Phospho-Mutant Activity Assays Provide Evidence for the Negative Regulation of Transcriptional Regulator PRE1 by Phosphorylation
Abstract
:1. Introduction
2. Results
2.1. Phosphorylation at Ser-67 Negatively Regulated the Functions of PRE1
2.2. Mutation of the 67th Serine to Glutamic Acid Reduced the Binding Ability of PRE1 to IBH1
2.3. PRE1S67E Failed to Abolish the Interaction between IBH1 and HBI1
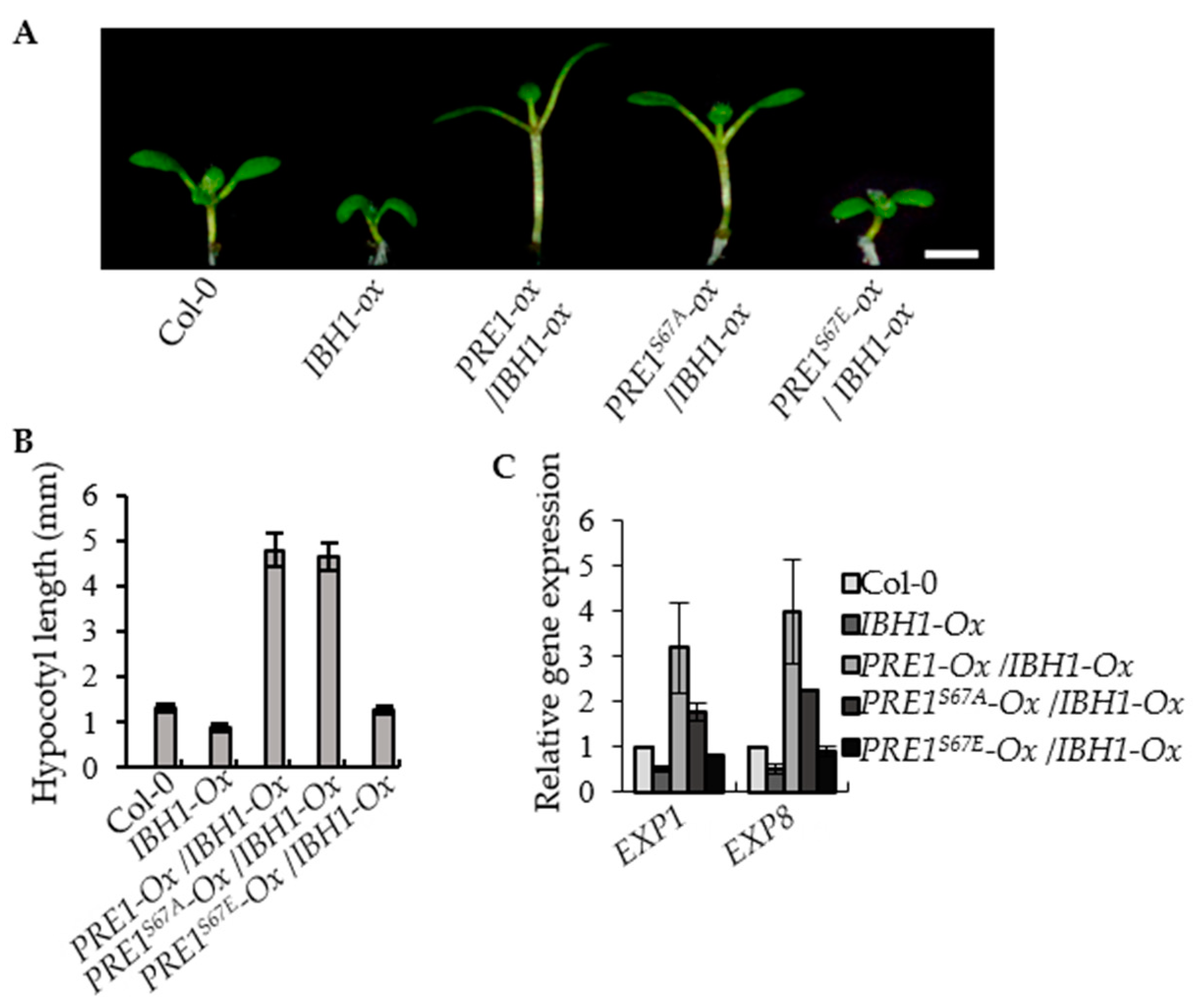
2.4. PRE1S67E Failed to Suppress the Dwarf Phenotypes of IBH1-Ox
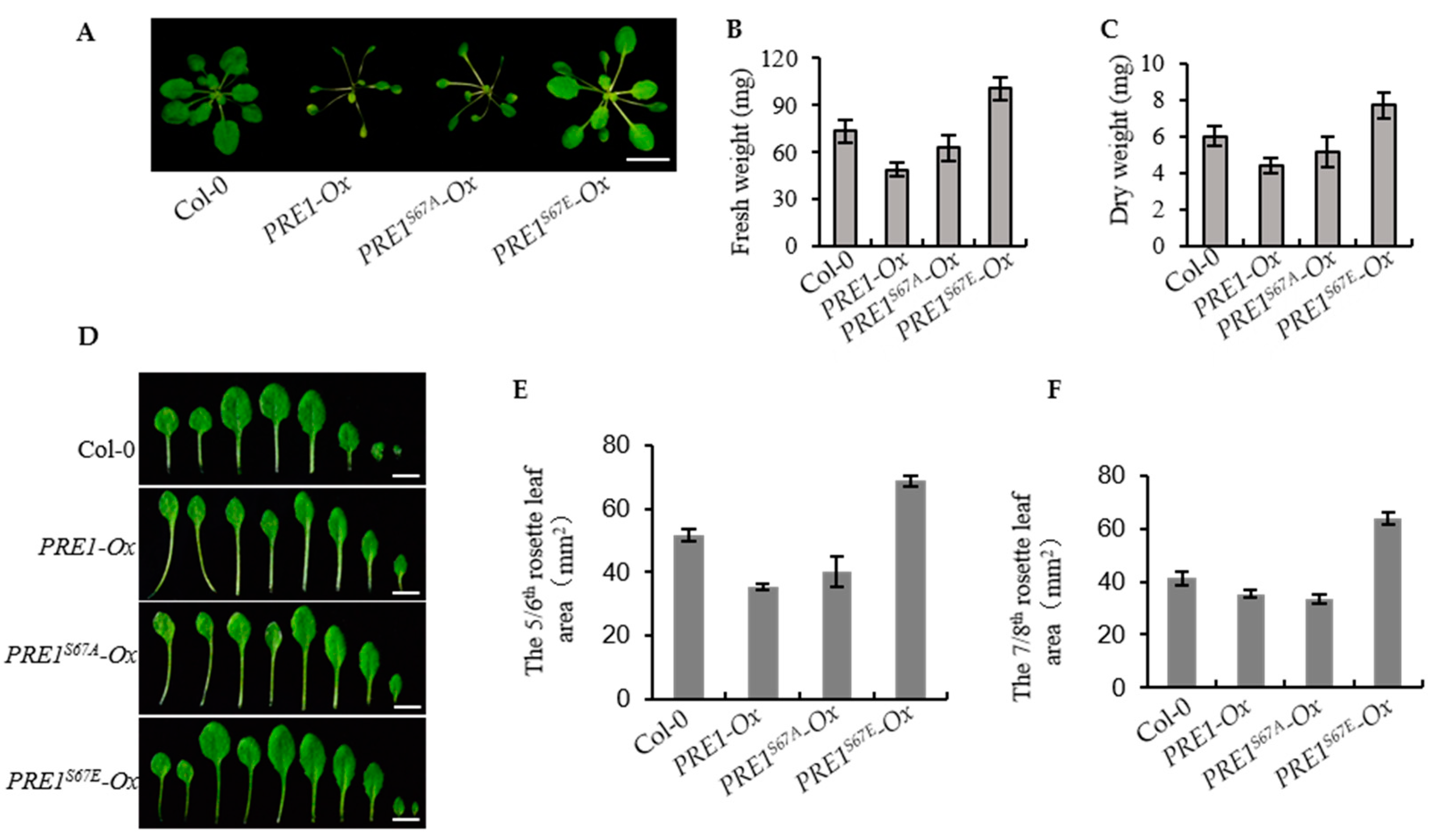
2.5. Overexpression of PRE1S67E Increase the Biomass of the Plant
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Vector Construction
4.3. Quantitative Real-Time PCR Analysis
4.4. Yeast Two-hybrid assays
4.5. β-gal Activity Analysis
4.6. Yeast Three-Hybrid Assays
4.7. Bimolecular Fluorescence Complementation (BiFC) Assays
4.8. Ratiometric Bimolecular Fluorescence Complementation (rBiFC) Assays
4.9. In Vitro Pull-down Assays
4.10. Coimmunoprecipitation Assays
4.11. In Vitro Kinase Assays
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| PRE | Paclobutrazol-resistance |
| Ser, S | Serine |
| Glu, E | glutamic acid |
| Ala, A | Alanine |
| BR | Brassinosteroid |
| GA | Gibberellin |
| ILI1 | Increased leaf inclination1 |
| IBH1 | ILI1 binding bHLH |
| HBI1 | Homolog of BEE 2 interacting with IBH1 |
| BIN2 | GSK3-like kinase brassinosteroid-insensitive 2 |
| PIFs | Phytochrome interacting factors |
| GST | Glutathione S-transferase |
| YFP | Yellow fluorescence protein |
| MBP | Maltose-binding protein |
| 3-AT | 3-amino-1,2,4-triazole |
| BIFC | Bimolecular fluorescence complementation |
| rBiFC | Ratiometric bimolecular fluorescence complementation |
| BD | Binding domain |
| AD | Acting domain |
| RT-PCR | Quantitative real-time PCR |
| COIP | Coimmunoprecipitation |
References
- Toledo-Ortiz, G.; Huq, E.; Quail, P.H. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 2003, 15, 1749–1770. [Google Scholar] [CrossRef] [Green Version]
- Ledent, V.; Vervoort, M. The basic helix-loop-helix protein family: Comparative genomics and phylogenetic analysis. Genome Res. 2001, 11, 754–770. [Google Scholar] [CrossRef] [Green Version]
