Abstract
Durum wheat (Triticum turgidum L. ssp. durum) is a minor crop grown on about 17 million hectares of land worldwide. Several grain characteristics determine semolina’s high end-use quality, such as grain protein content (GPC) which is directly related to the final products’ nutritional and technological values. GPC improvement could be pursued by considering a candidate gene approach. The glutamine synthetase (GS)/glutamate synthase (GOGAT) cycle represents a bottleneck in the first step of nitrogen assimilation. QTL for GPC have been located on all chromosomes, and several major ones have been reported on 2A and 2B chromosomes, where GS2 and Fd-GOGAT genes have been mapped. A useful and efficient method to validate a putative QTL is the constitution of near-isogenic lines (NILs) by using the marker found to be associated to that QTL. Here, we present the development of two distinct sets of heterogeneous inbred family (HIF)- based NILs segregating for GS2 and Fd-GOGAT genes obtained from heterozygous lines at those loci, as well as their genotypic and phenotypic characterizations. The results allow the validation of the previously identified GPC QTL on 2A and 2B chromosomes, along with the role of these key genes in GPC control.
1. Introduction
Durum wheat (Triticum turgidum var. durum Desf.) represents about 5% of the global wheat production and is mainly grown in three principal areas: the Mediterranean basin, the Northern Plains between USA and Canada, and within the desert areas of South West USA and Northern Mexico, with a global production which exceeded 38 million tons in the last cropping seasons. Among the Mediterranean countries, Italy is the major durum wheat producer with an annual average of almost 4.0 MMT (million metric tons) (International Grain Council, https://www.igc.int/en/default.aspx). Despite several local food products are obtained from durum wheat semolina (the coarse, purified durum wheat middlings), such as typical breads, couscous, or bulgur, this cereal crop is mainly used for high-quality pasta production. One of the most important characteristics determining semolina’s high end-use quality is grain protein content (GPC), as directly related to both nutritional and technological values of final food products [1,2]. The development of high GPC new varieties has been a constant priority in breeding programs, although it has been difficult to pursue, as GPC is a quantitative trait, regulated by a complex genetic system and affected by environmental factors, as well as genotype and management practices [3]. Moreover, one relevant aspect to be considered in GPC breeding programs is the well-known strong negative correlation with grain yield, which makes the simultaneous increase of both traits challenging to achieve [4,5]. Indeed, at a genetic level, both GPC and grain yield-related traits are determined by multiple quantitative trait loci (QTLs) interacting with each other and the environment. The negative correlation between GPC and yield-related traits has been observed in both segregating populations and germplasm collections [6,7]. Several studies have considered GPC and grain-yield components simultaneously assessed on the same population to identify GPC loci without pleiotropic effects and/or not closely linked to gene for low yield-related traits, and interesting results were reported both for 2A [8,9,10,11,12,13] and 2B chromosomes [12,14,15,16,17]. The identification of genetic sources of elevated protein content without negative pleiotropic effects would indeed be useful for improving GPC and GY simultaneously.
Improving GPC could be pursued by considering a candidate gene approach. Taking advantage of the recent advances in both bread and durum wheat genome sequencing and annotations [18,19], the identification of genes playing a key role in the Nitrogen Use Efficiency (NUE) process, such as N uptake from the soil, amino acid metabolism, or N transferring to the protein in the grain, has become an effective approach.
The glutamine synthetase (GS)/glutamate synthase (GOGAT) cycle represents a bottleneck in the first step of nitrogen assimilation, as these two enzymes work synergistically to incorporate the up-taken ammonium into organic molecules [20].
QTLs for GPC and NUE have been located on all chromosomes, and several authors reported major ones on 2A and 2B chromosomes of bi-parental mapping population, where GS2 and Fd-GOGAT genes have been mapped [8,9,10,11,14,15,16]. Nevertheless, QTLs validation is of primary importance for further breeding programs development or gene cloning. Single QTL effect on phenotype might be population-specific and/or overestimated, and this has been demonstrated for complex traits such as GPC [15]. The constitution of near-isogenic lines (NILs) for the two different alleles of a target QTL is a useful and efficient method to validate a putative QTL. NILs are lines segregating only at target QTL but homozygous at the rest of the genome. Principally, NILs can be obtained by two approaches: (a) by backcrossing lines carrying the QTL region from a donor to a receipt line several times [21,22]; and (b) by exploiting the residual heterozygous lines (RHLs) from inbred populations and generating heterogeneous inbred family (HIF) at a QTL region [23,24], with the latter being more efficient and more rapid for both major and minor QTL [25].
