Regulation of the First Committed Step in Lipopolysaccharide Biosynthesis Catalyzed by LpxC Requires the Essential Protein LapC (YejM) and HslVU Protease
Abstract
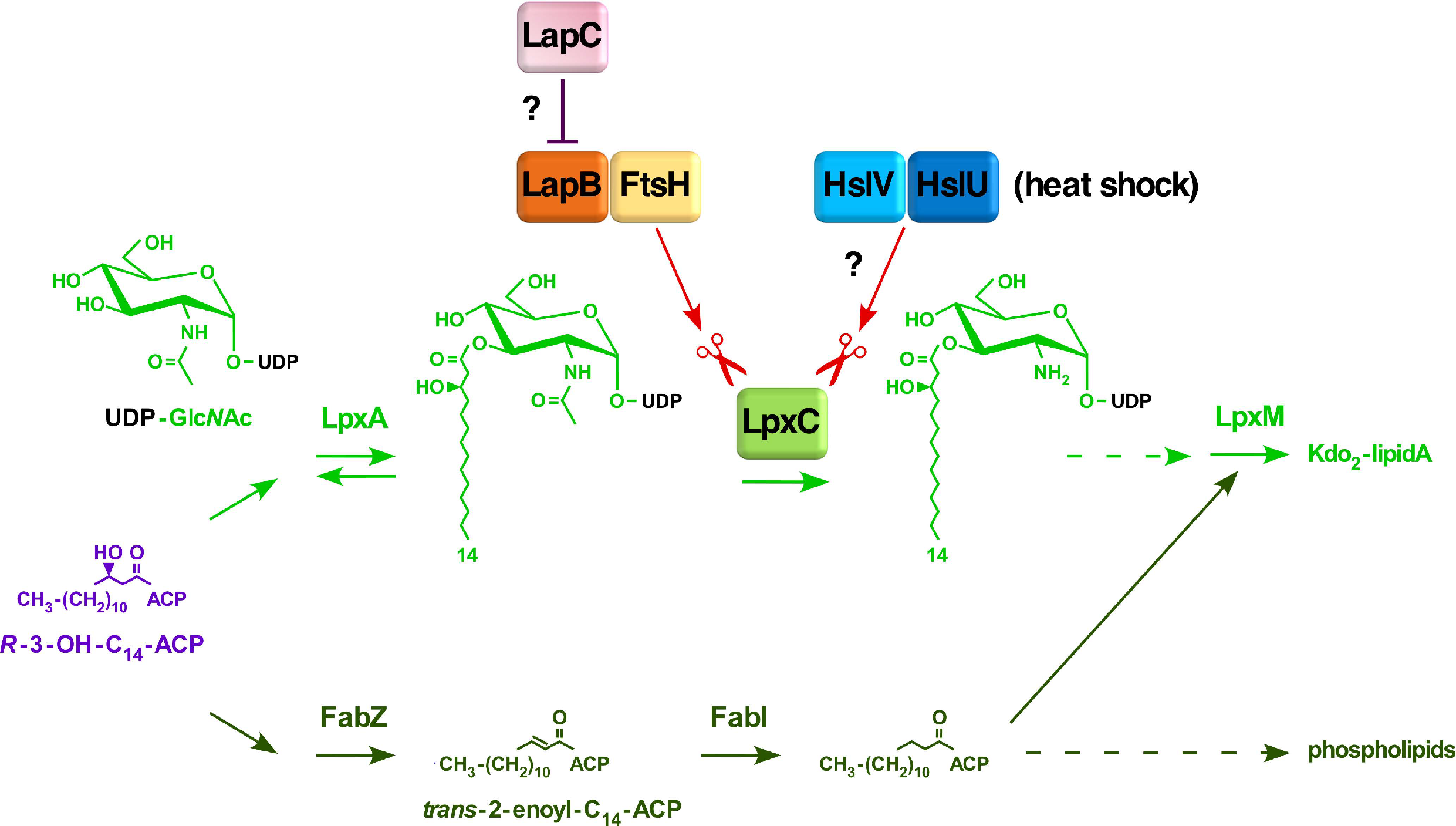
1. Introduction
2. Results
2.1. Rationale
2.2. Overexpression of Heat Shock Protein HslV Protease Can Bypass the Lethality Due to the Deletion of the lapB Gene, Whose Product Is Required for the Regulated Assembly of LPS
2.3. Overexpression of Either hslV or hslUV Rescue the Lethality of LapB Deletion by Enhancing LpxC Degradation in a LapB-Independent Manner
2.4. hslV and hslU Mutants Exhibit the Sensitivity to the LpxC Inhibitor CHIR090
2.5. An Extragenic Suppressor Mutation in the lapC Gene Restores the Normal LPS Synthesis and Prevents the Lethality in the Absence of LapB
2.6. The C-Terminal Truncation of the Periplasmic Globular Domain in the Essential LapC Protein Results in the Induction of RpoE-Dependent Envelope Stress Response, a Temperature-Sensitive Phenotype and the Sensitivity to the LpxC Inhibitor CHIR090
2.7. The lapC190 Mutation Resulting in the C-Terminal Truncation Causes the Reduction in Amounts of LpxC Leading to the Reduced Accumulation of LPS
2.8. Extragenic Suppressors That Restore LPS Levels in lapC190 Mutants Map to lpxC, lapA, lapB and ftsH
2.9. Suppressors of lapC190 Mutation Mapping to the lpxC Gene Prevent Its Degradation and Lead to the Restoration of LPS Amounts
2.10. A Range of Suppressors of lapC190 Map to the lapA/lapB Operon That Could Either Disturb LapB Protein Structure or Dampen Its Abundance by Preventing Its Synthesis
2.11. Suppressor Mutations in the Coding Region of the lapB Gene Map to Conserved Amino Acids in TPR Motifs that Result in the Reduced Abundance of LapB and Restoration of the LPS Synthesis
2.12. Suppressor Mutation in the FtsH Gene Maps to the Conserved Region Required for Its ATPase Activity
2.13. The lapB Gene Is Dispenable in lapC Loss-of-Function Mutations and Overexpression of lpxC Can Bypass the Essentiality of the lapC Gene
2.14. Co-Purification of LapB and LapC
2.15. The Expression of the lapC Gene Is Induced at High Temperatures
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Plasmids and Media
4.2. The Identification of Multicopy Suppressors Whose Overexpression Prevents Lethality of ΔlapA/B Bacteria
4.3. Identification of the lapC Gene Based on the Isolation of Mutations That Either Induce Transcription from the LPS-Responsive rpoEP3-lacZ Fusion or Suppressor a lapB Deletion
4.4. Mapping and Complementation of MacConkey-Sensitive, rpoEP3 Hyperactive and Temperature-Sensitive Mutations
4.5. The Isolation of Extragenic Suppressors of lapC Mutants and Their Mapping
4.6. Bacterial Growth Analysis and Measurement of β-Galactosidase Activity
4.7. LPS Extraction, Mass Spectrometry and Measurement of LPS Levels
4.8. Immunoblotting to Measure Amounts of LpxC, LapB and FtsH
4.9. Purification of LapC and LapB