- Bailey, P.C.; Martin, C.; Toledo-Ortiz, G.; Quail, P.H.; Huq, E.; Heim, M.A.; Weisshaar, B. Update on the basic helix-loop-helix transcription factor gene family in Arabidopsis thaliana. Plant Cell 2003, 15, 2497–2502. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Li, X.; Ma, L. Basic helix-loop-helix transcription factors and epidermal cell fate determination in Arabidopsis. Plant Signal Behav. 2012, 7, 1556–1560. [Google Scholar] [CrossRef] [Green Version]
- Sonnenfeld, M.J.; Delvecchio, C.; Sun, X. Analysis of the transcriptional activation domain of the Drosophila tango bHLH-PAS transcription factor. Dev. Genes Evol. 2005, 215, 221–229. [Google Scholar] [CrossRef]
- Murre, C.; McCaw, P.S.; Baltimore, D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell 1989, 56, 777–783. [Google Scholar] [CrossRef]
- Atchley, W.R.; Terhalle, W.; Dress, A. Positional dependence, cliques, and predictive motifs in the bHLH protein domain. J. Mol. Evol. 1999, 48, 501–516. [Google Scholar] [CrossRef]
- Massari, M.E.; Murre, C. Helix-loop-helix proteins: Regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 2000, 20, 429–440. [Google Scholar] [CrossRef] [Green Version]
- Ferre-D’Amare, A.R.; Pognonec, P.; Roeder, R.G.; Burley, S.K. Structure and function of the b/HLH/Z domain of USF. EMBO J. 1994, 13, 180–189. [Google Scholar] [CrossRef]
- Halliday, K.J.; Hudson, M.; Ni, M.; Qin, M.; Quail, P.H. poc1: An Arabidopsis mutant perturbed in phytochrome signaling because of a T DNA insertion in the promoter of PIF3, a gene encoding a phytochrome-interacting bHLH protein. Proc. Natl. Acad. Sci. USA 1999, 96, 5832–5837. [Google Scholar] [CrossRef] [Green Version]
- Huq, E.; Quail, P.H. PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 2002, 21, 2441–2450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leivar, P.; Tepperman, J.M.; Cohn, M.M.; Monte, E.; Al-Sady, B.; Erickson, E.; Quail, P.H. Dynamic antagonism between phytochromes and PIF family basic helix-loop-helix factors induces selective reciprocal responses to light and shade in a rapidly responsive transcriptional network in Arabidopsis. Plant Cell 2012, 24, 1398–1419. [Google Scholar] [CrossRef] [Green Version]
- Bernhardt, C.; Lee, M.M.; Gonzalez, A.; Zhang, F.; Lloyd, A.; Schiefelbein, J. The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 2003, 130, 6431–6439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oh, E.; Kim, J.; Park, E.; Kim, J.I.; Kang, C.; Choi, G. PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 2004, 16, 3045–3458. [Google Scholar] [CrossRef] [Green Version]