In previous studies, we identified GS2 and Fd-GOGAT genes as good candidates for GPC QTL with no pleiotropic effect on yield in durum wheat on 2B and 2A chromosomes, respectively [11,15,26,27]. Functional markers based on polymorphisms detected within the sequences of the two genes in the two parental lines were mapped in the durum wheat Svevo × Ciccio (S×C) RILs population [28]. GS2 gene co-localized with a GPC-QTL detected on chromosome arm 2BL, in the region flanked by the markers D_304657 and Xwmc332, significant in one environment and across environments with a LOD ≥ 3.0 [26]. Fd-GOGAT gene co-localized with a GPC QTL detected on chromosome arm 2AS, in the region comprised between the markers Xgwm372c and the EST-SSR TC82001 (including the two closer markers Xgwm339 and Xgwm95), significant in two environments and across environments. The Svevo allele (increasing the trait) had positive additive effects ranging from 0.13 to 0.27 with a mean R2 value of 0.24, and the percentage of phenotypic variance explained by the additive effects of the mapped QTL ranged from 6% to 19.4% between environments and the mean was 19.4% across environments [27].
QTL analysis performed with CIM (Composite Interval Mapping) confirmed the presence of these markers in two major QTL for grain protein content. Recently, these data were confirmed on a wider durum wheat collection consisting of 236 durum wheat genotypes [29,30]. In addition, we found a significant difference in both GS gene expression and enzymatic activities between the durum wheat cvs Ciccio and Svevo, consistently higher in the last one [31]. Following up these results, here we present the development of two distinct set of heterogeneous inbred family (HIF)-based NILs segregating for GS2 and Fd-GOGAT genes from heterozygous lines at those loci (previously identified in the S×C RIL population), and their genotypic and phenotypic characterizations, aimed to validate the previously identified GPC QTL on 2A and 2B chromosomes. Furthermore, we investigated the genomic characterization of the promoting regions of GS2 and Fd-GOGAT genes, as regulatory elements involved in the transcription processes might be the ones responsible for the different level of gene expression and might contribute in assessing the role of these key genes in GPC control.
2. Results
2.1. Development of NILs at the GPC QTL on Chromosomes 2A and 2B
The S × C mapping population as previously phenotyped for GPC and used to both genetically and physically map GS2 and Fd-GOGAT genes [26,27]. The RIL population consisted of 120 lines, two of which were found to be residual heterozygous lines at GS2 and Fd-GOGAT genes, respectively, and selected to develop the two NIL sets. Specifically, the RIL SC45 F6:F7 was found to be heterozygous for GS2 gene, and thus was allowed to self-pollinate to F8:F9 generation to develop GS2_NIL lines. The resulting 12 lines were tested with the previous reported functional marker GS-46 [26], as well as with four additional flanking markers from the integrated SNP map S×C [32] and six of them were excluded as still heterozygous at the locus. The final GS2_NIL genotypes consisted of six lines, three being homozygous for the high GPC allele of parental line Svevo (SC45-2, SC45-7 and SC45-12) and the remaining homozygous with the low GPC allele of parental line Ciccio (SC45-3, SC45-6 and SC45-11). Likewise, the Fd-GOGAT_NIL population was obtained from line SC107, heterozygous for Fd-GOGAT gene (Table 1). Although in this case the marker was dominant, the homozygosity and heterozygosity were confirmed by analyzing each line’s progeny. In addition to the GS2 NILs lines, four additional flanking markers from the integrated SNP map S×C [32] were used to genotype the Fd-GOGAT NILs. Out of 15 plants achieved from its self-pollination, only two were homozygous for the parental Ciccio allele (low GPC) (SC107-9 and SC107-15). Two lines homozygous for the parental Svevo allele (high GPC) were chosen among the available 13, based on their homozygosity at co-segregating markers (data not shown). The final Fd-GOGAT_NIL population consisted then of four lines. Nevertheless, one of them, SC107-15, was lost as no seeds were produced, therefore at the end the Fd-GOGAT NIL set comprised three lines, with two, SC107-1 and SC107-8, carrying the Svevo allele.

