DNA Double-Strand Breaks Are a Critical Regulator of Fear Memory Reconsolidation
Abstract
1. Introduction
2. Results
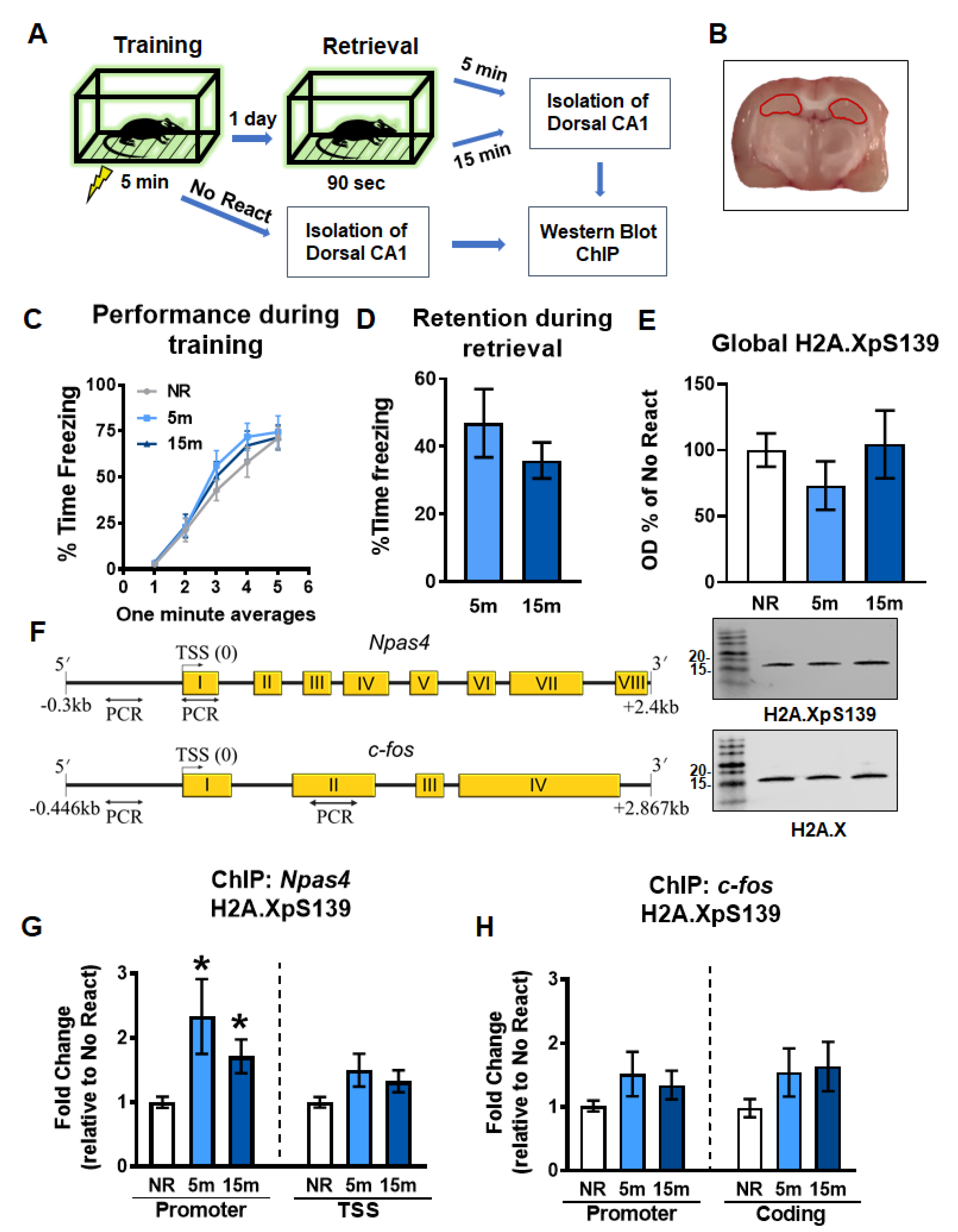
2.1. Gene-Specific Increases in H2A.XpS139 Levels in the Dorsal Hippocampus Following Memory Retrieval
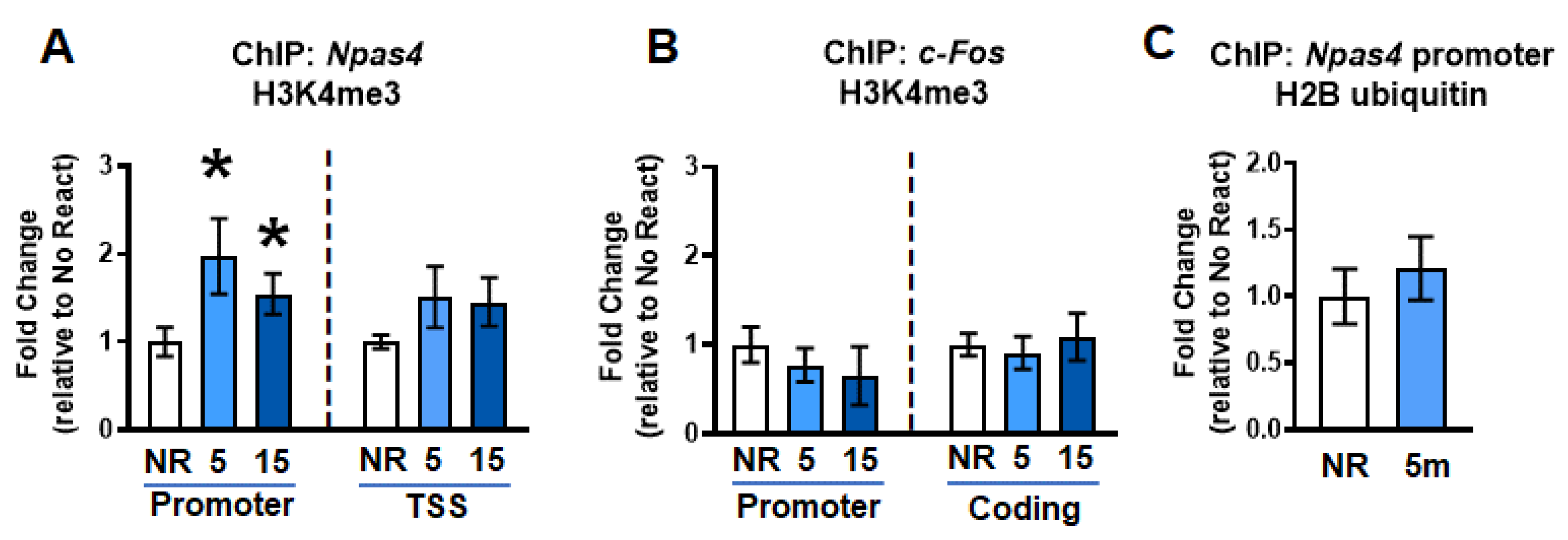
2.2. Gene-Specific H3K4me3 Levels Increase Simultaneously with H2A.XpS139 in the Dorsal Hippocampus Following Retrieval
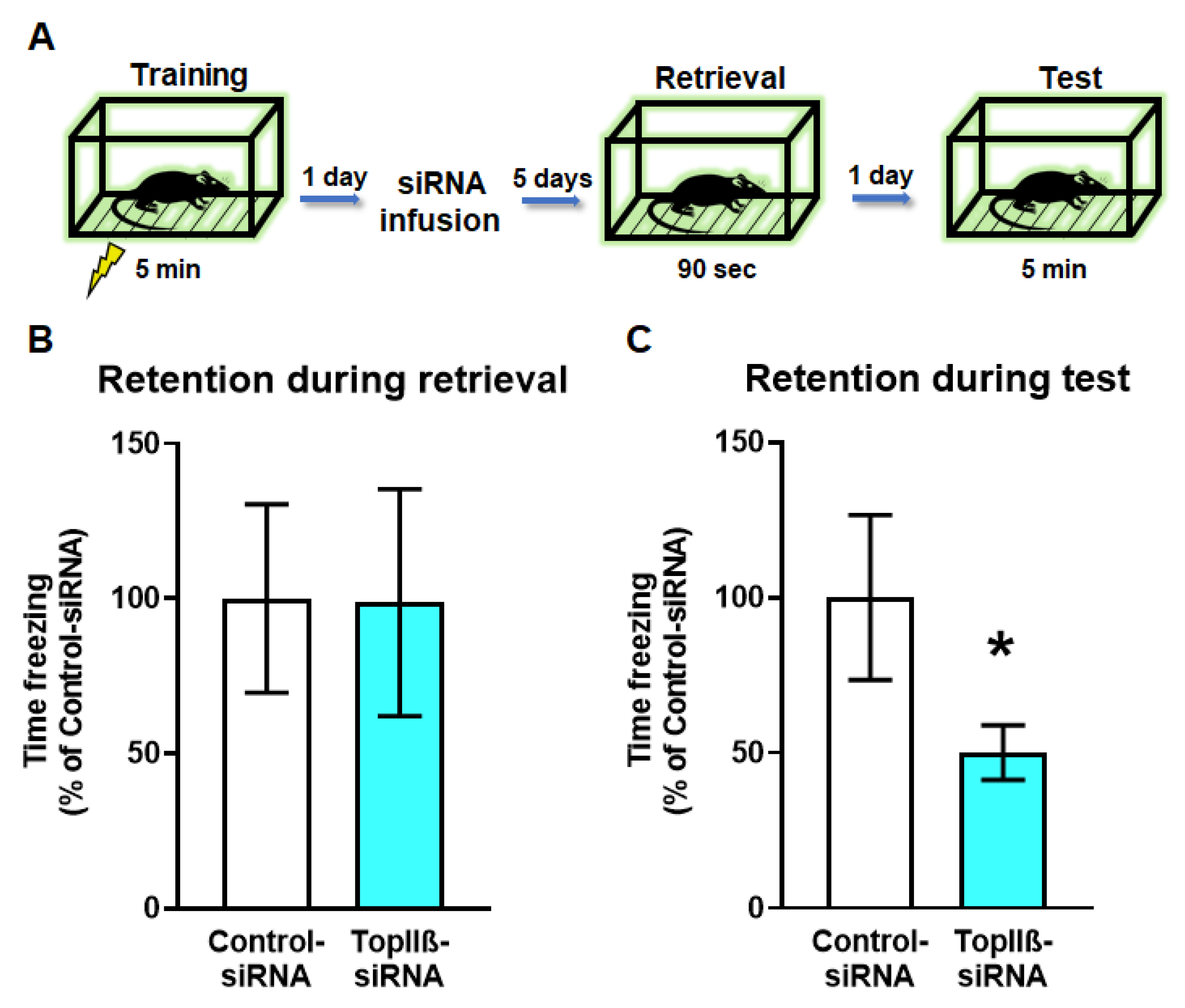
2.3. Knockdown of TopIIβ in the Dorsal Hippocampus Abolishes Retrieval-Induced Increases in Gene-Specific H2A.XpS139 and H3K4me3
2.4. Knockdown of TopIIβ in the Dorsal Hippocampus Prior to Retrieval Impairs Memory Reconsolidation
3. Discussion
4. Materials and Methods
4.1. Animals (Subjects)
4.2. Surgery and siRNA Delivery
4.3. Cell Culture
4.4. cDNA Synthesis and Quantitative Real-Time PCR
4.5. Apparatus
4.6. Behavioral Procedures
4.7. Tissue Collection and Protein Extraction
4.8. Histone Extraction
4.9. Antibodies
4.10. Western Blotting
4.11. Chromatin Immunoprecipitation (ChIP)
4.12. Statistical Analyses
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- McGaugh, J.L. Consolidating Memories. Annu. Rev. Psychol. 2015, 66, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Alberini, C.M.; LeDoux, J.E. Memory reconsolidation. Curr. Biol. 2013, 23, R746–R750. [Google Scholar] [CrossRef] [PubMed]
- Mamiya, N.; Fukushima, H.; Suzuki, A.; Matsuyama, Z.; Homma, S.; Frankland, P.W.; Kida, S. Brain region-specific gene expression activation required for reconsolidation and extinction of contextual fear memory. J. Neurosci. 2009, 29, 402–413. [Google Scholar] [CrossRef] [PubMed]
- Tronson, N.C.; Taylor, J.R. Molecular mechanisms of memory reconsolidation. Nat. Rev. Neurosci. 2007, 8, 262–275. [Google Scholar] [CrossRef]
- Nader, K.; Schafe, G.E.; Le Doux, J.E. Fear memories require protein synthesis in the amygdala for reconsolidation after retrieval. Nature 2000, 406, 722–726. [Google Scholar] [CrossRef]