- Park, J.; Lee, N.; Kim, W.; Lim, S.; Choi, G. ABI3 and PIL5 collaboratively activate the expression of SOMNUS by directly binding to its promoter in imbibed Arabidopsis seeds. Plant Cell 2011, 23, 1404–1415. [Google Scholar] [CrossRef] [Green Version]
- Pillitteri, L.J.; Torii, K.U. Breaking the silence: Three bHLH proteins direct cell-fate decisions during stomatal development. Bioessays 2007, 29, 861–870. [Google Scholar] [CrossRef]
- de Marcos, A.; Houbaert, A.; Trivino, M.; Delgado, D.; Martin-Trillo, M.; Russinova, E.; Fenoll, C.; Mena, M. A Mutation in the bHLH Domain of the SPCH Transcription Factor Uncovers a BR-Dependent Mechanism for Stomatal Development. Plant Physiol. 2017, 174, 823–842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, C.; Liu, Y.; Shi, W.; Qiao, Y.; Wang, L.; Tian, Y.; Fan, M.; Deng, Z.; Lau, O.S.; De Jaeger, G.; et al. KIN10 promotes stomatal development through stabilization of the SPEECHLESS transcription factor. Nat. Commun. 2020, 11, 4214. [Google Scholar] [CrossRef]
- Li, J.G.; Fan, M.; Hua, W.; Tian, Y.; Chen, L.G.; Sun, Y.; Bai, M.Y. Brassinosteroid and Hydrogen Peroxide Interdependently Induce Stomatal Opening by Promoting Guard Cell Starch Degradation. Plant Cell 2020, 32, 984–999. [Google Scholar] [CrossRef]
- Feng, Y.; Xu, P.; Li, B.; Li, P.; Wen, X.; An, F.; Gong, Y.; Xin, Y.; Zhu, Z.; Wang, Y.; et al. Ethylene promotes root hair growth through coordinated EIN3/EIL1 and RHD6/RSL1 activity in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, 13834–13839. [Google Scholar] [CrossRef] [Green Version]
- Castilhos, G.; Lazzarotto, F.; Spagnolo-Fonini, L.; Bodanese-Zanettini, M.H.; Margis-Pinheiro, M. Possible roles of basic helix-loop-helix transcription factors in adaptation to drought. Plant Sci. 2014, 223, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Hwang, Y.; Choi, H.S.; Cho, H.M.; Cho, H.T. Tracheophytes Contain Conserved Orthologs of a Basic Helix-Loop-Helix Transcription Factor That Modulate ROOT HAIR SPECIFIC Genes. Plant Cell 2017, 29, 39–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Q.; Ohashi, Y.; Kato, M.; Tsuge, T.; Gu, H.; Qu, L.J.; Aoyama, T. GLABRA2 Directly Suppresses Basic Helix-Loop-Helix Transcription Factor Genes with Diverse Functions in Root Hair Development. Plant Cell 2015, 27, 