Table 1.
Generation of heterogeneous inbred family (HIF)-NILs by self-crossing RILs from the Svevo × Ciccio mapping population having residual heterozygosity at GS2 and Fd-GOGAT candidate genes, respectively. In bold, the selected lines from which (HIF)-NILs were developed.
Microsatellite marker Xwmc332 and several SNP markers from the Svevo × Ciccio map [32] were analyzed in the NILs and this allowed us to reduce the GPC QTL on 2B to 4.7 cM. For QTL on 2A chromosome, EST-SSR TC82001 and several SNP markers were used to analyze the NILS, reducing the GPC QTL region to 4.9 cM. Those QTL interval regions were searched in the Svevo Genome Browser, and the gene lists within them were downloaded and screened. Candidate genes potentially involved in nitrogen and ammonium metabolism (genes that might play a key role in the N uptake from the soil, and of those controlling the enzymes of amino acid metabolism, potentially involved in N transferring to the protein in the grain) were looked for, but none was found.
2.2. Phenotypic Performance and Variation of the HIF Families
GS2_NIL and Fd-GOGAT_NIL lines were grown in five different site × season environments, and GPC, GYS and TKW data retrieved.
Concerning GS2_NIL, significant differences in GPC among parental lines and each derived NIL family were observed among the trials conducted in different locations, while Tukey Pairwise Comparisons showed a more variable situation for both GYS and TGW means of parental lines and NILs, most likely due to the environmental factors affecting those yield-related traits. Svevo had significantly higher GPC than Ciccio in all environments as well as the derived heterogeneous inbred family. Indeed, considering the mean across environments, the two parental lines showed a significant difference in their GPC values, as well as their derived NIL lines (SC45-2, SC45-7, SC45-12, and Svevo grouped together, as well as SC45-3, SC45-6, SC45-11, and Ciccio) (Table 2). Interestingly, the data also show a different situation for both GYS and TGW means, as no significant differences could be observed either between the two parental lines or their derived NILs sets, thus being a positive result considering the strongly negative correlation between GPC and yield.

Table 2.
Mean values for Grain protein content (GPC), grain yield per spike (GYS), and thousand grain weight (TGW) of heterogeneous inbred family (HIF)-based NILs segregating for GS2 and parental lines Svevo and Ciccio evaluated in five different environments. Different letters indicate significant differences (one-way ANOVA and Tukey’s tests; p < 0.05).
The same situation was observed when considering ANOVA and Tukey’s comparison of Fd-GOGAT HIF for GPC, GYS, and TKW. As seen for the GS2 HIF, Svevo had significantly higher GPC than Ciccio in all environments as well as its derived heterogeneous inbred family over the ones from Ciccio. Table 3 shows that, considering the mean across environments, SC 107-1 and SC 107-8 had higher GPC values, as well as Svevo. On the other side, SC 107-9 showed lower values, similar to the parental line Ciccio. No significant differences could be observed either between the two parental lines or their derived Fd-GOGAT HIF for GYS and TGW means across environments.

Table 3.
Mean values of Grain protein content (GPC), grain yield per spike (GYS), and thousand grain weight (TGW) of heterogeneous inbred family (HIF)-based NILs segregating for Fd-GOGAT and parental lines Svevo and Ciccio evaluated in five different environments. Different letters indicate significant differences (one-way ANOVA and Tukey’s tests; p < 0.05).
Data of yield of 1-m row were also analyzed to estimate yield on an area basis for both NILs groups. Within GS2 HIF derived NILs, values ranged from 0.78 (lineSC45-2) to 0.90 kg/m2 (line SC45-7). Likewise, Within Fd-GOGAT HIF derived NILs, values ranged from 0.53 (line SC107-1) to 0.63 kg/m2 (line SC107-9). No significant statistic difference was observed within the NILs.
2.3. Sequencing of the GS2 Gene Promoter Region
GS2 gene structure and characterization have been reported in both durum and bread wheat [26,31,33]. Both A and B homoeologous genes are organized into 13 exons and 12 introns, with two alternative splicing forms, while the D genome homoeologous has just one transcript form and one more exon at 5′ terminus, which is a UTR region (http://plants.ensembl.org/Triticum_aestivum/).