4.10. RNA Purification and qRT-PCR Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| L-Ara4N | 4-Amino-4-deoxy-L-arabinose |
| CHIR090 | N-Aroyl-L-threonine hydroxamic acid |
| ESI | Electrospray ionization |
| FabZ | R-3-Hydroxymyristoyl acyl carrier protein dehydratase |
| FT-ICR | Fourier transform-ion cyclotron |
| Kdo | 3-Deoxy-α-d-manno-oct-2-ulosonic acid |
| LPS | Lipopolysaccharide |
| LpxC | UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine deacetylase |
| P-EtN | Phosphoethanolamine |
| TPR | Tetratricopeptide repeat |
References
- Nikaido, H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 2003, 67, 593–656. [Google Scholar] [CrossRef] [PubMed]
- Raetz, C.R.H.; Whitfield, C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002, 71, 635–700. [Google Scholar] [CrossRef] [PubMed]
- Bohl, T.E.; Aihara, H. Current progress in the structural and biochemical characterization of proteins involved in the assembly of lipopolysaccharide. Int. J. Microbiol. 2018, 2018, 5319146. [Google Scholar] [CrossRef] [PubMed]
- Müller-Loennies, S.; Lindner, B.; Brade, H. Structural analysis of oligosaccharides from lipopolysaccharide (LPS) of Escherichia coli K12 strain W3100 reveals a link between inner and outer core LPS biosynthesis. J. Biol. Chem. 2003, 278, 34090–34101. [Google Scholar] [CrossRef]
- Klein, G.; Lindner, B.; Brade, H.; Raina, S. Molecular basis of lipopolysaccharide heterogeneity in Escherichia coli: Envelope stress-responsive regulators control the incorporation of glycoforms with a third 3-deoxy-α-D-manno-oct-2-ulosonic acid and rhamnose. J. Biol. Chem. 2011, 286, 42787–42807. [Google Scholar] [CrossRef]
- Klein, G.; Müller-Loennies, S.; Lindner, B.; Kobylak, N.; Brade, H.; Raina, S. Molecular and structural basis of inner core lipopolysaccharide alterations in Escherichia coli. Incorporation of glucuronic acid and phosphoethanolamine in the heptose region. J. Biol. Chem. 2013, 288, 8111–8127. [Google Scholar] [CrossRef]
- Klein, G.; Raina, S. Regulated control of the assembly and diversity of LPS by noncoding sRNAs. Biomed. Res. Int. 2015, 2015, 153561. [Google Scholar] [CrossRef]
- Raetz, C.R.; Reynolds, C.M.; Trent, M.S.; Bishop, R.E. Lipid A modification systems in Gram-negative bacteria. Annu. Rev. Biochem. 2007, 76, 295–329. [Google Scholar] [CrossRef]
- Klein, G.; Raina, S. Regulated assembly of LPS, its structural alterations and cellular response to LPS defects. Int. J. Mol. Sci. 2019, 20, 356. [Google Scholar] [CrossRef]
- Gronow, S.; Brade, H. Lipopolysaccharide biosynthesis: Which steps do bacteria need to survive? J. Endotoxin Res. 2001, 7, 3–23. [Google Scholar] [CrossRef]
- Klein, G.; Lindner, B.; Brabetz, W.; Brade, H.; Raina, S. Escherichia coli K-12 suppressor-free mutants lacking early glycosyltransferases and late acyltransferases: Minimal lipopolysaccharide structure and induction of envelope stress response. J. Biol. Chem. 2009, 284, 15369–15389. [Google Scholar] [CrossRef] [PubMed]