2894–2906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Li, L.; Xu, P.; Lian, H.; Wang, W.; Xu, F.; Mao, Z.; Zhang, T.; Yang, H. CRY1 interacts directly with HBI1 to regulate its transcriptional activity and photomorphogenesis in Arabidopsis. J. Exp. Bot. 2018, 69, 3867–3881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, M.; Liu, D.; Abdul, W.; Upreti, S.; Liu, Y.; Song, G.; Wu, J.; Liu, B.; Gan, Y. PIL5 represses floral transition in Arabidopsis under long day conditions. Biochem. Biophys. Res. Commun. 2018, 499, 513–518. [Google Scholar] [CrossRef] [PubMed]
- Ito, S.; Song, Y.H.; Josephson-Day, A.R.; Miller, R.J.; Breton, G.; Olmstead, R.G.; Imaizumi, T. FLOWERING BHLH transcriptional activators control expression of the photoperiodic flowering regulator CONSTANS in Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 3582–3587. [Google Scholar] [CrossRef] [Green Version]
- Zheng, K.; Wang, Y.; Zhang, N.; Jia, Q.; Wang, X.; Hou, C.; Chen, J.G.; Wang, S. Involvement of PACLOBUTRAZOL RESISTANCE6/KIDARI, an Atypical bHLH Transcription Factor, in Auxin Responses in Arabidopsis. Front. Plant Sci. 2017, 8, 1813. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.Y.; Bai, M.Y.; Wu, J.; Zhu, J.Y.; Wang, H.; Zhang, Z.; Wang, W.; Sun, Y.; Zhao, J.; Sun, X.; et al. Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 2009, 21, 3767–3780. [Google Scholar] [CrossRef] [Green Version]
- Tian, H.; Chen, S.; Yang, W.; Wang, T.; Zheng, K.; Wang, Y.; Cheng, Y.; Zhang, N.; Liu, S.; Li, D.; et al. A novel family of transcription factors conserved in angiosperms is required for ABA signalling. Plant Cell Environ. 2017, 40, 2958–2971. [Google Scholar] [CrossRef]
- Wang, H.; Zhu, Y.; Fujioka, S.; Asami, T.; Li, J.; Li, J. Regulation of Arabidopsis brassinosteroid signaling by atypical basic helix-loop-helix proteins. Plant Cell 2009, 21, 3781–3791. [Google Scholar] [CrossRef] [Green Version]
- Fan, M.; Bai, M.Y.; Kim, J.G.; Wang, T.; Oh, E.; Chen, L.; Park, C.H.; Son, S.H.; Kim, S.K.; Mudgett, M.B.; et al. The bHLH transcription factor HBI1 mediates the trade-off between growth and pathogen-associated molecular pattern-triggered immunity in Arabidopsis. Plant Cell 2014, 26, 828–841. [Google Scholar] [CrossRef] [Green Version]