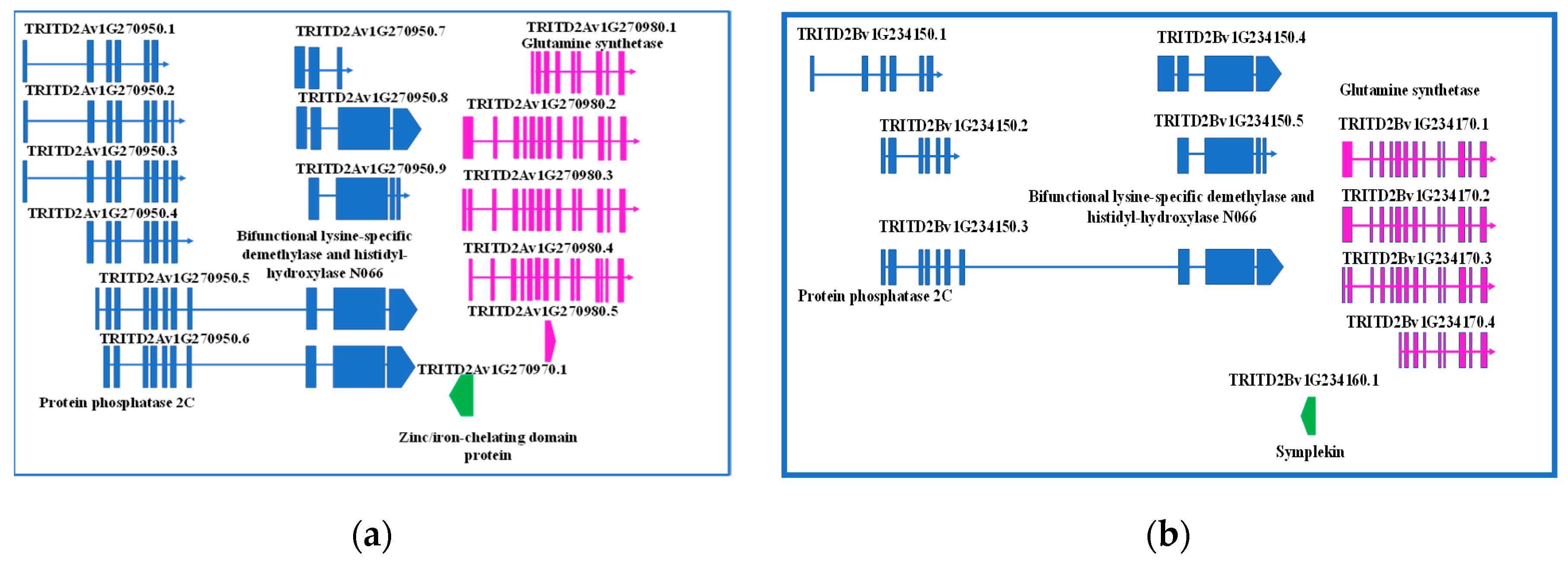
The fine investigation of this region was performed by searching the durum wheat genome browser, T. durum (Svevo.v1) [19]. The first exon of GS2-A2 gene was found to overlap with another element, a single exon gene encoding a zinc/iron-chelating domain protein G, oriented in the minus strand. Interestingly, upstream this gene, another one, having up to nine different splicing forms, was found, which encodes for a protein phosphatase 2C, a lysine-specific demethylase NO66, or a bifunctional lysine-specific demethylase and histidyl-hydroxylase NO66, depending on the splicing form. The region was also screened for regulatory elements, but none was found (Figure 1a).
Figure 1.
Detailed representation of genes located upstream GS2-2A (a) and GS-2B (b) genes, their structures, function, and eventual splicing isoforms.
A 333-bp element was found in the minus strand upstream GS2-B2 gene, but it was not overlapping. Sequence analysis determined it is a member of Symplekin gene family, which encodes a nuclear protein involved in the regulation of polyadenylation and promoting gene expression. As reviewed by Hunt (2008) [34], the protein forms a high-molecular weight complex with components of the polyadenylation machinery. It is thought to serve as a scaffold for recruiting regulatory factors to the polyadenylation complex. It also participates in 3’-end maturation of histone mRNAs, which do not undergo polyadenylation. As reported for the region upstream GS2-2A gene, also on the B genome a gene having five different splicing forms was found, encoding either for a protein phosphatase 2C or bifunctional lysine-specific demethylase and histidyl-hydroxylase NO66, depending on the splicing form.