- Raetz, C.R.; Brozek, K.A.; Clementz, T.; Coleman, J.D.; Galloway, S.M.; Golenbock, D.T.; Hampton, R.Y. Gram-negative endotoxin: A biologically active lipid. Cold Spring Harb. Symp. Quant. Biol. 1988, 53, 973–982. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.S.; Bulawa, C.E.; Raetz, C.R.H. The biosynthesis of Gram-negative endotoxin. Formation of lipid A precursors from UDP-GlcNAc in extracts of Escherichia coli. J. Biol. Chem. 1985, 260, 15536–15541. [Google Scholar] [PubMed]
- Anderson, M.S.; Bull, H.G.; Galloway, S.M.; Kelly, T.M.; Mohan, S.; Radika, K.; Raetz, C.R. UDP-N-acetylglucosamine acyltransferase of Escherichia coli. The first step of endotoxin biosynthesis is thermodynamically unfavourable. J. Biol. Chem. 1993, 268, 19858–19865. [Google Scholar] [PubMed]
- Zhou, P.; Zhao, J. Structure, inhibition, and regulation of essential lipid A enzymes. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2017, 1862, 1424–1438. [Google Scholar] [CrossRef]
- Reynolds, C.M.; Raetz, C.R.H. Replacement of lipopolysaccharide with free lipid A molecules in Escherichia coli mutants lacking all core sugars. Biochemistry 2009, 48, 9627–9640. [Google Scholar] [CrossRef]
- Klein, G.; Kobylak, N.; Lindner, B.; Stupak, A.; Raina, S. Assembly of lipopolysaccharide in Escherichia coli requires the essential LapB heat shock protein. J. Biol. Chem. 2014, 289, 14829–14853. [Google Scholar] [CrossRef]
- Klein, G.; Stupak, A.; Biernacka, D.; Wojtkiewicz, P.; Lindner, B.; Raina, S. Multiple transcriptional factors regulate transcription of the rpoE gene in Escherichia coli under different growth conditions and when the lipopolysaccharide biosynthesis is defective. J. Biol. Chem. 2016, 291, 22999–23019. [Google Scholar] [CrossRef]
- Galloway, S.M.; Raetz, C.R. A mutant of Escherichia coli defective in the first step of endotoxin biosynthesis. J. Biol. Chem. 1990, 265, 6394–6402. [Google Scholar]
- Ogura, T.; Inoue, K.; Tatsuta, T.; Suzaki, T.; Karata, K.; Young, K.; Su, L.H.; Fierke, C.A.; Jackman, J.E.; Raetz, C.R.; et al. Balanced biosynthesis of major membrane components through regulated degradation of the committed enzyme of lipid A biosynthesis by the AAA protease FtsH (HflB) in Escherichia coli. Mol. Microbiol. 1999, 31, 833–844. [Google Scholar] [CrossRef]
- Fuhrer, F.; Langklotz, S.; Narberhaus, F. The C-terminal end of LpxC is required for degradation by the FtsH protease. Mol. Microbiol. 2006, 59, 1025–1036. [Google Scholar] [CrossRef] [PubMed]
- Thomanek, N.; Arends, J.; Lindemann, C.; Barkovits, K.; Meyer, H.E.; Marcus, K.; Narberhaus, F. Intricate crosstalk between lipopolysaccharide, phospholipid and fatty acid metabolism in Escherichia coli modulates proteolysis of LpxC. Front. Microbiol. 2019, 9, 3285. [Google Scholar] [CrossRef] [PubMed]
- Mahalakshmi, S.; Sunayana, M.R.; SaiSree, L.; Reddy, M. yciM is an essential gene required for regulation of lipopolysaccharide synthesis in Escherichia coli. Mol. Microbiol. 2014, 91, 145–157. [Google Scholar] [CrossRef]