- Bai, M.Y.; Fan, M.; Oh, E.; Wang, Z.Y. A triple helix-loop-helix/basic helix-loop-helix cascade controls cell elongation downstream of multiple hormonal and environmental signaling pathways in Arabidopsis. Plant Cell 2012, 24, 4917–4929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hyun, Y.; Lee, I. KIDARI, encoding a non-DNA Binding bHLH protein, represses light signal transduction in Arabidopsis thaliana. Plant Mol. Biol. 2006, 61, 283–296. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Lee, S.; Yang, K.Y.; Kim, Y.M.; Park, S.Y.; Kim, S.Y.; Soh, M.S. Overexpression of PRE1 and its homologous genes activates Gibberellin-dependent responses in Arabidopsis thaliana. Plant Cell Physiol. 2006, 47, 591–600. [Google Scholar] [CrossRef] [PubMed]
- Mara, C.D.; Irish, V.F. Two GATA transcription factors are downstream effectors of floral homeotic gene action in Arabidopsis. Plant Physiol. 2008, 147, 707–718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schlereth, A.; Moller, B.; Liu, W.; Kientz, M.; Flipse, J.; Rademacher, E.H.; Schmid, M.; Jurgens, G.; Weijers, D. MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature 2010, 464, 913–916. [Google Scholar] [CrossRef]
- Lau, O.S.; Deng, X.W. Plant hormone signaling lightens up: Integrators of light and hormones. Curr. Opin. Plant Biol. 2010, 13, 571–577. [Google Scholar] [CrossRef]
- Chapman, E.J.; Greenham, K.; Castillejo, C.; Sartor, R.; Bialy, A.; Sun, T.P.; Estelle, M. Hypocotyl transcriptome reveals auxin regulation of growth-promoting genes through GA-dependent and -independent pathways. PLoS ONE 2012, 7, e36210. [Google Scholar] [CrossRef] [Green Version]
- Oh, E.; Zhu, J.Y.; Wang, Z.Y. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 2012, 14, 802–809. [Google Scholar] [CrossRef] [Green Version]
- Hao, Y.; Oh, E.; Choi, G.; Liang, Z.; Wang, Z.Y. Interactions between HLH and bHLH factors modulate light-regulated plant development. Mol. Plant 2012, 5, 688–697. [Google Scholar] [CrossRef] [Green Version]
- Ikeda, M.; Fujiwara, S.; Mitsuda, N.; Ohme-Takagi, M. A triantagonistic basic helix-loop-helix system regulates cell elongation in Arabidopsis. Plant Cell 2012, 24, 4483–4497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.Y.; Bai, M.Y.; Oh, E.; Zhu, J.Y. Brassinosteroid signaling network and regulation of photomorphogenesis. Annu. Rev. Genet. 2012, 46, 701–724. [Google Scholar] [CrossRef] [PubMed]
