viruses to antiviral defense. Ann. N. Y. Acad. Sci. 2015, 1341, 126–135. [Google Scholar] [CrossRef]
- Moelling, K.; Broecker, F.; Russo, G.; Sunagawa, S. RNase H as gene modifier, driver of evolution and antiviral defense. Front. Microbiol. 2017, 8, 1745. [Google Scholar] [CrossRef]
- Majorek, K.; Dunin-Horkawicz, S.; Steczkiewicz, K.; Muszewska, A.; Nowotny, M.; Ginalski, K.; Bujnicki, J. The RNase H-like superfamily: New members, comparative structural analysis and evolutionary classification. Nucleic Acids Res. 2014, 42, 4160–4179. [Google Scholar] [CrossRef]
- Tóth, K.; Pezic, D.; Stuwe, E.; Webster, A. The piRNA pathway guards the germline genome against transposable elements. Non-Coding RNA Reprod. Syst. 2015, 886, 51–77. [Google Scholar]
- Yamanaka, S.; Siomi, M.; Siomi, H. piRNA clusters and open chromatin structure. Mob. DNA 2014, 5, 22. [Google Scholar] [CrossRef] [PubMed]
- Horie, M.; Honda, T.; Suzuki, Y.; Kobayashi, Y.; Daito, T.; Oshida, T.; Ikuta, K.; Jern, P.; Gojobori, T.; Coffin, J.; et al. Endogenous non-retroviral RNA virus elements in mammalian genomes. Nature 2010, 463, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Ophinni, Y.; Palatini, U.; Hayashi, Y.; Parrish, N. piRNA-guided CRISPR-like immunity in eukaryotes. Trends Immunol. 2019, 40, 998–1010. [Google Scholar] [CrossRef] [PubMed]
- Aravin, A.; Bourc’his, D. Small RNA guides for de novo DNA methylation in mammalian germ cells. Genes Dev. 2008, 22, 970–975. [Google Scholar] [CrossRef]
- Gallie, D. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991, 5, 2108–2116. [Google Scholar] [CrossRef]
- Kushner, S. mRNA Decay in prokaryotes and eukaryotes: Different approaches to a similar problem. Int. Union Biochem. Mol. Biol. Life 2004, 56, 585–594. [Google Scholar] [CrossRef]
- Laalami, S.; Zig, L.; Putzer, H. Initiation of mRNA decay in bacteria. Cell. Mol. Life Sci. 2013, 71, 1799–1828. [Google Scholar] [CrossRef]
- Deana, A.; Celesnik, H.; Belasco, J. The bacterial enzyme RppH triggers messenger RNA degradation by 5′ pyrophosphate removal. Nature 2008, 451, 355–358. [Google Scholar] [CrossRef]
- Dunckley, T.; Parker, R. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 1999, 18, 5411–5422. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, D.; Guan, Z.; Li, D.; Pei, K.; Liu, J.; Zou, T.; Yin, P. DapF stabilizes the substrate-favoring conformation of RppH to stimulate its RNA-pyrophosphohydrolase activity in Escherichia Coli. Nucleic Acids Res. 2018, 46, 6880–6892. [Google Scholar] [CrossRef]
- Rehwinkel, J.; Behm-Ansmant, I.; Gatfield, D.; Izaurralde, E. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA 2005, 11, 1640–1647. [Google Scholar] [CrossRef] [PubMed]
- Hoekzema, M.; Romilly, C.; Holmqvist, E.; Wagner, E. Hfq-dependent mRNA unfolding promotes sRNA -based inhibition of translation. EMBO J. 2019, 38, e101199. [Google Scholar] [CrossRef] [PubMed]
- Møller, T.; Franch, T.; Højrup, P.; Keene, D.; Bächinger, H.; Brennan, R.; Valentin-Hansen, P. Hfq: A bacterial Sm-like protein that mediates RNA-RNA interaction. Mol. Cell 2002, 9, 23–30. [Google Scholar] [CrossRef]
- Sauter, C.; Basquin, J.; Suck, D. Sm-like proteins in Eubacteria: The crystal structure of the Hfq protein from Escherichia Coli. Nucleic Acids Res. 2003, 31, 4091–4098. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, A.; Tharun, S. Activation of decapping involves binding of the mRNA and facilitation of the post-binding steps by the Lsm1-7-Pat1 complex. RNA 2009, 15, 1837–1848. [Google Scholar] [CrossRef][Green Version]