- Dillon, D.A.; Wu, W.I.; Riedel, B.; Wissing, J.B.; Dowhan, W.; Carman, G.M. The Escherichia coli pgpB gene encodes for a diacyl- glycerol pyrophosphate phosphatase activity. J. Biol. Chem. 1996, 271, 30548–30553. [Google Scholar] [CrossRef] [PubMed]
- Prince, C.; Jia, Z. An unexpected duo: Rubredoxin binds nine TPR motifs to form LapB, an essential regulator of lipopolysaccharide synthesis. Structure 2015, 23, 1500–1506. [Google Scholar] [CrossRef] [PubMed]
- Kanemori, M.; Nishihara, K.; Yanagi, H.; Yura, T. Synergistic roles of HslVU and other ATP-dependent proteases in controlling in vivo turnover of σ32 and abnormal proteins in Escherichia coli. J. Bacteriol. 1997, 179, 7219–7225. [Google Scholar] [CrossRef]
- Missiakas, D.; Schwager, F.; Betton, J.M.; Georgopoulos, C.; Raina, S. Identification and characterization of HslV HslU (ClpQ ClpY) proteins involved in overall proteolysis of misfolded proteins in Escherichia coli. EMBO J. 1996, 15, 6899–68909. [Google Scholar] [CrossRef]
- Tomoyasu, T.; Gamer, J.; Bukau, B.; Kanemori, M.; Mori, H.; Rutman, A.J.; Oppenheim, A.B.; Yura, T.; Yamanaka, K.; Niki, H.; et al. Escherichia coli FtsH is a membrane-bound, ATP-dependent protease which degrades the heat-shock transcription factor σ32. EMBO J. 1995, 14, 2551–2560. [Google Scholar] [CrossRef]
- Kanemori, M.; Yanagi, H.; Yura, T. The ATP-dependent HslVU/ClpQY protease participates in turnover of cell division inhibitor SulA in Escherichia coli. J. Bacteriol. 1999, 181, 3674–3680. [Google Scholar] [CrossRef]
- Wu, W.F.; Zhou, Y.N.; Gottesman, S. Redundant in vivo proteolytic activities of Escherichia coli Lon and the ClpYQ (HslUV) protease. J. Bacteriol. 1999, 181, 3681–3687. [Google Scholar] [CrossRef]
- De Lay, N.R.; Cronan, J.E. Genetic interaction between the Escherichia coli AcpT phosphopantetheinyl transferase and the YejM inner membrane protein. Genetics 2008, 178, 1327–1337. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Clairfeuille, T.; Buchholz, K.R.; Li, Q.; Verschueren, E.; Liu, P.; Sangaraju, D.; Park, S.; Noland, C.L.; Storek, K.M.; Nickerson, N.N.; et al. Structure of the essential inner membrane lipopolysaccharide-PbgA complex. Nature 2020, 584. [Google Scholar] [CrossRef] [PubMed]
- Cian, M.B.; Giordano, N.P.; Masilamani, R.; Minor, K.E.; Delabroux, Z.D. Salmonella enterica serovar Typhimurium uses PdgA/YejM to regulate lipopolysaccharide assembly during bacteremia. Infect. Immun. 2020, 88, e00758Ce19. [Google Scholar] [CrossRef]
- Gabale, U.; Palomino, P.A.P.; Kim, H.A.; Chen, W.; Ressi, S. The essential inner membrane protein YejM is a metalloenzyme. Sci. Rep. 2020, 10, 17794. [Google Scholar] [CrossRef]
- Guest, R.L.; Samé Guerra, D.; Wissler, M.; Grimm, J.; Silhavy, T.J. YejM modulates activity of the YciM/FtsH protease complex to prevent lethal accumulation of lipopolysaccharide. mBio 2020, 11, e00598-20. [Google Scholar] [CrossRef] [PubMed]
