Secretome Analysis of Inductive Signals for BM-MSC Transdifferentiation into Salivary Gland Progenitors
Abstract
1. Introduction
2. Results
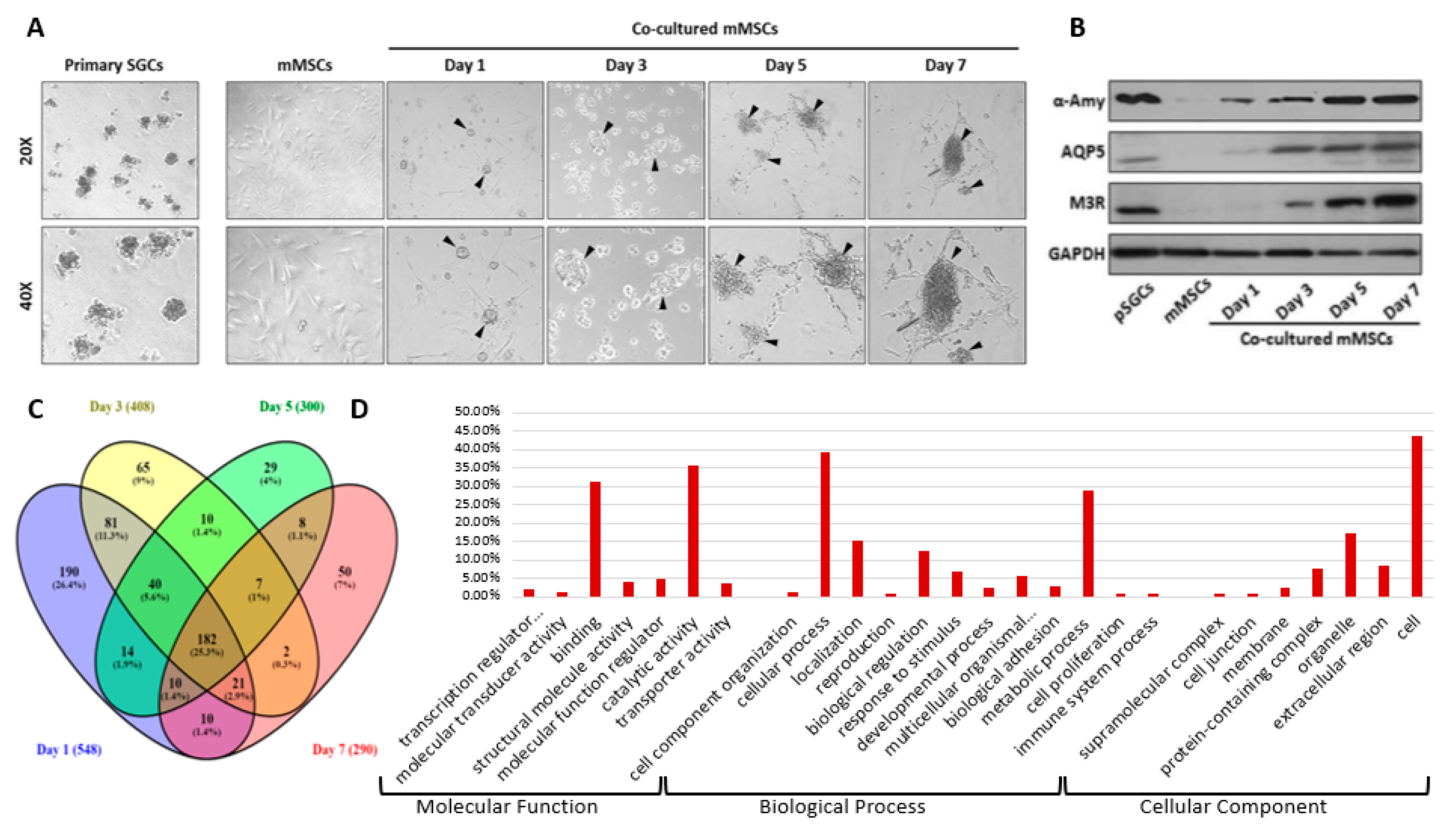
2.1. Secretome Data Analysis and Gene Ontology (GO) Classification
2.2. Protein Clusters and Cellular Function Analysis of Newly Secreted Proteins in the Conditioned Media
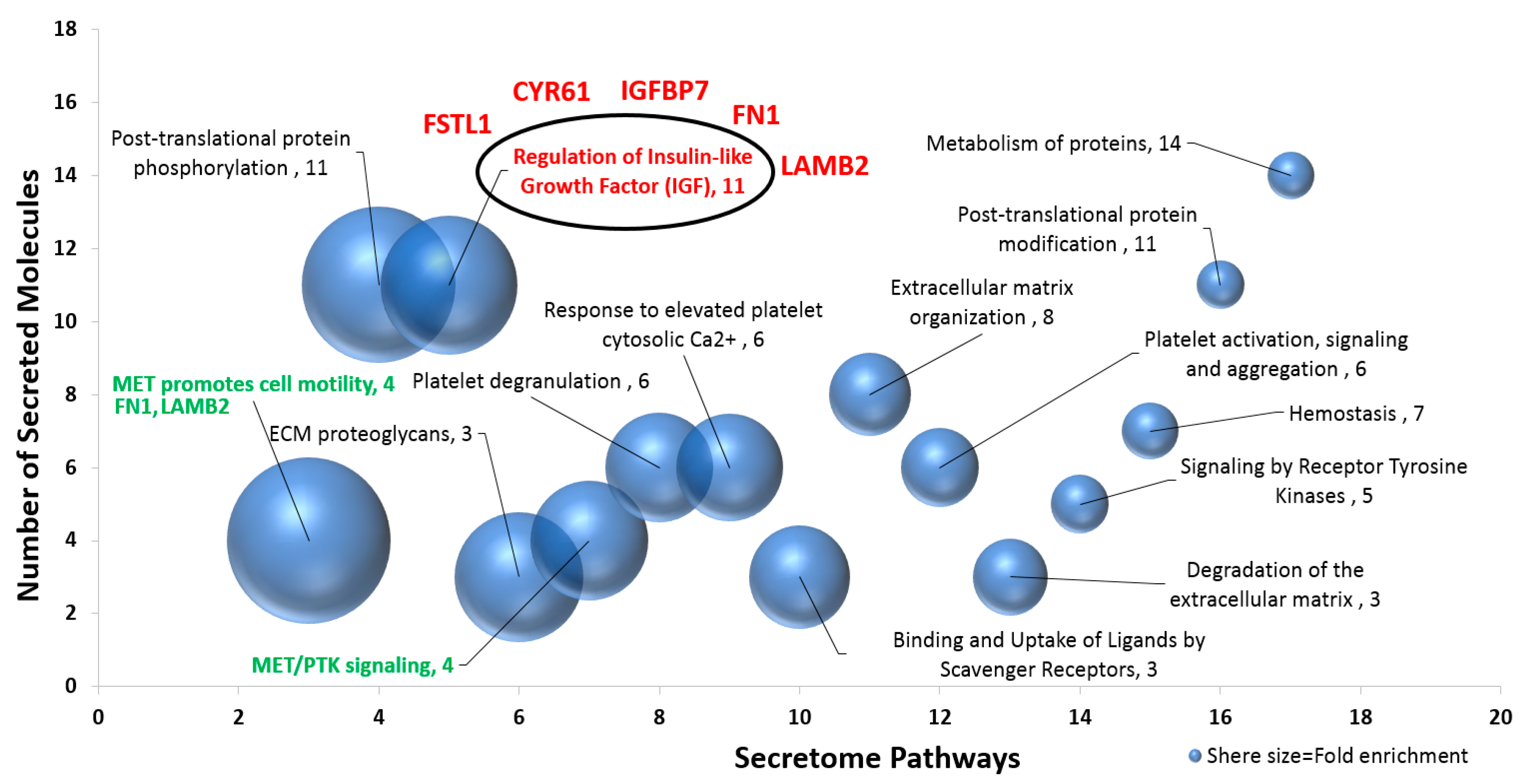
2.3. Pathway Enrichment of Newly Expressed Proteins in the Conditioned Media
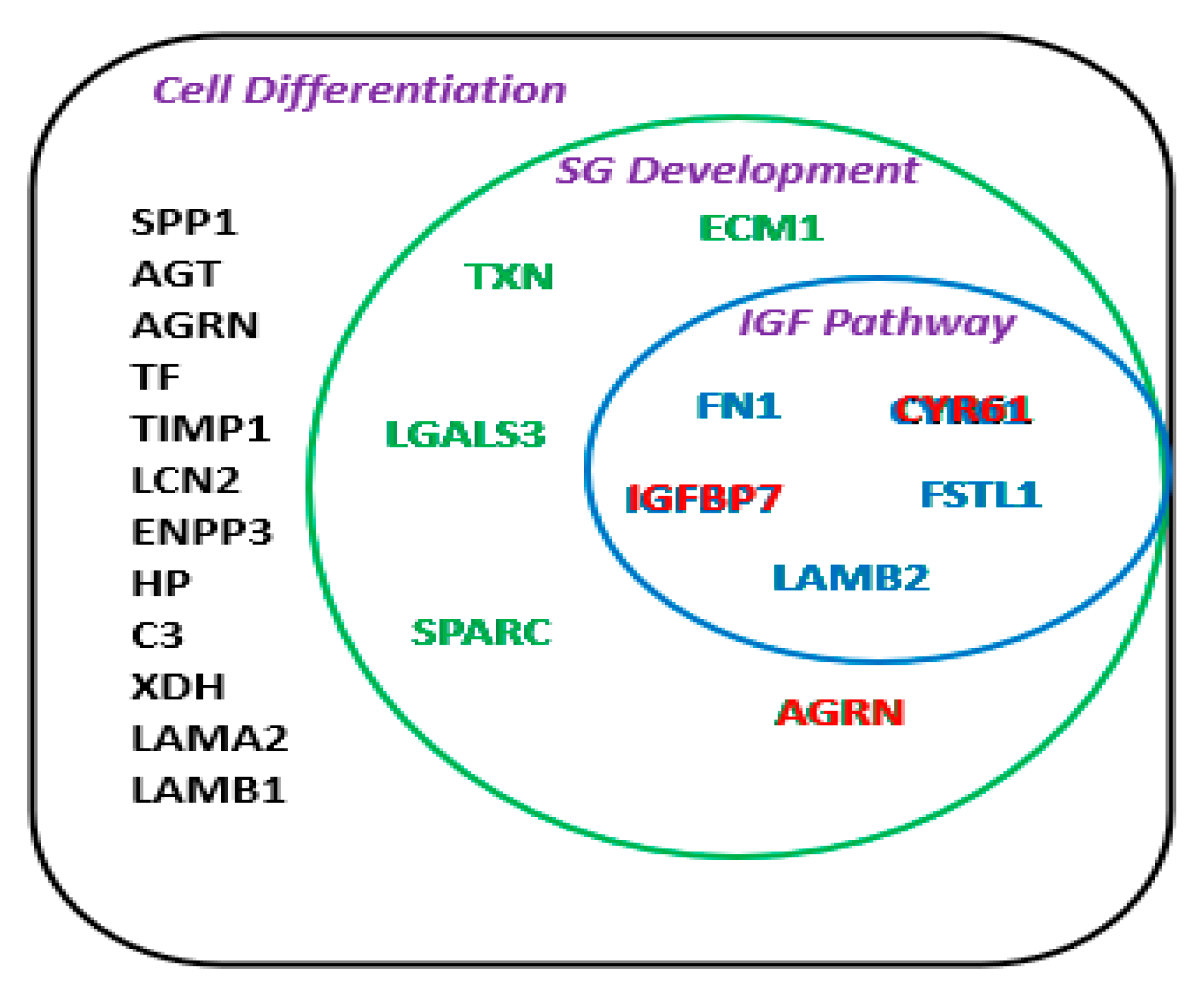
2.4. Intracellular and Extracellular Interactome
3. Discussion
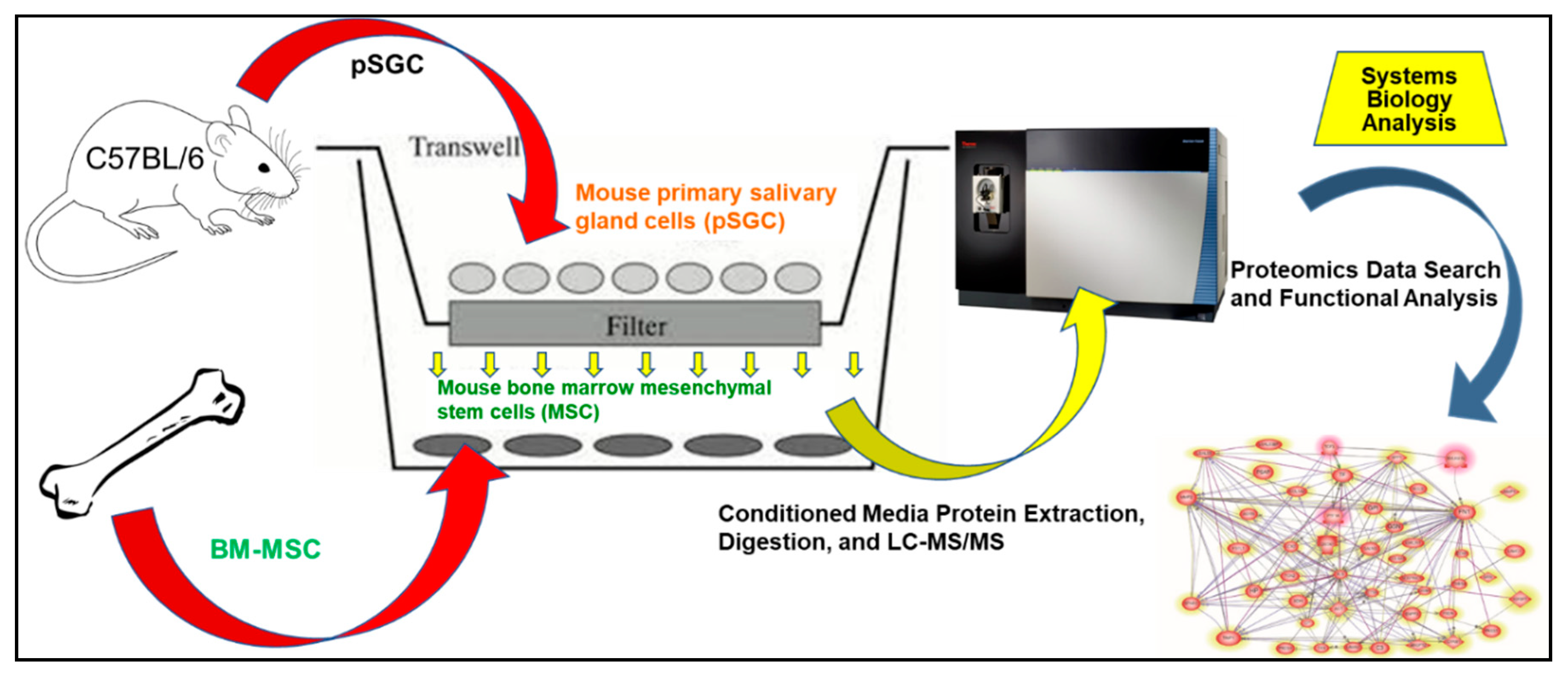
4. Materials and Methods
4.1. Animals
4.2. Mouse Bone Marrow-Derived Mesenchymal Stem Cell Culture
4.3. Co-Culture of mMSC and pSGC
4.4. Protein Extraction, Digestion, and LC-MS/MS
4.5. Proteomics Data Search and Analysis
4.6. Functional and Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| MSC | Mesenchymal stem cell |
| mMSC | mouse bone marrow-derived stem cells |
| GO | Gene Ontology |
| LC-MS/MS | Liquid chromatography tandem mass spectrometry |
| FN1 | Fibronectin 1 |
| AGRN | Agrin |
| LGALS3 | Lectin, galactoside-binding, soluble, 3 |
| CYR61 | Cysteine-rich, angiogenic inducer, 61 |
| IGFBP7 | Insulin-like growth factor binding protein 7 |
| FSTL1 | Follistatin-like 1 |
| LAMB2 | Laminin, beta 2 |
References
- Patel, V.N.; Hoffman, M.P. Salivary gland development: A template for regeneration. Semin. Cell Dev. Biol. 2013, 25, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Lombaert, I.M.; Knox, S.M.; Hoffman, M.P. Salivary gland progenitor cell biology provides a rationale for therapeutic salivary gland regeneration. Oral Dis. 2011, 17, 445–449. [Google Scholar] [CrossRef] [PubMed]
- Bakhtiari, S.; Sehatpour, M.; Mortazavi, H.; Bakhshi, M. Orofacial manifestations of adverse drug reactions: A review study. Clujul. Med. 2018, 91, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Maciejczyk, M.; Szulimowska, J.; Taranta-Janusz, K.; Wasilewska, A.; Zalewska, A. Salivary Gland Dysfunction, Protein Glycooxidation and Nitrosative Stress in Children with Chronic Kidney Disease. J. Clin. Med. 2020, 9, 1285. [Google Scholar] [CrossRef]
- Aljerf, L.; Alhaffar, I. Salivary Distinctiveness and Modifications in Males with Diabetes and Behçet’s Disease. Biochem. Res. Int. 2017, 2017, 1–12. [Google Scholar] [CrossRef]
- Skutnik-Radziszewska, A.; Maciejczyk, M.; Flisiak, I.; Krahel, J.A.; Kołodziej, J.K.U.; Kotowska-Rodziewicz, A.; Klimiuk, A.; Zalewska, A. Enhanced Inflammation and Nitrosative Stress in the Saliva and Plasma of Patients with Plaque Psoriasis. J. Clin. Med. 2020, 9, 745. [Google Scholar] [CrossRef]
- Vissink, A.; Mitchell, J.B.; Baum, B.J.; Limesand, K.H.; Jensen, S.B.; Fox, P.C.; Elting, L.S.; Langendijk, J.A.; Coppes, R.P.; Reyland, M.E. Clinical Management of Salivary Gland Hypofunction and Xerostomia in Head-and-Neck Cancer Patients: Successes and Barriers. Int. J. Radiat. Oncol. 2010, 78, 983–991. [Google Scholar] [CrossRef]
- Burlage, F.R.; Faber, H.; Kampinga, H.H.; Langendijk, J.A.; Vissink, A.; Coppes, R.P.; Coppes, R.P. Enhanced proliferation of acinar and progenitor cells by prophylactic pilocarpine treatment underlies the observed amelioration of radiation injury to parotid glands. Radiother. Oncol. 2009, 90, 253–256. [Google Scholar] [CrossRef]
- Feng, J.; Van Der Zwaag, M.; Stokman, M.A.; Van Os, R.; Coppes, R.P. Isolation and characterization of human salivary gland cells for stem cell transplantation to reduce radiation-induced hyposalivation. Radiother. Oncol. 2009, 92, 466–471. [Google Scholar] [CrossRef]
- Pringle, S.; Van Os, R.; Coppes, R.P. Concise Review: Adult Salivary Gland Stem Cells and a Potential Therapy for Xerostomia. Stem Cells 2013, 31, 613–619. [Google Scholar] [CrossRef]