- Woith, E.; Fuhrmann, G.; Melzig, M. Extracellular vesicles—Connecting Kingdoms. Int. J. Mol. Sci. 2019, 20, 5695. [Google Scholar] [CrossRef]
- Gill, S.; Catchpole, R.; Forterre, P. Extracellular membrane vesicles in the three domains of life and beyond. Fems Microbiol. Rev. 2018, 43, 273–303. [Google Scholar] [CrossRef]
- Kawamura, Y.; Yamamoto, Y.; Sato, T.; Ochiya, T. Extracellular vesicles as trans-genomic agents: Emerging roles in disease and evolution. Cancer Sci. 2017, 108, 824–830. [Google Scholar] [CrossRef]
- Renelli, M.; Matias, V.; Lo, R.; Beveridge, T. DNA-containing membrane vesicles of Pseudomonas aeruginosa PAO1 and their genetic transformation potential. Microbiology 2004, 150, 2161–2169. [Google Scholar] [CrossRef]
- Yaron, S.; Kolling, G.; Simon, L.; Matthews, K. Vesicle-mediated transfer of virulence genes from Escherichia coli O157:H7 to other enteric bacteria. Appl. Environ. Microbiol. 2000, 66, 4414–4420. [Google Scholar] [CrossRef]
- Malabirade, A.; Habier, J.; Heintz-Buschart, A.; May, P.; Godet, J.; Halder, R.; Etheridge, A.; Galas, D.; Wilmes, P.; Fritz, J. The RNA complement of outer membrane vesicles from Salmonella enterica serovar typhimurium under distinct culture conditions. Front. Microbiol. 2018, 9, 2015. [Google Scholar] [CrossRef] [PubMed]
- Resch, U.; Tsatsaronis, J.; Le Rhun, A.; Stübiger, G.; Rohde, M.; Kasvandik, S.; Holzmeister, S.; Tinnefeld, P.; Wai, S.; Charpentier, E. A two-component regulatory system impacts extracellular membrane-derived vesicle production in group A streptococcus. mBio 2016, 7, e00207-16. [Google Scholar] [CrossRef] [PubMed]
- Buck, A.; Coakley, G.; Simbari, F.; McSorley, H.; Quintana, J.; Le Bihan, T.; Kumar, S.; Abreu-Goodger, C.; Lear, M.; Harcus, Y.; et al. Exosomes secreted by nematode parasites transfer small RNAs to mammalian cells and modulate innate immunity. Nat. Commun. 2014, 5, 5488. [Google Scholar] [CrossRef] [PubMed]
- Shears, R.; Bancroft, A.; Hughes, G.; Grencis, R.; Thornton, D. Extracellular vesicles induce protective immunity against Trichuris muris. Parasite Immunol. 2018, 40, e12536. [Google Scholar] [CrossRef]
- Wang, L.; Li, Z.; Shen, J.; Liu, Z.; Liang, J.; Wu, X.; Sun, X.; Wu, Z. Exosome-like vesicles derived by Schistosoma japonicum adult worms mediates M1 type immune- activity of macrophage. Parasitol. Res. 2015, 114, 1865–1873. [Google Scholar] [CrossRef]
- Zhang, L.; Hou, D.; Chen, X.; Li, D.; Zhu, L.; Zhang, Y.; Li, J.; Bian, Z.; Liang, X.; Cai, X.; et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: Evidence of cross-kingdom regulation by microRNA. Cell Res. 2011, 22, 107–126. [Google Scholar] [CrossRef]
- Liang, G.; Zhu, Y.; Sun, B.; Shao, Y.; Jing, A.; Wang, J.; Xiao, Z. Assessing the survival of exogenous plant microRNA in mice. Food Sci. Nutr. 2014, 2, 380–388. [Google Scholar] [CrossRef]
- Liang, H.; Zhang, S.; Fu, Z.; Wang, Y.; Wang, N.; Liu, Y.; Zhao, C.; Wu, J.; Hu, Y.; Zhang, J.; et al. Effective detection and quantification of dietetically absorbed plant microRNAs in human plasma. J. Nutr. Biochem. 2015, 26, 505–512. [Google Scholar] [CrossRef]
- Li, Z.; Xu, R.; Li, N. MicroRNAs from plants to animals, do they define a new messenger for communication? Nutr. Metab. 2018, 15, 68. [Google Scholar] [CrossRef]
- Mu, J.; Zhuang, X.; Wang, Q.; Jiang, H.; Deng, Z.; Wang, B.; Zhang, L.; Kakar, S.; Jun, Y.; Miller, D.; et al. Interspecies communication between plant and mouse gut host cells through edible plant derived exosome-like nanoparticles. Mol. Nutr. Food Res. 2014, 58, 1561–1573. [Google Scholar] [CrossRef]