- Fivenson, E.M.; Bernhardt, T.G. An essential membrane protein modulates the proteolysis of LpxC to control lipopolysaccharide synthesis in Escherichia coli. mBio 2020, 11, e00939-20. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.; Kelly, K.; Qiu, N.; Misra, R. YejM controls LpxC levels by regulating protease activity of the FtsH/YciM complex of Escherichia coli. J. Bacteriol. 2020, 202, e00303–e00320. [Google Scholar] [CrossRef]
- Zheng, D.; Zhao, J.; Chung, H.S.; Guan, Z.; Raetz, C.R.H.; Zhou, P. Mutants resistant to LpxC inhibitors by rebalancing cellular homeostasis. J. Biol. Chem. 2013, 288, 5475–5486. [Google Scholar] [CrossRef]
- Barb, A.W.; Zhou, P. Mechanism and inhibition of LpxC. An essential zinc-dependent deacetylase of bacterial lipid A synthesis. Curr. Pharm. Biotech. 2008, 9, 9–15. [Google Scholar] [CrossRef]
- Kitagawa, M.; Ara, T.; Arifuzzaman, M.; Ioka-Nakamichi, T.; Inamoto, E.; Toyonaga, H.; Mori, H. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): Unique resources for biological research. DNA Res. 2005, 12, 291–299. [Google Scholar] [CrossRef]
- Rohrwild, M.; Coux, O.; Huang, H.C.; Moerschell, R.P.; Yoo, S.J.; Seol, J.H.; Chung, C.H.; Goldberg, A.L. HslV-HslU: A novel ATP-dependent protease complex in Escherichia coli related to the eukaryotic proteasome. Proc. Natl. Acad. Sci. USA 1996, 93, 5808–5813. [Google Scholar] [CrossRef]
- Chuang, S.E.; Blattner, F.R. Characterization of twenty-six new heat shock genes of Escherichia coli. J. Bacteriol. 1993, 175, 5242–5252. [Google Scholar] [CrossRef] [PubMed]
- Rechsteiner, M.; Hoffman, L.; Dubiel, W. The multicatalytic and 26 S proteases. J. Biol. Chem. 1993, 268, 6065–6068. [Google Scholar] [PubMed]
- Bochtler, M.; Ditzel, L.; Groll, M.; Huber, R. Crystal structure of heat shock locus V (HslV) from Escherichia coli. Proc. Natl. Acad. Sci. USA 1997, 94, 6070–6074. [Google Scholar] [CrossRef] [PubMed]
- Raina, S.; Georgopoulos, C. A new Escherichia coli heat shock gene, htrC, whose product is essential for viability only at high temperatures. J. Bacteriol. 1990, 172, 3417–3426. [Google Scholar] [CrossRef]
- Hirvas, L.; Nurminen, M.; Helander, I.M.; Vuorio, R.; Vaara, M. The lipid A biosynthesis deficiency of the Escherichia coli antibiotic-supersensitive mutant LH530 is suppressed by a novel locus, ORF195. Microbiology 1997, 143, 73–81. [Google Scholar] [CrossRef]
- Dong, H.; Zhang, Z.; Tang, X.; Huang, S.; Li, H.; Peng, B.; Dong, C. Structural insights into cardiolipin transfer from the inner membrane to the outer membrane by PbgA in Gram-negative bacteria. Sci. Rep. 2016, 6, 30815. [Google Scholar] [CrossRef]
- Fan, J.; Petersen, E.M.; Hinds, T.R.; Zheng, N.; Miller, S.I. Structure of an inner membrane protein required for PhoPQ-regulated increases in outer membrane cardiolipin. mBio 2020, 11, e03277-19. [Google Scholar] [CrossRef]
- Lee, C.J.; Liang, X.; Gopalaswamy, R.; Najeeb, J.; Ark, E.D.; Toone, E.J.; Zhou, P. Structural basis of the promiscuous inhibitor susceptibility of Escherichia coli LpxC. ACS Chem. Biol. 2014, 9, 237–246. [Google Scholar] [CrossRef]