- Nakagami, H.; Sugiyama, N.; Mochida, K.; Daudi, A.; Yoshida, Y.; Toyoda, T.; Tomita, M.; Ishihama, Y.; Shirasu, K. Large-scale comparative phosphoproteomics identifies conserved phosphorylation sites in plants. Plant Physiol. 2010, 153, 1161–1174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ni, W.; Xu, S.L.; Gonzalez-Grandio, E.; Chalkley, R.J.; Huhmer, A.F.R.; Burlingame, A.L.; Wang, Z.Y.; Quail, P.H. PPKs mediate direct signal transfer from phytochrome photoreceptors to transcription factor PIF3. Nat. Commun. 2017, 8, 15236. [Google Scholar] [CrossRef]
- Ni, W.; Xu, S.L.; Chalkley, R.J.; Pham, T.N.; Guan, S.; Maltby, D.A.; Burlingame, A.L.; Wang, Z.Y.; Quail, P.H. Multisite light-induced phosphorylation of the transcription factor PIF3 is necessary for both its rapid degradation and concomitant negative feedback modulation of photoreceptor phyB levels in Arabidopsis. Plant Cell 2013, 25, 2679–2698. [Google Scholar] [CrossRef] [Green Version]
- Yang, K.; Wang, L.; Le, J.; Dong, J. Cell polarity: Regulators and mechanisms in plants. J. Integr. Plant Biol. 2020, 62, 132–147. [Google Scholar] [CrossRef] [Green Version]
- Gudesblat, G.E.; Schneider-Pizon, J.; Betti, C.; Mayerhofer, J.; Vanhoutte, I.; van Dongen, W.; Boeren, S.; Zhiponova, M.; de Vries, S.; Jonak, C.; et al. SPEECHLESS integrates brassinosteroid and stomata signalling pathways. Nat. Cell Biol. 2012, 14, 548–554. [Google Scholar] [CrossRef]
- Lampard, G.R.; Macalister, C.A.; Bergmann, D.C. Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science 2008, 322, 1113–1116. [Google Scholar] [CrossRef] [Green Version]
- Lu, K.J.; De Rybel, B.; van Mourik, H.; Weijers, D. Regulation of intercellular TARGET OF MONOPTEROS 7 protein transport in the Arabidopsis root. Development 2018, 145. [Google Scholar] [CrossRef] [Green Version]
- Sugiyama, N.; Nakagami, H.; Mochida, K.; Daudi, A.; Tomita, M.; Shirasu, K.; Ishihama, Y. Large-scale phosphorylation mapping reveals the extent of tyrosine phosphorylation in Arabidopsis. Mol. Syst. Biol. 2008, 4, 193. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Loosening of plant cell walls by expansins. Nature 2000, 407, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.Y.; Nakano, T.; Gendron, J.; He, J.; Chen, M.; Vafeados, D.; Yang, Y.; Fujioka, S.; Yoshida, S.; Asami, T.; et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2002, 2, 505–513. [Google Scholar] [CrossRef] [Green Version]