- Debiec, J.; LeDoux, J.E.; Nader, K.J.N. Cellular and systems reconsolidation in the hippocampus. Neuron 2002, 36, 527–538. [Google Scholar] [CrossRef]
- Lubin, F.D.; Sweatt, J.D. The IkappaB kinase regulates chromatin structure during reconsolidation of conditioned fear memories. Neuron 2007, 55, 942–957. [Google Scholar] [CrossRef] [PubMed]
- Kida, S.; Josselyn, S.A.; de Ortiz, S.P.; Kogan, J.H.; Chevere, I.; Masushige, S.; Silva, A.J. CREB required for the stability of new and reactivated fear memories. Nat. Neurosci. 2002, 5, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Webb, W.M.; Sanchez, R.G.; Perez, G.; Butler, A.A.; Hauser, R.M.; Rich, M.C.; O’Bierne, A.L.; Jarome, T.J.; Lubin, F.D. Dynamic association of epigenetic H3K4me3 and DNA 5hmC marks in the dorsal hippocampus and anterior cingulate cortex following reactivation of a fear memory. Neurobiol. Learn. Mem. 2017, 142, 66–78. [Google Scholar] [CrossRef] [PubMed]
- Ploski, J.E.; Monsey, M.S.; Nguyen, T.; Dileone, R.J.; Schafe, G.E. The Neuronal PAS Domain Protein 4 (Npas4) Is Required for New and Reactivated Fear Memories. PLoS ONE 2011, 6, e23760. [Google Scholar] [CrossRef] [PubMed]
- Weng, F.-J.; Garcia, R.I.; Lutzu, S.; Alviña, K.; Zhang, Y.; Dushko, M.; Ku, T.; Zemoura, K.; Rich, D.; Garcia-Dominguez, D.; et al. Npas4 Is a Critical Regulator of Learning-Induced Plasticity at Mossy Fiber-CA3 Synapses during Contextual Memory Formation. Neuron 2018, 97, 1137–1152.e5. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Lin, Y. Npas4: Linking neuronal activity to memory. Trends Neurosci. 2016, 39, 264–275. [Google Scholar] [CrossRef] [PubMed]
- Madabhushi, R.; Gao, F.; Pfenning, A.R.; Pan, L.; Yamakawa, S.; Seo, J.; Rueda, R.; Phan, T.X.; Yamakawa, H.; Pao, P.-C.; et al. Activity-Induced DNA Breaks Govern the Expression of Neuronal Early-Response Genes. Cell 2015, 161, 1592–1605. [Google Scholar] [CrossRef] [PubMed]