- Müller-Loennies, S.; Brade, L.; MacKenzie, C.R.; Di Padova, F.E.; Brade, H. Identification of a cross-reacting epitope widely present in lipopolysaccharide from enterobacteria and recognized by the cross-protective monoclonal antibody WN1 222-5. J. Biol. Chem. 2003, 278, 25618–25627. [Google Scholar] [CrossRef]
- Langklotz, S.; Schäkermann, M.; Narberhaus, F. Control of lipopolysaccharide biosynthesis by FtsH-mediated proteolysis of LpxC is conserved in enterobacteria but not in all gram-negative bacteria. J. Bacteriol. 2011, 193, 1090–1097. [Google Scholar] [CrossRef] [PubMed]
- Mostafavi, M.; Wang, L.; Xie, L.; Takeoka, K.T.; Richie, D.L.; Casey, F.; Ruzin, A.; Sawyer, W.S.; Rath, C.M.; Wei, J.R.; et al. Interplay of Klebsiella pneumoniae fabZ and lpxC mutations leads to LpxC inhibitor-dependent growth resulting from loss of membrane homeostasis. mSphere 2018, 3, e00508–e00518. [Google Scholar] [CrossRef] [PubMed]
- Karata, K.; Inagawa, T.; Wilkinson, A.J.; Tatsuta, T.; Ogura, T. Dissecting the role of a conserved motif (the second region of homology) in the AAA family of ATPases. Site-directed mutagenesis of the ATP-dependent protease FtsH. J. Biol. Chem. 1999, 274, 26225–26232. [Google Scholar] [CrossRef] [PubMed]
- Krzywda, S.; Brzozowski, A.M.; Verma, C.; Karata, K.; Ogura, T.; Wilkinson, A.J. The crystal structure of the AAA domain of the ATP-dependent protease FtsH of Escherichia coli at 1.5 Å resolution. Structure 2002, 10, 1073–1083. [Google Scholar] [CrossRef]
- Emiola, A.; Andrews, S.S.; Heller, C.; George, J. Crosstalk between the lipopolysaccharide and phospholipid pathways during outer membrane biogenesis in Escherichia coli. Proc. Natl. Acad. Sci. USA 2016, 113, 3108–3113. [Google Scholar] [CrossRef]
- Schäkermann, M.; Langklotz, S.; Narberhaus, F. FtsH-mediated coordination of lipopolysaccharide biosynthesis in Escherichia coli correlates with the growth rate and the alarmone (p)ppGpp. J. Bacteriol. 2013, 195, 1912–1919. [Google Scholar] [CrossRef]
- Basta, D.W.; Angeles-Albores, D.; Spero, M.A.; Ciemniecki, J.A.; Newman, D.K. Heat-shock proteases promote survival of Pseudomonas aeruginosa during growth arrest. Proc. Natl. Acad. Sci. USA 2020, 117, 4358–4367. [Google Scholar] [CrossRef]
- Fanelli, F.; Di Pinto, A.; Mottola, A.; Mule, G.; Chieffi, D.; Baruzzi, F.; Tantillo, G.; Fusco, V. Genomic characterization of Arcobacter butzleri isolated from shellfish: Novel insight into antibiotic resistance and virulence determinants. Front. Microbiol. 2019, 10, 670. [Google Scholar] [CrossRef]
- Betton, J.M.; Boscus, D.; Missiakas, D.; Raina, S.; Hofnung, M. Probing the structural role of an αβ loop of maltose-binding protein by mutagenesis: Heat-shock induction by loop variants of the maltose-binding protein that form periplasmic inclusion bodies. J. Mol. Biol. 1996, 262, 140–150. [Google Scholar] [CrossRef]