- Islam, W.; Noman, A.; Qasim, M.; Wang, L. Plant responses to pathogen attack: Small RNAs in focus. Int. J. Mol. Sci. 2018, 19, 515. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.; Feng, T.; Yao, S.; Wolf, K.; Liu, C.; Liu, X.; Elson, C.; Cong, Y. Microbiota downregulates dendritic cell expression of miR-10a, which targets IL-12/IL-23p40. J. Immunol. 2011, 187, 5879–5886. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Al-Maghout, T.; Cao, H.; Pelzl, L.; Salker, M.; Veldhoen, M.; Cheng, A.; Lang, F.; Singh, Y. Gut bacterial metabolite Urolithin A (UA) mitigates Ca2+ entry in T cells by regulating miR-10a-5p. Front. Immunol. 2019, 10, 1737. [Google Scholar] [CrossRef] [PubMed]
- Dang, T.; Tyagi, S.; D’Cunha, G.; Bhave, M.; Crawford, R.; Ivanova, E. Computational prediction of microRNAs in marine bacteria of the genus Thalassospira. PloS ONE 2019, 14, e0212996. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Choi, J.; Lee, Y.; Hong, S.; Lee, H. Identification of microRNA-size, small RNAs in Escherichia coli. Curr. Microbiol. 2013, 67, 609–613. [Google Scholar] [CrossRef]
- Lee, H.; Hong, S. Analysis of microRNA-size, small RNAs in Streptococcus mutans by deep sequencing. FEMS Microbiol. Lett. 2011, 326, 131–136. [Google Scholar] [CrossRef]
- Furuse, Y.; Finethy, R.; Saka, H.; Xet-Mull, A.; Sisk, D.; Smith, K.; Lee, S.; Coers, J.; Valdivia, R.; Tobin, D.; et al. Search for MicroRNAs expressed by intracellular bacterial pathogens in infected mammalian cells. PLoS ONE 2014, 9, e106434. [Google Scholar] [CrossRef]
- Choi, J.; Kim, S.; Hong, S.; Lee, H. Secretable small RNAs via outer membrane vesicles in periodontal pathogens. J. Dent. Res. 2017, 96, 458–466. [Google Scholar] [CrossRef]
- Cekici, A.; Kantarci, A.; Hasturk, H.; Van Dyke, T. Inflammatory and immune pathways in the pathogenesis of periodontal disease. Periodontology 2000 2013, 64, 57–80. [Google Scholar] [CrossRef]
- Raman, P.; Zaghab, S.; Traver, E.; Jose, A. The double-stranded RNA binding protein RDE-4 can act cell autonomously during feeding RNAi in C. elegans. Nucleic Acids Res. 2017, 45, 8463–8473. [Google Scholar] [CrossRef]
- Liu, J.; Bittker, J.; Lonshteyn, M.; Liu, D. Functional dissection of sRNA translational regulators by nonhomologous random recombination and In Vivo selection. Chem. Biol. 2005, 12, 757–767. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kuo, H.; Chao, H.; Liao, P.; Hsu, L.; Chang, K.; Tung, C.; Chen, C.; Liou, M. Functional characterization of Acinetobacter baumannii lacking the RNA chaperone Hfq. Front. Microbiol. 2017, 8, 2068. [Google Scholar] [CrossRef] [PubMed]
- Chow, F.; Koutsovoulos, G.; Ovando-Vázquez, C.; Neophytou, K.; Bermúdez-Barrientos, J.; Laetsch, D.; Robertson, E.; Kumar, S.; Claycomb, J.; Blaxter, M.; et al. Secretion of an Argonaute protein by a parasitic nematode and the evolution of its siRNA guides. Nucleic Acids Res. 2019, 47, 3594–3606. [Google Scholar] [CrossRef] [PubMed]
- Allegra, A.; Musolino, C.; Tonacci, A.; Pioggia, G.; Gangemi, S. Interactions between the MicroRNAs and microbiota in cancer development: Roles and therapeutic opportunities. Cancers 2020, 12, 805. [Google Scholar] [CrossRef]
- Ambrozkiewicz, F.; Karczmarski, J.; Kulecka, M.; Paziewska, A.; Niemira, M.; Zeber-Lubecka, N.; Zagorowicz, E.; Kretowski, A.; Ostrowski, J. In search for interplay between stool microRNAs, microbiota and short chain fatty acids in Crohn’s disease—A preliminary study. BMC Gastroenterol. 2020, 20, 1–18. [Google Scholar] [CrossRef]