- Ramamoorthi, K.; Fropf, R.; Belfort, G.M.; Fitzmaurice, H.L.; McKinney, R.M.; Neve, R.L.; Otto, T.; Lin, Y. Npas4 regulates a transcriptional program in CA3 required for contextual memory formation. Science 2011, 334, 1669–1675. [Google Scholar] [CrossRef] [PubMed]
- Bredy, T.W.; Barad, M. The histone deacetylase inhibitor valproic acid enhances acquisition, extinction, and reconsolidation of conditioned fear. Learn. Mem. 2008, 15, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Jarome, T.J.; Lubin, F.D. Epigenetic mechanisms of memory formation and reconsolidation. Neurobiol. Learn. Mem. 2014, 115, 116–127. [Google Scholar] [CrossRef]
- Jarome, T.J.; Perez, G.A.; Hauser, R.M.; Hatch, K.M.; Lubin, F.D. EZH2 Methyltransferase Activity Controls Pten Expression and mTOR Signaling during Fear Memory Reconsolidation. J. Neurosci. 2018, 38, 7635–7648. [Google Scholar] [CrossRef]
- Bae, S.; Lesch, B.J. H3K4me1 Distribution Predicts Transcription State and Poising at Promoters. Front. Cell Dev. Biol. 2020, 8, 289. [Google Scholar] [CrossRef]
- Barski, A.; Cuddapah, S.; Cui, K.; Roh, T.-Y.; Schones, D.E.; Wang, Z.; Wei, G.; Chepelev, I.; Zhao, K. High-resolution profiling of histone methylations in the human genome. Cell 2007, 129, 823–837. [Google Scholar] [CrossRef]
- Santos-Rosa, H.; Schneider, R.; Bannister, A.J.; Sherriff, J.; Bernstein, B.E.; Emre, N.C.T.; Schreiber, S.L.; Mellor, J.; Kouzarides, T. Active genes are tri-methylated at K4 of histone H3. Nature 2002, 419, 407–411. [Google Scholar] [CrossRef]
- Gupta, S.; Kim, S.Y.; Artis, S.; Molfese, D.L.; Schumacher, A.; Sweatt, J.D.; Paylor, R.E.; Lubin, F.D. Histone Methylation Regulates Memory Formation. J. Neurosci. 2010, 30, 3589–3599. [Google Scholar] [CrossRef] [PubMed]
- Kerimoglu, C.; Agis-Balboa, R.C.; Kranz, A.; Stilling, R.; Bahari-Javan, S.; Benito-Garagorri, E.; Halder, R.; Burkhardt, S.; Stewart, A.F.; Fischer, A. Histone-Methyltransferase MLL2 (KMT2B) Is Required for Memory Formation in Mice. J. Neurosci. 2013, 33, 3452. [Google Scholar] [CrossRef] [PubMed]