- Lombaert, I.; Movahednia, M.M.; Adine, C.; Ferreira, J.N. Concise Review: Salivary Gland Regeneration: Therapeutic Approaches from Stem Cells to Tissue Organoids. Stem Cells 2017, 35, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Chen, L.; Scott, P.G.; Tredget, E.E. Mesenchymal Stem Cells Enhance Wound Healing Through Differentiation and Angiogenesis. Stem Cells 2007, 25, 2648–2659. [Google Scholar] [CrossRef]
- Sumita, Y.; Liu, Y.; Khalili, S.; Maria, O.M.; Xia, D.; Key, S.; Cotrim, A.P.; Mezey, E.; Tran, S.D. Bone marrow-derived cells rescue salivary gland function in mice with head and neck irradiation. Int. J. Biochem. Cell Biol. 2011, 43, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-Y.; Chang, F.-H.; Chen, C.-Y.; Huang, C.-Y.; Hu, F.-C.; Huang, W.-K.; Ju, S.-S.; Chen, M.-H. Cell Therapy for Salivary Gland Regeneration. J. Dent. Res. 2011, 90, 341–346. [Google Scholar] [CrossRef] [PubMed]
- Mead, B.; Berry, M.; Logan, A.; Scott, R.A.; Leadbeater, W.; Scheven, B.A. Stem cell treatment of degenerative eye disease. Stem Cell Res. 2015, 14, 243–257. [Google Scholar] [CrossRef] [PubMed]
- Kichenbrand, C.; Velot, E.; Menu, P.; Moby, V. Dental Pulp Stem Cell-Derived Conditioned Medium: An Attractive Alternative for Regenerative Therapy. Tissue Eng. Part B Rev. 2019, 25, 78–88. [Google Scholar] [CrossRef]
- Geranmayeh, M.H.; Nourazarian, A.; Avci, Ç.B.; Rahbarghazi, R.; Farhoudi, M. Stem Cells as a Promising Tool for the Restoration of Brain Neurovascular Unit and Angiogenic Orientation. Mol. Neurobiol. 2016, 54, 7689–7705. [Google Scholar] [CrossRef]
- Eisen, J.S. Faculty Opinions recommendation of Glial origin of mesenchymal stem cells in a tooth model system. Fac. Opin. Post Publ. Peer Rev. Biomed. Lit. 2014, 513, 551–554. [Google Scholar] [CrossRef]
- Baglioni, S.; Francalanci, M.; Squecco, R.; Lombardi, A.; Cantini, G.; Angeli, R.; Gelmini, S.; Guasti, D.; Benvenuti, S.; Annunziato, F.; et al. Characterization of human adult stem-cell populations isolated from visceral and subcutaneous adipose tissue. FASEB J. 2009, 23, 3494–3505. [Google Scholar] [CrossRef]
- Donnelly, L.E.; Durham, A. F1000Prime recommendation of Evidence for human lung stem cells. F1000 Post Publ. Peer Rev. Biomed. Lit. 2011, 364, 1795–1806. [Google Scholar] [CrossRef]
- Pellegrini, G.; E Traverso, C.; Franzi, A.T.; Zingirian, M.; Cancedda, R.; De Luca, M. Long-term restoration of damaged corneal surfaces with autologous cultivated corneal epithelium. Lancet 1997, 349, 990–993. [Google Scholar] [CrossRef]
- Khalili, S.; Liu, Y.; Kornete, M.; Roescher, N.; Kodama, S.; Peterson, A.; Piccirillo, C.A.; Tran, S.D. Mesenchymal Stromal Cells Improve Salivary Function and Reduce Lymphocytic Infiltrates in Mice with Sjögren’s-Like Disease. PLoS ONE 2012, 7, e38615. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.-Y.; Yi, T.; Choi, J.-S.; Jang, Y.H.; Lee, S.; Kim, H.J.; Song, S.U.; Kim, Y.-M. Intraglandular transplantation of bone marrow-derived clonal mesenchymal stem cells for amelioration of post-irradiation salivary gland damage. Oral Oncol. 2013, 49, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Coppes, R.P.; Stokman, M.A. Stem cells and the repair of radiation-induced salivary gland damage. Oral Dis. 2011, 17, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.-J.; Koh, J.; Kwon, J.T.; Park, Y.-S.; Yang, L.; Cha, S. Uncovering stem cell differentiation factors for salivary gland regeneration by quantitative analysis of differential proteomes. PLoS ONE 2017, 12, e0169677. [Google Scholar] [CrossRef]
- Park, Y.-J.; Koh, J.; Gauna, A.E.; Chen, S.; Cha, S. Identification of Regulatory Factors for Mesenchymal Stem Cell-Derived Salivary Epithelial Cells in a Co-Culture System. PLoS ONE 2014, 9, e112158. [Google Scholar] [CrossRef]
- Vizcaino, J.A.; Deutsch, E.W.; Wang, R.; Csordas, A.; Reisinger, F.; Ríos, D.; Dianes, J.A.; Sun, Z.; Farrah, T.; Bandeira, N.; et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 2014, 32, 223–226. [Google Scholar] [CrossRef]