Interestingly, a region of 160 bp of the Symplekin gene was identified as a MITE element. In addition, 312 bp upstream this region, a 94-bp conserved element was found, which has been identified several times in coding regions of bread wheat, once reported as ncRNA (Figure 1b).
Two sets of genome specific primers were then designed and used to amplify the genomic region upstream GS2 homoeologous genes in the two durum wheat parental lines Svevo and Ciccio and the HIF-NIL lines (primer sequencing and amplification condition are reported in Table S1).
Sequencing of approximatively 1 kb upstream the first exon of GS-A2 gene revealed an identity of 100% between Svevo and Ciccio sequences, with no gaps or single nucleotide polymorphisms detected. Instead, the sequencing and analysis of GS-B2 gene promoter region highlighted a slightly different situation. The alignment of GS2-B2 gene promoters of Svevo and Ciccio revealed a 132-bp deletion in Ciccio cv (Figure 2a). The deletion in Ciccio cv entailed the loss of 83 bp of the previously reported conserved element, including a 55 bp tandem repeat, whose 27-bp seed was repeated twice.
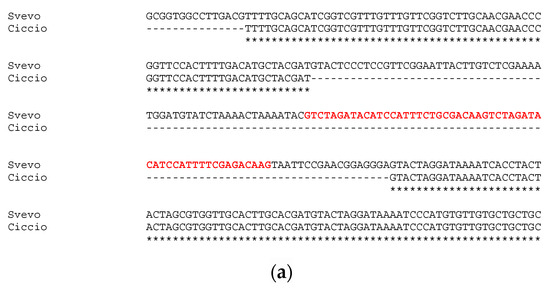
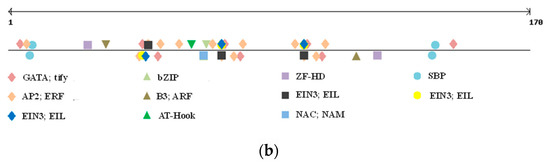
Figure 2.
Alignment of GS2B gene’s promoter sequenced in Svevo and Ciccio cvs. with the tandem repeat reported in red bold (a); and details of TF families’ position found in the indel (b).
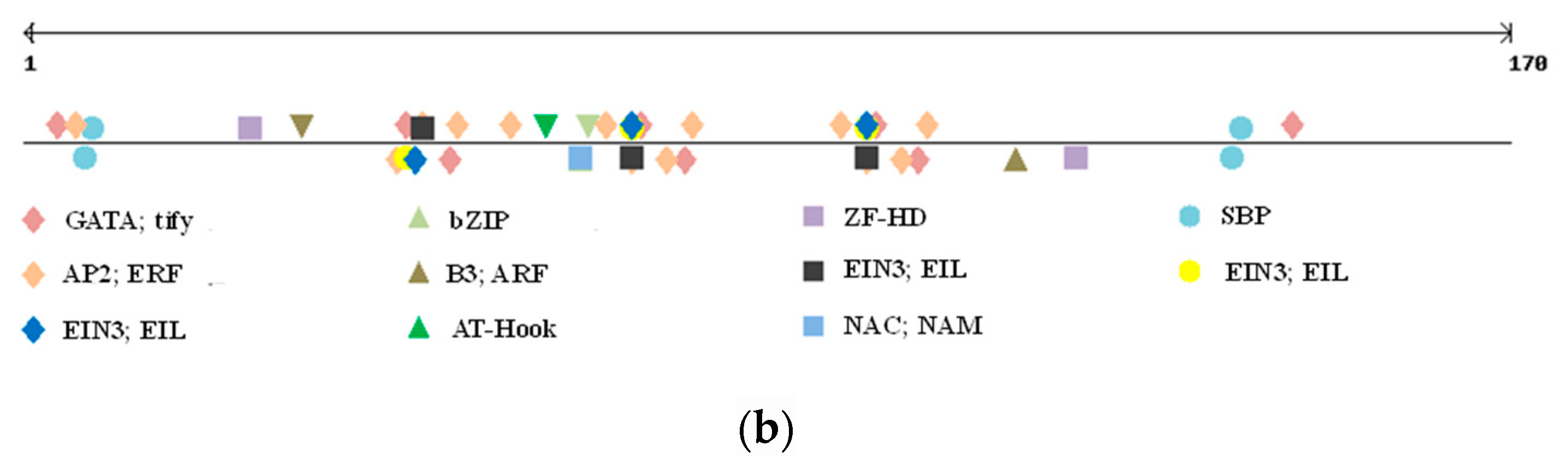
The polymorphic region was screened for eventual TFBSs, as well as GpC islands and repetitive motifs by searching PlantPAN3.0 database for A. thaliana, Oryza sativa, Brachypodum distachyon, and Zea mais. While no GpC island was located in the polymorphic indel, a number of TFBSs were detected (Figure 2b).