- Champoux, J.J. DNA topoisomerases: Structure, function, and mechanism. Annu. Rev. Biochem. 2001, 70, 369–413. [Google Scholar] [CrossRef] [PubMed]
- Calderwood, S.K. A critical role for topoisomerase IIb and DNA double strand breaks in transcription. Transcription 2016, 7, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Nitiss, J.L. DNA topoisomerase II and its growing repertoire of biological functions. Nat. Rev. Cancer 2009, 9, 327–337. [Google Scholar] [CrossRef]
- Gale, K.; Osheroff, N. Intrinsic intermolecular DNA ligation activity of eukaryotic topoisomerase II. Potential roles in recombination. J. Biol. Chem. 1992, 267, 12090–12097. [Google Scholar]
- Morimoto, S.; Tsuda, M.; Bunch, H.; Sasanuma, H.; Austin, C.A.; Takeda, S. Type II DNA Topoisomerases Cause Spontaneous Double-Strand Breaks in Genomic DNA. Genes 2019, 10, 868. [Google Scholar] [CrossRef]
- Puc, J.; Aggarwal, A.K.; Rosenfeld, M.G. Physiological functions of programmed DNA breaks in signal-induced transcription. Nat. Rev. Mol. Cell Biol. 2017, 18, 471–476. [Google Scholar] [CrossRef]
- Lyu, Y.L.; Lin, C.-P.; Azarova, A.M.; Cai, L.; Wang, J.C.; Liu, L.F. Role of topoisomerase IIbeta in the expression of developmentally regulated genes. Mol. Cell Biol. 2006, 26, 7929–7941. [Google Scholar] [CrossRef]
- Ju, B.-G.; Lunyak, V.V.; Perissi, V.; Garcia-Bassets, I.; Rose, D.W.; Glass, C.K.; Rosenfeld, M.G. A topoisomerase IIß-mediated dsDNA break required for regulated transcription. Science 2006, 312, 1798–1802. [Google Scholar] [CrossRef]
- Tiwari, V.K.; Burger, L.; Nikoletopoulou, V.; Deogracias, R.; Thakurela, S.; Wirbelauer, C.; Kaut, J.; Terranova, R.; Hoerner, L.; Mielke, C.; et al. Target genes of Topoisomerase IIβ regulate neuronal survival and are defined by their chromatin state. Proc. Natl. Acad. Sci. USA 2012, 109, E934–E943. [Google Scholar] [CrossRef] [PubMed]