- Saarilahti, K.; Kouri, M.; Collan, J.; Hämäläinen, T.; Atula, T.; Joensuu, H.; Tenhunen, M. Intensity modulated radiotherapy for head and neck cancer: Evidence for preserved salivary gland function. Radiother. Oncol. 2005, 74, 251–258. [Google Scholar] [CrossRef]
- Greenspan, D.; Daniels, T.E. Effectiveness of pilocarpine in postradiation xerostomia. Cancer 1987, 59, 1123–1125. [Google Scholar] [CrossRef]
- Zheng, C.; Cotrim, A.P.; Rowzee, A.; Swaim, W.; Sowers, A.; Mitchell, J.B.; Baum, B.J. Prevention of Radiation-Induced Salivary Hypofunction Following hKGF Gene Delivery to Murine Submandibular Glands. Clin. Cancer Res. 2011, 17, 2842–2851. [Google Scholar] [CrossRef]
- Lombaert, I.M.A.; Brunsting, J.F.; Wierenga, P.K.; Faber, H.; Stokman, M.A.; Kok, T.; Visser, W.H.; Kampinga, H.H.; De Haan, G.; Coppes, R.P. Rescue of Salivary Gland Function after Stem Cell Transplantation in Irradiated Glands. PLoS ONE 2008, 3, e2063. [Google Scholar] [CrossRef]
- Yaniv, A.U.; Neumann, Y.; David, R.; Stiubea-Cohen, R.; Orbach, Y.; Lang, S.; Rotter, N.; Dvir-Ginzberg, M.; Aframian, D.J.; Palmon, A. Establishment of Immortal Multipotent Rat Salivary Progenitor Cell Line Toward Salivary Gland Regeneration. Tissue Eng. Part C Methods 2011, 17, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Van Wijk, B.; Gunst, Q.D.; Moorman, A.F.M.; Hoff, M.J.B.V.D. Cardiac Regeneration from Activated Epicardium. PLoS ONE 2012, 7, e44692. [Google Scholar] [CrossRef] [PubMed]
- Yoo, C.; Vines, J.; Alexander, G.C.; Murdock, K.; Hwang, P.; Jun, H.-W. Adult stem cells and tissue engineering strategies for salivary gland regeneration: A review. Biomater. Res. 2014, 18, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Leri, A.; Kajstura, J.; Anversa, P. Cardiac Stem Cells and Mechanisms of Myocardial Regeneration. Physiol. Rev. 2005, 85, 1373–1416. [Google Scholar] [CrossRef]
- Mimeault, M.; Batra, S.K. Concise Review: Recent Advances on the Significance of Stem Cells in Tissue Regeneration and Cancer Therapies. Stem Cells 2006, 24, 2319–2345. [Google Scholar] [CrossRef]
- Passier, R. Origin and use of embryonic and adult stem cells in differentiation and tissue repair. Cardiovasc. Res. 2003, 58, 324–335. [Google Scholar] [CrossRef]
- Bryder, D.; Weissman, I.L. Hematopoietic stem cells: The paradigmatic tissue-specific stem cell. Am. J. Pathol. 2006, 169, 338–346. [Google Scholar] [CrossRef]
- Zhang, Y.; Fonslow, B.R.; Shan, B.; Baek, M.-C.; Yates, J.R. Protein Analysis by Shotgun/Bottom-up Proteomics. Chem. Rev. 2013, 113, 2343–2394. [Google Scholar] [CrossRef]
- Gundry, R.L.; Tchernyshyov, I.; Sheng, S.; Tarasova, Y.; Raginski, K.; Boheler, K.R.; Van Eyk, J.E. Expanding the mouse embryonic stem cell proteome: Combining three proteomic approaches. Proteomics 2010, 10, 2728–2732. [Google Scholar] [CrossRef]
- Mizuno, M.; Banzai, Y. Calcium ion release from calcium hydroxide stimulated fibronectin gene expression in dental pulp cells and the differentiation of dental pulp cells to mineralized tissue forming cells by fibronectin. Int. Endod. J. 2008, 41, 933–938. [Google Scholar] [CrossRef] [PubMed]
- Bachem, M.G.; Schneider, E.; Groß, H.; Weidenbach, H.; Schmid, R.M.; Menke, A.; Siech, M.; Beger, H.; Grünert, A.; Adler, G. Identification, culture, and characterization of pancreatic stellate cells in rats and humans. Gastroenterology 1998, 115, 421–432. [Google Scholar] [CrossRef]
- Mona, M.; Miller, R.; Li, H.; Park, Y.-J.; Zaman, R.; Yang, L.; Cha, S. MIST1, an Inductive Signal for Salivary Amylase in Mesenchymal Stem Cells. Int. J. Mol. Sci. 2019, 20, 767. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y. New Insights into Epithelial-Mesenchymal Transition in Kidney Fibrosis. J. Am. Soc. Nephrol. 2010, 21, 212–222. [Google Scholar] [CrossRef]