- Kimura, K.; Hohjoh, H.; Yamamura, T. The role for exosomal microRNAs in disruption of regulatory T cell homeostasis in multiple sclerosis. J. Exp. Neurosci. 2018, 12, 117906951876489. [Google Scholar] [CrossRef]
- Ragusa, M.; Santagati, M.; Mirabella, F.; Lauretta, G.; Cirnigliaro, M.; Brex, D.; Barbagallo, C.; Domini, C.; Gulisano, M.; Barone, R.; et al. Potential associations among alteration of salivary miRNAs, saliva microbiome structure, and cognitive impairments in Autistic children. Int. J. Mol. Sci. 2020, 21, 6203. [Google Scholar] [CrossRef]
| Eukaryotic RNA | Length | Functions | Mechanisms | Distribution | Bacterial RNA | Length | Functions | Mechanisms | Distribution |
|---|---|---|---|---|---|---|---|---|---|
| miRNA | 21–25 nucleotides |
| Imperfect complementarity to target RNase III enzymes for biogenesis (Dicer and Drosha) Typically bind the 3’UTR of target mRNA AGO-dependent degradation and translational repression Chromatin modification through targeting epigenetic factors | Dicer and Drosha-independent mechanisms also exist Found in most eukaryotes | Trans-acting asRNA | Typically long (100s–1000s of nucleotides) |
| Imperfect complementarity to target Usually derived from intergenic regions Can target mRNA or protein Hfq as a chaperone RNase E, RNase Y or RNase III-mediated cleavage | Hfq is only present in some bacteria RNase E is only present in Gram-negative bacteria RNase Y is only present in Gram-positive bacteria |
| siRNA | 22–25 nucleotides |
| Perfect complementarity to target RNase III enzymes for biogenesis (Dicer) AGO-dependent degradation Histone modification | Found in most eukaryotes | Cis-acting asRNA | Mostly short |
| Perfect complementarity to target Located opposite the mRNA it regulates Can be structural inhibition of transcription/translation RNase E, RNase Y or RNase III-mediated cleavage | RNase E is only present in Gram-negative bacteria RNase Y is only present in Gram-positive bacteria |
| piRNA | 24–31 nucleotides |
| Perfect complementarity to target for canonical piRNAs Imperfect complementarity to target for other piRNAs Often derived from piRNA clusters PIWI-dependent degradation | Found in metazoans Biogenesis is highly variable amongst different organisms | crRNA tracrRNA | 20 nucleotide spacers in crRNA Approx. 100 nucleotides in tracrRNA |
| crRNAs have perfect complementarity to target tracrRNAs have imperfect complementarity Derived from CRISPR loci Cas9-dependent degradation | Found in 50% of bacteria 90% of archaea |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Layton, E.; Fairhurst, A.-M.; Griffiths-Jones, S.; Grencis, R.K.; Roberts, I.S. Regulatory RNAs: A Universal Language for Inter-Domain Communication. Int. J. Mol. Sci. 2020, 21, 8919. https://doi.org/10.3390/ijms21238919
Layton E, Fairhurst A-M, Griffiths-Jones S, Grencis RK, Roberts IS. Regulatory RNAs: A Universal Language for Inter-Domain Communication. International Journal of Molecular Sciences. 2020; 21(23):8919. https://doi.org/10.3390/ijms21238919
Chicago/Turabian StyleLayton, Emma, Anna-Marie Fairhurst, Sam Griffiths-Jones, Richard K. Grencis, and Ian S. Roberts. 2020. "Regulatory RNAs: A Universal Language for Inter-Domain Communication" International Journal of Molecular Sciences 21, no. 23: 8919. https://doi.org/10.3390/ijms21238919
APA StyleLayton, E., Fairhurst, A.-M., Griffiths-Jones, S., Grencis, R. K., & Roberts, I. S. (2020). Regulatory RNAs: A Universal Language for Inter-Domain Communication. International Journal of Molecular Sciences, 21(23), 8919. https://doi.org/10.3390/ijms21238919