- Sedelnikova, O.A.; Rogakou, E.P.; Panyutin, I.G.; Bonner, W.M. Quantitative detection of 125IdU-induced DNA double-strand breaks with γ-H2AX antibody. Radiat. Res. 2002, 158, 486–492. [Google Scholar] [CrossRef]
- Burma, S.; Chen, B.P.; Murphy, M.; Kurimasa, A.; Chen, D.J. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 2001, 276, 42462–42467. [Google Scholar] [CrossRef]
- Kuo, L.J.; Yang, L.-X. γ-H2AX-a novel biomarker for DNA double-strand breaks. In Vivo 2008, 22, 305–309. [Google Scholar]
- Ji, J.; Zhang, Y.; Redon, C.E.; Reinhold, W.C.; Chen, A.P.; Fogli, L.K.; Holbeck, S.L.; Parchment, R.E.; Hollingshead, M.; Tomaszewski, J.E.; et al. Phosphorylated fraction of H2AX as a measurement for DNA damage in cancer cells and potential applications of a novel assay. PLoS ONE 2017, 12, e0171582. [Google Scholar] [CrossRef] [PubMed]
- Faucher, D.; Wellinger, R.J. Methylated H3K4, a transcription-associated histone modification, is involved in the DNA damage response pathway. PLoS Genet 2010, 6, e1001082. [Google Scholar] [CrossRef]
- Gafford, G.M.; Parsons, R.G.; Helmstetter, F.J. Consolidation and reconsolidation of contextual fear memory requires mammalian target of rapamycin-dependent translation in the dorsal hippocampus. Neuroscience 2011, 182, 98–104. [Google Scholar] [CrossRef]
- Celeste, A.; Fernandez-Capetillo, O.; Kruhlak, M.J.; Pilch, D.R.; Staudt, D.W.; Lee, A.; Bonner, R.F.; Bonner, W.M.; Nussenzweig, A. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat. Cell Biol. 2003, 5, 675–679. [Google Scholar] [CrossRef]
- Jarome, T.J.; Butler, A.A.; Nichols, J.N.; Pacheco, N.L.; Lubin, F.D. NF-κB mediates Gadd45β expression and DNA demethylation in the hippocampus during fear memory formation. Front. Mol. Neurosci. 2015, 8, 54. [Google Scholar] [CrossRef]
- Monsey, M.S.; Ota, K.T.; Akingbade, I.F.; Hong, E.S.; Schafe, G.E. Epigenetic Alterations Are Critical for Fear Memory Consolidation and Synaptic Plasticity in the Lateral Amygdala. PLoS ONE 2011, 6, e19958. [Google Scholar] [CrossRef]