- Tremblay, M.; Herblot, S.; Lecuyer, E.; Hoang, T. Regulation of pT alpha gene expression by a dosage of E2A, HEB, and SCL. J. Biol. Chem. 2003, 278, 12680–12687. [Google Scholar] [CrossRef] [PubMed]
- Rovira, M.; Delaspre, F.; Massumi, M.; Serra, S.A.; Valverde, M.A.; Lloreta, J.; Dufresne, M.; Payré, B.; Konieczny, S.F.; Savatier, P.; et al. Murine Embryonic Stem Cell–Derived Pancreatic Acinar Cells Recapitulate Features of Early Pancreatic Differentiation. Gastroenterology 2008, 135, 1301–1310. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kantharidis, P.; Wang, B.; Carew, R.M.; Lan, H.Y. Diabetes Complications: The MicroRNA Perspective. Diabetes 2011, 60, 1832–1837. [Google Scholar] [CrossRef]
- Toshima, J.; Toshima, J.Y.; Armano, T.; Yang, N.; Narumiya, S.; Mizuno, K. Cofilin phosphorylation by protein kinase testicular protein kinase 1 and its role in integrin-mediated actin reorganization and focal adhesion formation. Mol. Biol. Cell 2001, 12, 1131–1145. [Google Scholar] [CrossRef]
- Feng, P.; Wang, B.; Ren, E.C. Cyr61/CCN1 is a tumor suppressor in human hepatocellular carcinoma and involved in DNA damage response. Int. J. Biochem. Cell Biol. 2008, 40, 98–109. [Google Scholar] [CrossRef]
- Murph, M.M.; Liu, W.; Yu, S.; Lu, Y.; Hall, H.; Hennessy, B.T.; Lahad, J.; Schaner, M.; Helland, Å.; Kristensen, G.; et al. Lysophosphatidic Acid-Induced Transcriptional Profile Represents Serous Epithelial Ovarian Carcinoma and Worsened Prognosis. PLoS ONE 2009, 4, e5583. [Google Scholar] [CrossRef]
- Komiya, E.; Furuya, M.; Watanabe, N.; Miyagi, Y.; Higashi, S.; Miyazaki, K. Elevated expression of angiomodulin (AGM/IGFBP-rP1) in tumor stroma and its roles in fibroblast activation. Cancer Sci. 2012, 103, 691–699. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.H.; Liu, L.; Zhang, H.Y.; Zhang, Q.Q.; Li, Y.; Tian, X.X.; Qiu, Z.H. Insulin-like growth factor binding protein-related protein 1 contributes to hepatic fibrogenesis. J. Dig. Dis. 2014, 15, 202–210. [Google Scholar] [CrossRef] [PubMed]
- Bixby, J.L.; La Torre, K.B.-D.; Wang, C.; Rathjen, F.G.; Rüegg, M.A. A neuronal inhibitory domain in the N-terminal half of agrin. J. Neurobiol. 2002, 50, 164–179. [Google Scholar] [CrossRef] [PubMed]
- Hopf, C.; Hoch, W. Dimerization of the Muscle-specific Kinase Induces Tyrosine Phosphorylation of Acetylcholine Receptors and Their Aggregation on the Surface of Myotubes. J. Biol. Chem. 1998, 273, 6467–6473. [Google Scholar] [CrossRef] [PubMed]
- Godfrey, E.W.; Schwarte, R.C. Nitric oxide and cyclic GMP regulate early events in agrin signaling in skeletal muscle cells. Exp. Cell Res. 2010, 316, 1935–1945. [Google Scholar] [CrossRef]
- Campagna, J.A.; Ruegg, M.A.; Bixby, J.L. Evidence that agrin directly influences presynaptic differentiation at neuromuscular junctions in vitro. Eur. J. Neurosci. 1997, 9, 2269–2283. [Google Scholar] [CrossRef]
- Gautam, M.; DeChiara, T.M.; Glass, D.J.; Yancopoulos, G.D.; Sanes, J.R. Distinct phenotypes of mutant mice lacking agrin, MuSK, or rapsyn. Dev. Brain Res. 1999, 114, 171–178. [Google Scholar] [CrossRef]
- Banerji, S.; Mehta, S.B. Posterior Restorations. Pract. Proced. Aesthetic Dent. 2017, 31, 128–133. [Google Scholar]
- Amenta, A.R.; Creely, H.E.; Mercado, M.L.T.; Hagiwara, H.; McKechnie, B.A.; Lechner, B.E.; Rossi, S.G.; Wang, Q.; Owens, R.T.; Marrero, E.; et al. Biglycan Is an Extracellular MuSK Binding Protein Important for Synapse Stability. J. Neurosci. 2012, 32, 2324–2334. [Google Scholar] [CrossRef]
- Lacazette, E.; Le Calvez, S.; Gajendran, N.; Brenner, H.R. A novel pathway for MuSK to induce key genes in neuromuscular synapse formation. J. Cell Biol. 2003, 161, 727–736. [Google Scholar] [CrossRef]