- Schweiger, R.; Soll, J.; Jung, K.; Heermann, R.; Schwenkert, S. Quantification of interaction strengths between chaperones and tetratricopeptide repeat domain-containing membrane proteins. J. Biol. Chem. 2013, 288, 30614–30625. [Google Scholar] [CrossRef]
- Zeytuni, N.; Zarivach, R. Structural and functional discussion of the tetra-trico-peptide repeat, a protein interaction module. Structure 2012, 20, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riba, A.; Itzhaki, L.S. The tetratricopeptide-repeat motif is a versatile platform that enables diverse modes of molecular recognition. Curr. Opin. Struct. Biol. 2019, 54, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Williamson, J.C.; Edwards, A.V.; Verano-Braga, T.; Schwämmle, V.; Kjeldsen, F.; Jensen, O.N.; Larsen, M.R. High-performance hybrid Orbitrap mass spectrometers for quantitative proteome analysis: Observation and implications. Proteomics 2016, 16, 907–914. [Google Scholar] [CrossRef] [PubMed]
- Guzmán-Flores, J.E.; Steinemann-Hernández, L.; González de la Vara, L.E.; Gavilanes-Ruiz, M.; Romeo, T.; Alvarez, A.F.; Georgellis, D. Proteomic analysis of Escherichia coli detergent-resistant membranes (DRM). PLoS ONE 2019, 14, e0223794. [Google Scholar] [CrossRef]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef]
- Dartigalongue, C.; Loferer, H.; Raina, S. EcfE, a new essential inner membrane protease: Its role in the regulation of heat shock response in Escherichia coli. EMBO J. 2001, 20, 5908–5918. [Google Scholar] [CrossRef]
- Raina, S.; Georgopoulos, C. The htrM gene, whose product is essential for Escherichia coli viability only at elevated temperatures, is identical to the rfaD gene. Nucleic Acids Res. 1991, 19, 3811–3819. [Google Scholar] [CrossRef]
- Galanos, C.; Lüderitz, O.; Westphal, O. A new method for the extraction of R lipopolysaccharides. Eur. J. Biochem. 1969, 9, 245–249. [Google Scholar] [CrossRef]
- Kondakova, A.; Lindner, B. Structural characterization of complex bacterial glycolipids by Fourier-transform mass spectrometry. Eur. J. Mass Spectrom. 2005, 11, 535–546. [Google Scholar] [CrossRef]
- Raina, S.; Missiakas, D.; Baird, L.; Kumar, S.; Georgopoulos, C. Identification and transcriptional analysis of the Escherichia coli htrE operon which is homologous to pap and related pilin operons. J. Bacteriol. 1993, 175, 5009–5021. [Google Scholar] [CrossRef]
- Wojtkiewicz, P.; Biernacka, D.; Gorzelak, P.; Stupak, A.; Klein, G.; Raina, S. Multicopy suppressor analysis of strains lacking cytoplasmic peptidyl-prolyl cis/trans isomerases identifies three new PPIase activities in Escherichia coli that includes the DksA transcription factor. Int. J. Mol. Sci. 2020, 21, 5843. [Google Scholar] [CrossRef] [PubMed]















| Number of Transductants with Selection for Kanamycin Resistance on Minimal Medium | |||
|---|---|---|---|
| Donor | Recipient | ||
| BW25113 + vector | BW25113 + phslV+ | BW25113 + phslUV+ | |