We only considered TFBSs having a perfect match with the ones already reported in the abovementioned species (score = 1). As shown in Table 4, nine different TFBSs were detected within the indel sequence, and all of them, except for the SBP binding factor, were located within the tandem repeat region. Positive regulator to ethylene response pathway, zinc finger domain, and bZIP binding sites were found, but the most interesting one was represented by a NAC; NAM binding site. Indeed, while all the other TFs binding to the identified sites are involved in generic plant stress response and metabolism, this specific one has been reported as affecting grain protein content and micronutrient amount [35,36,37,38].

Table 4.
Transcription factor (TF) binding sites detected in the polymorphic indel between the two parental lines and NIL families for GS-B2 gene.
2.4. Sequencing of the Fd-GOGAT Gene Promoter Region
Fd-GOGAT gene structures have been extensively reported in durum wheat [27]; this large gene (33 exons and 32 introns) spans over about 15 kbp. Moreover, several different splicing isoforms have been found for homoeologous genes, as well as precisely four for 2A and eight for 2B homoeologous chromosomes. The regions upstream Fd-GOGAT genes were investigated to detect eventual close and/or overlapping genes in the promoter regions as observed for the GS2 genes, but none was found.
As done for GS2 genes, a set of genome specific primer was also designed to amplify and sequence both Fd-GOGAT-2A and Fd-GOGAT-2B promoters in Ciccio, Svevo, and their derived HIF-NILs lines. While no polymorphism was detected for the 2B promoter, some were found in the 2A homoeologous gene. Specifically, three SNPs were detected between cv Ciccio and Svevo: two transversions (G:T and C:G) and one transition (A:G). The promoter regions were investigated and screened in both parental lines for eventual differences in TFBS sites due to the nucleotide polymorphisms mentioned above, and some interesting differences were outlined, as shown in Table 5. The detected SNPs determined the occurrence or loss of some specific TFBSs at that site, resulting in additional BSs or, in some cases, in specific and unique BSs for one of the two parental lines. Specifically, in cv Svevo, a TCP and a Myb/SANT; G2-like TFBSs were detected at high similarity score (>0.9) in the regions where SNPs were located. Moreover, these polymorphisms determined the occurrence of additional TFBSs to those already reported for both cvs, and specifically a Trihelix, a bHLH, and a NAC; NAM site.

Table 5.
Specific and additional Transcription factor (TF) binding sites detected in parental line Svevo and its derived HIF for Fd-GOGAT gene due to SNPs polymorphism in the promoter region.
A gene group analysis performed simultaneously on Svevo’s GS-B2 and Fd-GOGAT_2A promoter regions determined the co-occurrence of a number of TFBSs. Specifically, 252 common sites were identified, with most of them belonging to specific TF families, such as AP2, ERF, bHLH, bZIP, Dof, MYB, NAM, and WRKY.
3. Discussion
Here, we report the development of two distinct sets of HIF-based NILs, segregating for GS2 and Fd-GOGAT genes, from heterozygous lines at those loci, previously identified in a durum wheat RIL population, as well as their genotypic and phenotypic characterization.
The HIF method [24] used in this work allowed us to generate useful NILs at a single locus from a single cross (RIL Svevo × Ciccio [28]) instead of using markers flanking a QTL [39]. Single functional markers, for GS2 and Fd-GOGAT genes, respectively, both linked with the GPC QTL of interest, were used for generating the two NILs sets. Compared to the traditional method, which is based on the selection of two flanking markers delimiting a QTL of interest, the HIF method allows the production of NILs with reduced sizes of the “non-desirable” chromosomal segment discriminating the isolines.