- Eldridge, S.; Nalesso, G.; Ismail, H.; Vicente-Greco, K.; Kabouridis, P.; Ramachandran, M.; Niemeier, A.; Herz, J.; Pitzalis, C.; Perretti, M.; et al. Agrin mediates chondrocyte homeostasis and requires both LRP4 and α-dystroglycan to enhance cartilage formation in vitro and in vivo. Ann. Rheum. Dis. 2015, 75, 1228–1235. [Google Scholar] [CrossRef] [PubMed]
- Coppock, D.L.; Kopman, C.; Scandalis, S.; Gilleran, S. Preferential gene expression in quiescent human lung fibroblasts. Cell Growth Differ. Mol. Boil. J. Am. Assoc. Cancer Res. 1993, 4, 483–493. [Google Scholar]
- Xu, J.; Qi, X.; Gong, J.; Yu, M.; Zhang, F.; Sha, H.; Gao, X. Fstl1 Antagonizes BMP Signaling and Regulates Ureter Development. PLoS ONE 2012, 7, e32554. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liu, Y.; Li, X.; Zhao, J.; Geng, Y.; Ning, W. Follistatin like-1 (Fstl1) is required for the normal formation of lung airway and vascular smooth muscle at birth. PLoS ONE 2017, 12, e0177899. [Google Scholar] [CrossRef]
- Su, J.-L.; Chiou, J.; Tang, C.-H.; Zhao, M.; Tsai, C.-H.; Chen, P.-S.; Chang, Y.-W.; Chien, M.-H.; Peng, C.-Y.; Hsiao, M.; et al. CYR61 Regulates BMP-2-dependent Osteoblast Differentiation through the αvβ3Integrin/Integrin-linked Kinase/ERK Pathway. J. Biol. Chem. 2010, 285, 31325–31336. [Google Scholar] [CrossRef]
- Chen, Y.; Du, X.-Y. Functional properties and intracellular signaling of CCN1/Cyr61. J. Cell. Biochem. 2007, 100, 1337–1345. [Google Scholar] [CrossRef]
- Gaffen, S.L. Faculty Opinions recommendation of A critical role of Cyr61 in interleukin-17-dependent proliferation of fibroblast-like synoviocytes in rheumatoid arthritis. Fac. Opin. Post Publ. Peer Rev. Biomed. Lit. 2010, 60, 3602–3612. [Google Scholar] [CrossRef]
- Kulkarni, R.N.; Bakker, A.D.; Everts, V.; Klein-Nulend, J. Mechanical loading prevents the stimulating effect of IL-1β on osteocyte-modulated osteoclastogenesis. Biochem. Biophys. Res. Commun. 2012, 420, 11–16. [Google Scholar] [CrossRef]
- Wu, D.D.; Zhang, F.; Hao, F.; Chun, J.; Xu, X.; Cui, M.-Z. Matricellular Protein Cyr61 Bridges Lysophosphatidic Acid and Integrin Pathways Leading to Cell Migration. J. Biol. Chem. 2013, 289, 5774–5783. [Google Scholar] [CrossRef]
- Latinkic, B.V.; Mo, F.E.; Greenspan, J.A.; Copeland, N.G.; Gilbert, D.J.; Jenkins, N.A.; Ross, S.R.; Lau, L.F. Promoter function of the angiogenic inducer Cyr61gene in transgenic mice: Tissue specificity, inducibility during wound healing, and role of the serum response element. Endocrinology 2001, 142, 2549–2557. [Google Scholar] [CrossRef]
- O’Brien, T.P.; Lau, L.F. Expression of the growth factor-inducible immediate early gene cyr61 correlates with chondrogenesis during mouse embryonic development. Cell Growth Differ. Mol. Boil. J. Am. Assoc. Cancer Res. 1992, 3, 645–654. [Google Scholar]
- Wong, M.; Kireeva, M.L.; Kolesnikova, T.V.; Lau, L.F. Cyr61, Product of a Growth Factor-Inducible Immediate-Early Gene, Regulates Chondrogenesis in Mouse Limb Bud Mesenchymal Cells. Dev. Biol. 1997, 192, 492–508. [Google Scholar] [CrossRef] [PubMed]
- Si, W.; Kang, Q.; Luu, H.H.; Park, J.K.; Luo, Q.; Song, W.-X.; Jiang, W.; Luo, X.; Li, X.; Yin, H.; et al. CCN1/Cyr61 Is Regulated by the Canonical Wnt Signal and Plays an Important Role in Wnt3A-Induced Osteoblast Differentiation of Mesenchymal Stem Cells. Mol. Cell. Biol. 2006, 26, 2955–2964. [Google Scholar] [CrossRef] [PubMed]
- Lienau, J.; Schell, H.; Epari, D.R.; Schütze, N.; Jakob, F.; Duda, G.N.; Bail, H.J. CYR61 (CCN1) Protein Expression during Fracture Healing in an Ovine Tibial Model and Its Relation to the Mechanical Fixation Stability. J. Orthop. Res. 2006, 24, 254–262. [Google Scholar] [CrossRef] [PubMed]
- Adam, O.; Lavall, D.; Theobald, K.; Hohl, M.; Grube, M.; Ameling, S.; Sussman, M.A.; Rosenkranz, S.; Kroemer, H.K.; Schäfers, H.-J.; et al. Rac1-Induced Connective Tissue Growth Factor Regulates Connexin 43 and N-Cadherin Expression in Atrial Fibrillation. J. Am. Coll. Cardiol. 2010, 55, 469–480. [Google Scholar] [CrossRef] [PubMed]