- Kim, S.; Kaang, B.-K. Epigenetic regulation and chromatin remodeling in learning and memory. Exp. Mol. Med. 2017, 49, e281. [Google Scholar] [CrossRef] [PubMed]
- Levenson, J.M.; Sweatt, J.D. Epigenetic mechanisms in memory formation. Nat. Rev. Neurosci. 2005, 6, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Jarome, T.J.; Thomas, J.S.; Lubin, F.D. Chapter One—The Epigenetic Basis of Memory Formation and Storage. In Progress in Molecular Biology and Translational Science; Akbarian, S., Lubin, F., Eds.; Academic Press: Cambridge, MA, USA, 2014; Volume 128, pp. 1–27. [Google Scholar]
- Suberbielle, E.; Sanchez, P.E.; Kravitz, A.V.; Wang, X.; Ho, K.; Eilertson, K.; Devidze, N.; Kreitzer, A.C.; Mucke, L. Physiologic brain activity causes DNA double-strand breaks in neurons, with exacerbation by amyloid-β. Nat. Neurosci. 2013, 16, 613–621. [Google Scholar] [CrossRef] [PubMed]
- Shanbhag, N.M.; Evans, M.D.; Mao, W.; Nana, A.L.; Seeley, W.W.; Adame, A.; Rissman, R.A.; Masliah, E.; Mucke, L. Early neuronal accumulation of DNA double strand breaks in Alzheimer’s disease. Acta Neuropathol. Commun. 2019, 7, 77. [Google Scholar] [CrossRef] [PubMed]
- Crowe, S.L.; Movsesyan, V.A.; Jorgensen, T.J.; Kondratyev, A.J. Rapid phosphorylation of histone H2A.X following ionotropic glutamate receptor activation. Eur. J. Neurosci. 2006, 23, 2351–2361. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Marshall, P.R.; Leighton, L.J.; Zajaczkowski, E.L.; Wang, Z.; Madugalle, S.U.; Yin, J.; Bredy, T.W.; Wei, W. The DNA Repair-Associated Protein Gadd45γ Regulates the Temporal Coding of Immediate Early Gene Expression within the Prelimbic Prefrontal Cortex and Is Required for the Consolidation of Associative Fear Memory. J. Neurosci. 2018, 39, 970–983. [Google Scholar] [CrossRef] [PubMed]