- Vert, G.; Walcher, C.L.; Chory, J.; Nemhauser, J.L. Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proc. Natl. Acad. Sci. USA 2008, 105, 9829–9834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, J.X.; Gendron, J.M.; Yang, Y.; Li, J.; Wang, Z.Y. The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2002, 99, 10185–10190. [Google Scholar] [CrossRef] [Green Version]
- Bernardo-Garcia, S.; de Lucas, M.; Martinez, C.; Espinosa-Ruiz, A.; Daviere, J.M.; Prat, S. BR-dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth. Genes Dev. 2014, 28, 1681–1694. [Google Scholar] [CrossRef] [Green Version]
- Tian, Y.; Fan, M.; Qin, Z.; Lv, H.; Wang, M.; Zhang, Z.; Zhou, W.; Zhao, N.; Li, X.; Han, C.; et al. Hydrogen peroxide positively regulates brassinosteroid signaling through oxidation of the BRASSINAZOLE-RESISTANT1 transcription factor. Nat. Commun. 2018, 9, 1063. [Google Scholar] [CrossRef]
- Kashina, A.S. Assaying ATE1 Activity in Yeast by beta-Gal Degradation. Methods Mol. Biol. 2015, 1337, 59–65. [Google Scholar] [CrossRef]
- Yin, Y.; Wang, Z.Y.; Mora-Garcia, S.; Li, J.; Yoshida, S.; Asami, T.; Chory, J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 2002, 109, 181–191. [Google Scholar] [CrossRef] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, M.; Tian, Y.; Han, C.; Zhou, C.; Bai, M.-Y.; Fan, M. Phospho-Mutant Activity Assays Provide Evidence for the Negative Regulation of Transcriptional Regulator PRE1 by Phosphorylation. Int. J. Mol. Sci. 2020, 21, 9183. https://doi.org/10.3390/ijms21239183
Wang M, Tian Y, Han C, Zhou C, Bai M-Y, Fan M. Phospho-Mutant Activity Assays Provide Evidence for the Negative Regulation of Transcriptional Regulator PRE1 by Phosphorylation. International Journal of Molecular Sciences. 2020; 21(23):9183. https://doi.org/10.3390/ijms21239183
Chicago/Turabian StyleWang, Minmin, Yanchen Tian, Chao Han, Chuanen Zhou, Ming-Yi Bai, and Min Fan. 2020. "Phospho-Mutant Activity Assays Provide Evidence for the Negative Regulation of Transcriptional Regulator PRE1 by Phosphorylation" International Journal of Molecular Sciences 21, no. 23: 9183. https://doi.org/10.3390/ijms21239183
APA StyleWang, M., Tian, Y., Han, C., Zhou, C., Bai, M.-Y., & Fan, M. (2020). Phospho-Mutant Activity Assays Provide Evidence for the Negative Regulation of Transcriptional Regulator PRE1 by Phosphorylation. International Journal of Molecular Sciences, 21(23), 9183. https://doi.org/10.3390/ijms21239183