- Nousbeck, J.; Sarig, O.; Avidan, N.; Indelman, M.; Bergman, R.; Ramon, M.; Enk, C.D.; Sprecher, E. Insulin-Like Growth Factor-Binding Protein 7 Regulates Keratinocyte Proliferation, Differentiation and Apoptosis. J. Investig. Dermatol. 2010, 130, 378–387. [Google Scholar] [CrossRef]
- Nousbeck, J.; Ishidayamamoto, A.; Bidder, M.; Fuchs, D.; Eckl, K.; Hennies, H.C.; Sagiv, N.; Gat, A.; Gini, M.; Filip, I.; et al. IGFBP7 as a Potential Therapeutic Target in Psoriasis. J. Investig. Dermatol. 2011, 131, 1767–1770. [Google Scholar] [CrossRef]
- Heesch, S.; Schlee, C.; Neumann, M.; Stroux, A.; Kühnl, A.; Schwartz, S.; Haferlach, T.; Goekbuget, N.; Hoelzer, D.; Thiel, E.; et al. BAALC-associated gene expression profiles define IGFBP7 as a novel molecular marker in acute leukemia. Leukemia 2010, 24, 1429–1436. [Google Scholar] [CrossRef]
- Jeschke, U.; Mayr, D.; Schiessl, B.; Mylonas, I.; Schulze, S.; Kuhn, C.; Friese, K.; Walzel, H. Expression of galectin-1, 3 (gal-1, gal-3) and the Thomsen-Friedenreich (TF) antigen in normal, IUGR, preeclamptic and HELLP placentas. Placenta 2007, 28, 1165–1173. [Google Scholar] [CrossRef]
- Li, F.; Kato, I.; Kawaguchi, H.; Takasawa, K.; Hibino, Y.; Hiraga, K. The galectin-3 gene promoter binding proteins in the liver of rats 48-h post-treatment with CCl4. Gene 2006, 367, 46–55. [Google Scholar] [CrossRef]
- Tripathi, R.; Saini, H.K.; Rad, R.; Abreu-Goodger, C.; Van Dongen, S.; Enright, A.J. Messenger RNA and microRNA profiling during early mouse EB formation. Gene Expr. Patterns 2011, 11, 334–344. [Google Scholar] [CrossRef] [PubMed]
- Grant, D.S.; Tashiro, K.; Segui-Real, B.; Yamada, Y.; Martin, G.R.; Kleinman, H.K. Two different laminin domains mediate the differentiation of human endothelial cells into capillary-like structures in vitro. Cell 1989, 58, 933–943. [Google Scholar] [CrossRef]
- Morgan, J.E.; Zammit, P.S. Direct effects of the pathogenic mutation on satellite cell function in muscular dystrophy. Exp. Cell Res. 2010, 316, 3100–3108. [Google Scholar] [CrossRef]
- Yuasa, K.; Fukumoto, S.; Kamasaki, Y.; Yamada, A.; Fukumoto, E.; Kanaoka, K.; Saito, K.; Harada, H.; Arikawa-Hirasawa, E.; Miyagoe-Suzuki, Y.; et al. Laminin α2 Is Essential for Odontoblast Differentiation Regulating Dentin Sialoprotein Expression. J. Biol. Chem. 2004, 279, 10286–10292. [Google Scholar] [CrossRef]
- Meyer, S.; Chibly, A.; Burd, R.; Limesand, K. Insulin-Like Growth Factor-1–Mediated DNA Repair in Irradiated Salivary Glands Is Sirtuin-1 Dependent. J. Dent. Res. 2016, 96, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, S.; Kamino, Y.; Hiratsuka, K.; Kiyama-Kishikawa, M.; Abiko, Y. Age-related changes in IGF-1 expression in submandibular glands of senescence-accelerated mice. J. Oral Sci. 2004, 46, 119–125. [Google Scholar] [CrossRef][Green Version]
- Limesand, K.H.; Said, S.; Anderson, S.M. Suppression of Radiation-Induced Salivary Gland Dysfunction by IGF-1. PLoS ONE 2009, 4, e4663. [Google Scholar] [CrossRef]
- Grundmann, O.; Fillinger, J.L.; Victory, K.R.; Burd, R.; Limesand, K.H. Restoration of radiation therapy-induced salivary gland dysfunction in mice by post therapy IGF-1 administration. BMC Cancer 2010, 10, 417. [Google Scholar] [CrossRef]
- Pringle, S.; Nanduri, L.S.Y.; Marianne, V.D.Z.; Ronald, V.O.; Coppes, R.P. Isolation of Mouse Salivary Gland Stem Cells. J. Vis. Exp. 2011, 2011, e2484. [Google Scholar] [CrossRef]
- Zhang, T.; Schneider, J.D.; Lin, C.; Geng, S.; Ma, T.; Lawrence, S.R.; Dufresne, C.P.; Harmon, A.C.; Chen, S. MPK4 Phosphorylation Dynamics and Interacting Proteins in Plant Immunity. J. Proteome Res. 2019, 18, 826–840. [Google Scholar] [CrossRef]
- Bonnet, A.; Lagarrigue, S.; Liaubet, L.; Robert-Granié, C.; SanCristobal, M.; Tosser-Klopp, G. Pathway results from the chicken data set using GOTM, Pathway Studio and Ingenuity softwares. BMC Proc. 2009, 3, S11. [Google Scholar] [CrossRef] [PubMed]