- Gupta-Agarwal, S.; Franklin, A.V.; DeRamus, T.; Wheelock, M.; Davis, R.L.; McMahon, L.L.; Lubin, F.D. G9a/GLP histone lysine dimethyltransferase complex activity in the hippocampus and the entorhinal cortex is required for gene activation and silencing during memory consolidation. J. Neurosci. 2012, 32, 5440–5453. [Google Scholar] [CrossRef]
- Muerdter, F.; Stark, A. Gene regulation: Activation through space. Curr. Biol. 2016, 26, R895–R898. [Google Scholar] [CrossRef][Green Version]
- Merino, A.; Madden, K.R.; Lane, W.S.; Champoux, J.J.; Reinberg, D. DNA topoisomerase I is involved in both repression and activation of transcription. Nature 1993, 365, 227–232. [Google Scholar] [CrossRef]
- Shykind, B.M.; Kim, J.; Stewart, L.; Champoux, J.J.; Sharp, P.A. Topoisomerase I enhances TFIID-TFIIA complex assembly during activation of transcription. Genes Dev. 1997, 11, 397–407. [Google Scholar] [CrossRef]
- Durand-Dubief, M.; Persson, J.; Norman, U.; Hartsuiker, E.; Ekwall, K. Topoisomerase I regulates open chromatin and controls gene expression in vivo. EMBO J. 2010, 29, 2126–2134. [Google Scholar] [CrossRef] [PubMed]
- Kouzine, F.; Gupta, A.; Baranello, L.; Wojtowicz, D.; Ben-Aissa, K.; Liu, J.; Przytycka, T.M.; Levens, D.J. Transcription-dependent dynamic supercoiling is a short-range genomic force. Nat. Struct. Mol. Biol. 2013, 20, 396–403. [Google Scholar] [CrossRef] [PubMed]
- Dunaway, M.; Ostrander, E.A. Local domains of supercoiling activate a eukaryotic promoter in vivo. Nature 1993, 361, 746–748. [Google Scholar] [CrossRef]
- Pommier, Y.; Sun, Y.; Huang, S.-Y.N.; Nitiss, J.L. Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat. Rev. Mol. Cell Biol. 2016, 17, 703–721. [Google Scholar] [CrossRef] [PubMed]
- Baranello, L.; Wojtowicz, D.; Cui, K.; Devaiah, B.N.; Chung, H.-J.; Chan-Salis, K.Y.; Guha, R.; Wilson, K.; Zhang, X.; Zhang, H.; et al. RNA Polymerase II Regulates Topoisomerase 1 Activity to Favor Efficient Transcription. Cell 2016, 165, 357–371. [Google Scholar] [CrossRef]