| P1 SR17187 ΔlapA/B KanR | 35 KanR small colonies | 1112 KanR | 3230 KanR |
| P1 SR7753 ΔlapB KanR | 43 KanR small colonies | 1230 KanR | 2980 KanR |
| Gene | Mutation Position | Amounts of Isolates |
|---|---|---|
| lpxC | V37G (GTC → GGC) | 4 |
| V37L (GTC → CTC) | 1 | |
| R230C (CGT → TGT) | 3 | |
| K270T (AAA → ACA) | 1 | |
| a frame-shift by the deletion of 2 nt TA from the stop codon resulting into the addition of 20 aa at the C-terminus | 1 | |
| lapB | A88V (GCT → GTT) TPR2 | 2 |
| R115H (CGT → CAT) TPR3 | 1 | |
| D124Y (GAC → TAC) TPR3 | 1 | |
| R125L (CGC → CTC) TPR3 | 1 | |
| H181R (CAT → CGT) TPR5 | 1 | |
| H325L (CAC → CTC) TPR9 | 1 | |
| H325P (CAC → CCC) TPR9 | 1 | |
| lapA | IS element after 34 nt | 2 |
| IS element after 103 nt | 1 | |
| IS element after -107 nt - 2 nt after P2hs promoter of the lapA gene | 1 | |
| a frame-shift by the insertion of G after 69 nt (23 aa from LapA wt and 7 aa new followed by the stop codon) | 1 | |
| a frame-shift by the deletion of 137 nt C (45 aa from LapA wt and 12 aa new followed by the stop codon) | 1 | |
| LapA L8 (TTA → TGA) stop codon | 1 | |
| ftsH | A296V (GCG → GTG) in the SRH domain | 1 |
| Number of Transductants with Selection for Kanamycin Resistance on Minimal Medium at 30 °C | |||||
|---|---|---|---|---|---|
| Donor | Recipient | ||||
| BW25113 | BW25113 lapC190 | BW25113 lapC377fs | BW25113 + plpxC+ | ||
| -IPTG | 50 μM IPTG | ||||
| ΔlapA/BKan | 33 KanR small colonies | 2930 KanR normal size | 3470 KanR normal size | ND | ND |
| ΔlapBKan | 36 KanR small colonies | 3154 KanR normal size | 3696 KanR normal size | ND | ND |
| ΔlapCKan | None | ND | ND | None | 754 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Biernacka, D.; Gorzelak, P.; Klein, G.; Raina, S. Regulation of the First Committed Step in Lipopolysaccharide Biosynthesis Catalyzed by LpxC Requires the Essential Protein LapC (YejM) and HslVU Protease. Int. J. Mol. Sci. 2020, 21, 9088. https://doi.org/10.3390/ijms21239088
Biernacka D, Gorzelak P, Klein G, Raina S. Regulation of the First Committed Step in Lipopolysaccharide Biosynthesis Catalyzed by LpxC Requires the Essential Protein LapC (YejM) and HslVU Protease. International Journal of Molecular Sciences. 2020; 21(23):9088. https://doi.org/10.3390/ijms21239088
Chicago/Turabian StyleBiernacka, Daria, Patrycja Gorzelak, Gracjana Klein, and Satish Raina. 2020. "Regulation of the First Committed Step in Lipopolysaccharide Biosynthesis Catalyzed by LpxC Requires the Essential Protein LapC (YejM) and HslVU Protease" International Journal of Molecular Sciences 21, no. 23: 9088. https://doi.org/10.3390/ijms21239088
APA StyleBiernacka, D., Gorzelak, P., Klein, G., & Raina, S. (2020). Regulation of the First Committed Step in Lipopolysaccharide Biosynthesis Catalyzed by LpxC Requires the Essential Protein LapC (YejM) and HslVU Protease. International Journal of Molecular Sciences, 21(23), 9088. https://doi.org/10.3390/ijms21239088