- Yuryev, A.; Kotelnikova, E.; Daraselia, N. Ariadne’s ChemEffect and Pathway Studio knowledge base. Expert Opin. Drug Discov. 2009, 4, 1307–1318. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Huang, X.; Ebert, D.; Mills, C.; Guo, X.; Thomas, P.D. Protocol Update for large-scale genome and gene function analysis with the PANTHER classification system (version 14.0). Nat. Protoc. 2019, 14, 703–721. [Google Scholar] [CrossRef] [PubMed]
| No. | Protein Name | Gene Name | UniProt |
|---|---|---|---|
| 1 | Ceruloplasmin (ferroxidase) | CP | G3X8Q5_MOUSE |
| 2 | Transferrin | TF | Q542D9_MOUSE |
| 3 | Glucose-6-phosphate isomerase | GPI | G6PI_MOUSE |
| 4 | Aldo-keto reductase family 1, member B10 (aldose reductase) | AKR1B10 | Q5U415_MOUSE |
| 5 | Ectonucleotide pyrophosphatase/phosphodiesterase 3 | ENPP3 | ENPP3_MOUSE |
| 6 | Galactosamine (N-acetyl)-6-sulfate sulfatase | GALNS | GALNS_MOUSE |
| 7 | Xanthine dehydrogenase | XDH | B2RUJ7_MOUSE |
| 8 | Superoxide dismutase 3, extracellular | SOD3 | Q64466_MOUSE |
| 9 | Thioredoxin | TXN | THIO_MOUSE |
| 10 | Quiescin Q6 sulfhydryl oxidase 1 | QSOX1 | QSOX1_MOUSE |
| 11 | Lectin, galactoside-binding, soluble, 3 | LGALS3 | LEG3_MOUSE |
| 12 | Lectin, galactoside-binding, soluble, 3 binding protein | LGALS3BP | Q07797_MOUSE |
| 13 | Fibronectin 1 | FN1 | Q9Z1Z8_MOUSE |
| 14 | Basal cell adhesion molecule (Lutheran blood group) | BCAM | Q99K86_MOUSE |
| 15 | Gelsolin | GSN | Q3TGJ9_MOUSE |
| 16 | Agrin | AGRN | AGRIN_MOUSE |
| 17 | Elastin microfibril interfacer 1 | EMILIN1 | Q3U254_MOUSE |
| 18 | Secreted protein, acidic, cysteine-rich (osteonectin) | SPARC | Q5NCU4_MOUSE |
| 19 | Collagen, type I, alpha 2 | COL1A2 | Q3TP88_MOUSE |
| 20 | Collagen, type VI, alpha 3 | COL6A3 | O88493_MOUSE |
| 21 | Thrombospondin 4 | THBS4 | B2RTL6_MOUSE |
| 22 | Hemicentin 1 | HMCN1 | D3YXG0_MOUSE |
| 23 | Angiopoietin 2 | ANGPT2 | ANGP2_MOUSE |
| 24 | Granulin | GRN | H3BJ90_MOUSE |
| 25 | Aminoacyl tRNA synthetase complex-interacting multifunctional protein 1 | AIMP1 | Q3UZG4_MOUSE |
| 26 | Cysteine-rich, angiogenic inducer, 61 | CYR61 | CYR61_MOUSE |
| 27 | Secreted phosphoprotein 1 | SPP1 | Q3UZY3_MOUSE |
| 28 | Angiotensinogen | AGT | Q8VCN0_MOUSE |
| 29 | Insulin-like growth factor binding protein 7 | IGFBP7 | Q3UFA6_MOUSE |
| 30 | Matrix metallopeptidase 2 | MMP2 | Q3UG07_MOUSE |
| 31 | Cathepsin B | CTSB | CATB_MOUSE |
| 32 | Lipocalin 2 | LCN2 | NGAL_MOUSE |
| 33 | Peroxiredoxin 4 | PRDX4 | PRDX4_MOUSE |
| 34 | Prosaposin | PSAP | Q3UE29_MOUSE |
| 35 | TIMP metallopeptidase inhibitor 1 | TIMP1 | TIMP1_MOUSE |
| 36 | Haptoglobin | HP | HPT_MOUSE |
| 37 | Laminin, beta 1 | LAMB1 | LAMB1_MOUSE |
| 38 | Chitinase, acidic | CHIA | CHIA_MOUSE |
| 39 | Complement component 3 | C3 | CO3_MOUSE |
| 40 | ISG15 ubiquitin-like modifier | ISG15 | ISG15_MOUSE |
| 41 | Peroxidasin homolog (Drosophila) | PXDN | PXDN_MOUSE |
| 42 | Extracellular matrix protein 1 | ECM1 | Q9Z2R8_MOUSE |
| 43 | Sphingomyelin phosphodiesterase, acid-like 3B | SMPDL3B | ASM3B_MOUSE |
| 44 | ADP-dependent glucokinase | ADPGK | Q3UDS7_MOUSE |
| 45 | Insulin-degrading enzyme | IDE | F6RPJ9_MOUSE |
| 46 | Serpin peptidase inhibitor, clade C (antithrombin) | SERPINC1 | ANT3_MOUSE |
| 47 | Protease, serine, 1 (trypsin 1) | PRSS1 | E9QPR6_MOUSE |
| 48 | Transcobalamin II | TCN2 | TCO2_MOUSE |
| 49 | Laminin, alpha 2 | LAMA2 | LAMA2_MOUSE |
| 50 | Laminin, beta 2 | LAMB2 | LAMB2_MOUSE |
| 51 | Follistatin-like 1 | FSTL1 | FSTL1_MOUSE |
| 52 | Family with sequence similarity 3, member D | FAM3D | FAM3D_MOUSE |
| 53 | Inter-alpha-trypsin inhibitor heavy chain family, member 4 | ITIH4 | ITIH4_MOUSE |
| 54 | Protease, serine, 22 | PRSS22 | Q7TML0_MOUSE |
| 55 | NHL repeat containing 3 | NHLRC3 | NHLRC3_MOUSE |