- Joshi, R.S.; Pina, B.; Roca, J.J. Topoisomerase II is required for the production of long Pol II gene transcripts in yeast. Nucleic Acids Res. 2012, 40, 7907–7915. [Google Scholar] [CrossRef]
- Husain, A.; Begum, N.A.; Taniguchi, T.T.H.; Taniguchi, H.; Kobayashi, M.; Honjo, T. Chromatin remodeller SMARCA4 recruits topoisomerase 1 and suppresses transcription-associated genomic instability. Nat. Commun. 2016, 7, 10549. [Google Scholar] [CrossRef]
- Bekinschtein, P.; Cammarota, M.; Igaz, L.M.; Bevilaqua, L.R.; Izquierdo, I.; Medina, J.H. Persistence of long-term memory storage requires a late protein synthesis- and BDNF- dependent phase in the hippocampus. Neuron 2007, 53, 261–277. [Google Scholar] [CrossRef]
- Orsi, S.A.; Devulapalli, R.K.; Nelsen, J.L.; McFadden, T.; Surineni, R.; Jarome, T.J. Distinct subcellular changes in proteasome activity and linkage-specific protein polyubiquitination in the amygdala during the consolidation and reconsolidation of a fear memory. Neurobiol. Learn. Mem. 2019, 157, 1–11. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Navabpour, S.; Rogers, J.; McFadden, T.; Jarome, T.J. DNA Double-Strand Breaks Are a Critical Regulator of Fear Memory Reconsolidation. Int. J. Mol. Sci. 2020, 21, 8995. https://doi.org/10.3390/ijms21238995
Navabpour S, Rogers J, McFadden T, Jarome TJ. DNA Double-Strand Breaks Are a Critical Regulator of Fear Memory Reconsolidation. International Journal of Molecular Sciences. 2020; 21(23):8995. https://doi.org/10.3390/ijms21238995
Chicago/Turabian StyleNavabpour, Shaghayegh, Jessie Rogers, Taylor McFadden, and Timothy J. Jarome. 2020. "DNA Double-Strand Breaks Are a Critical Regulator of Fear Memory Reconsolidation" International Journal of Molecular Sciences 21, no. 23: 8995. https://doi.org/10.3390/ijms21238995
APA StyleNavabpour, S., Rogers, J., McFadden, T., & Jarome, T. J. (2020). DNA Double-Strand Breaks Are a Critical Regulator of Fear Memory Reconsolidation. International Journal of Molecular Sciences, 21(23), 8995. https://doi.org/10.3390/ijms21238995