| 56 | Submandibular gland protein C | CP | B9EHK5_MOUSE |
| 57 | Submaxillary gland androgen regulated protein 3A | TF | TRFE_MOUSE |
| No. | Protein Name | Gene Name | UniProt | Level of Evidence * |
|---|---|---|---|---|
| 13 | Fibronectin 1 | FN1 | Q9Z1Z8_MOUSE | <100 |
| 27 | Secreted phosphoprotein 1 | SPP1 | Q3UZY3_MOUSE | <100 |
| 28 | Angiotensinogen | AGT | Q8VCN0_MOUSE | <100 |
| 16 | Agrin | AGRN | AGRIN_MOUSE | 90 |
| 57 | Submaxillary gland androgen regulated protein 3A | TF | TRFE_MOUSE | 76 |
| 18 | Secreted protein, acidic, cysteine-rich (osteonectin) | SPARC | Q5NCU4_MOUSE | 65 |
| 11 | Lectin, galactoside-binding, soluble, 3 | LGALS3 | LEG3_MOUSE | 57 |
| 35 | TIMP metallopeptidase inhibitor 1 | TIMP1 | TIMP1_MOUSE | 52 |
| 26 | Cysteine-rich, angiogenic inducer, 61 | CYR61 | CYR61_MOUSE | 46 |
| 29 | Insulin-like growth factor binding protein 7 | IGFBP7 | Q3UFA6_MOUSE | 23 |
| 32 | Lipocalin 2 | LCN2 | NGAL_MOUSE | 23 |
| 9 | Thioredoxin | TXN | THIO_MOUSE | 19 |
| 5 | Ectonucleotide pyrophosphatase/phosphodiesterase 3 | ENPP3 | ENPP3_MOUSE | 12 |
| 36 | Haptoglobin | HP | HPT_MOUSE | 10 |
| 39 | Complement component 3 | C3 | CO3_MOUSE | 10 |
| 51 | Follistatin-like 1 | FSTL1 | FSTL1_MOUSE | 10 |
| 42 | Extracellular matrix protein 1 | ECM1 | Q9Z2R8_MOUSE | 7 |
| 7 | Xanthine dehydrogenase | XDH | B2RUJ7_MOUSE | 6 |
| 49 | Laminin, alpha 2 | LAMA2 | LAMA2_MOUSE | 5 |
| 50 | Laminin, beta 2 | LAMB2 | LAMB2_MOUSE | 5 |
| 37 | Laminin, beta 1 | LAMB1 | TRFE_MOUSE | 4 |
| No. | Protein Name | Gene Name | UniProt | Cell Type * | Epithelium * | Notes * | ||
|---|---|---|---|---|---|---|---|---|
| Epithelium | Mesenchyme | End Bud | Duct | |||||
| 13 | Fibronectin 1 | FN1 | Q9Z1Z8_MOUSE | Y | Highly expressed early in development | |||
| 16 | Agrin | AGRN | AGRIN_MOUSE | Y | Y | Y | Y | Higher expression in epithelium |
| 18 | Osteonectin | SPARC | Q5NCU4_MOUSE | Y | Y | Y | Y | |
| 11 | Lectin, galactoside-binding, soluble, 3 | LGALS3 | LEG3 _MOUSE | Y | ||||
| 26 | Cysteine-rich, angiogenic inducer, 61 | CYR61 | CYR61_MOUSE | Y | Y | Y | 2x in mesenchyme | |
| 29 | Insulin-like growth factor binding protein 7 | IGFBP7 | Q3UFA6_MOUSE | Y | Expressed late in development | |||
| 9 | Thioredoxin | TXN | THIO_MOUSE | Y | Y | Y | Y | |
| 51 | Follistatin-like 1 | FSTL1 | FSTL1_MOUSE | Y | Y | Y | Y | 3X in mesenchyme and 2X more in duct |
| 42 | Extracellular matrix protein 1 | ECM1 | Q9Z2R8_MOUSE | Y | Expressed late in development | |||
| 50 | Laminin, beta 2 | LAMB2 | LAMB2_MOUSE | Y | Stronger expression late in development | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mona, M.; Kobeissy, F.; Park, Y.-J.; Miller, R.; Saleh, W.; Koh, J.; Yoo, M.-J.; Chen, S.; Cha, S. Secretome Analysis of Inductive Signals for BM-MSC Transdifferentiation into Salivary Gland Progenitors. Int. J. Mol. Sci. 2020, 21, 9055. https://doi.org/10.3390/ijms21239055
Mona M, Kobeissy F, Park Y-J, Miller R, Saleh W, Koh J, Yoo M-J, Chen S, Cha S. Secretome Analysis of Inductive Signals for BM-MSC Transdifferentiation into Salivary Gland Progenitors. International Journal of Molecular Sciences. 2020; 21(23):9055. https://doi.org/10.3390/ijms21239055
Chicago/Turabian StyleMona, Mahmoud, Firas Kobeissy, Yun-Jong Park, Rehae Miller, Wafaa Saleh, Jin Koh, Mi-Jeong Yoo, Sixue Chen, and Seunghee Cha. 2020. "Secretome Analysis of Inductive Signals for BM-MSC Transdifferentiation into Salivary Gland Progenitors" International Journal of Molecular Sciences 21, no. 23: 9055. https://doi.org/10.3390/ijms21239055
APA StyleMona, M., Kobeissy, F., Park, Y.-J., Miller, R., Saleh, W., Koh, J., Yoo, M.-J., Chen, S., & Cha, S. (2020). Secretome Analysis of Inductive Signals for BM-MSC Transdifferentiation into Salivary Gland Progenitors. International Journal of Molecular Sciences, 21(23), 9055. https://doi.org/10.3390/ijms21239055