Traditionally, each NIL set has its own genetic composition, which determines unique morphological and phenological characteristics influencing the expression of a quantitative trait [38].
The assessment of the effect of a particular gene on a specific trait should be more accurate by using a NIL pair obtained by the HIF method, as fewer isolines would be needed for investigating the effect of a QTL/gene.
Thus far, HIF-based NILs have been intensively used for investigating the effects of various QTL and traits of interest in wheat, such as dormancy QTL [40], pre-harvest sprouting [41], and spikelet number per spike [42].
During the past decades, the increase in GPC has mainly been achieved by intensifying nitrogen (N) fertilization. Considering the high costs of N fertilizers and the harmful environmental impacts of nitrate loss from the soil, decreasing the amount of N applied to cereal cropping systems while maintaining high productivity of modern cultivars has therefore become a breeding priority. The identification of candidate genes and their allelic variants affecting this trait is an effective method to develop new wheat varieties with high GPC. Nitrogen remobilization efficiency is an important factor increasing grain protein content (GPC) and consequently, the nutritional and technological properties of flour and semolina [43,44]. Several authors have focused on deciphering GPC and NUE QTLs, and genetic diversity at candidate genes have recently been considered for this purpose. Indeed, in our previous studies, we identified both GS2 and Fd-GOGAT genes as good candidates in GPC study in the S×C RIL population [26,27]. Specifically, these two genes work synergistically in the GS/GOGAT cycle, which has a key role in nitrogen assimilation and recycling in young leaves within the chloroplast, where nitrite reduction occurs, and ammonium is assimilated [45]. Both genes are located on the chromosome 2 homoeologous group, whose influence on GPC control was reported in different genetic materials, thus suggesting its key role in the control of the character [11,15,46,47,48].
Interestingly, Kichey et al. (2005) and Habash et al. (2007) [9,49] showed that QTLs for GS activity co-localized with QTL for grain N on chromosome 2A and hinted this may be coincident with QTL on 2B and 2D homeologs for soluble protein content, and that increased activity was associated with higher grain N. This finding was confirmed on different genetic material [50]. In addition, in maize, the GS2 locus was found to be coincident with a leaf senescence QTL [51], but, unlike in maize, Fontaine et al. (2009) [50] did not find a correlation between GS activity and yield components in wheat, this being in agreement with our data. Indeed, as previously reported [11,15], QTL for GPC were found on 2A and 2B chromosomes, in the same region where Fd-GOGAT and GS2 gene were mapped in Svevo × Ciccio RIL population, respectively [26,27]. Both authors reported that these QTLs showed no negative effects on grain yield related traits, making them good candidate for marker-assisted selection to improve GPC and grain yield simultaneously. However, considering that a QTL usually spans several cM, it is necessary to perform more detailed genetic analyses on larger populations or specific genetic material, such as NILs, in order to assess whether a single gene with pleiotropic effects or different loci within the linkage group are responsible for the different traits. The near-isogenic lines we developed at both GS2 and Fd-GOGAT loci showed that, despite having significant differences in GPC, no significant ones were observed in GYS. This could be either explained with both GS2 and Fd-GOGAT having no negative pleiotropic effects on yield components, or that a potential effect is actually masked by environment. Despite in both HIF-NILs families it was noticed that lines having the Svevo allele showed higher GPC, it was also outlined that the differences observed within NILs were highly statistically significant especially for GS HIF-derived NILs, as Fd-GOGAT HIF-derived ones showed a lower value of significant difference, as shown in Table 2 and Table 3. In both cases, we could assume that the Svevo allele is the one increasing the GPC trait.
The differences in GPC between the isolines developed for two key genes involved in nitrogen metabolism Glutamine synthetase and Glutamate synthase, not only further confirm the significance of these two genes in the processes determining the final grain protein content but would also facilitate the exploitation of these HIF NILs families in further characterizing the major GPC QTL on 2A and 2B chromosomes, studying their interaction with other traits of agronomic importance, and developing functional markers that can be reliably used to follow these major loci.
For GS2 genes, a different gene expression was reported between the two cultivars that could affect the GPC content [31]. In order to detect eventual polymorphism potentially involved in a differential gene expression, the promoter regions of both GS2 and Fd-GOGAT genes were screened in Svevo and Ciccio parental lines.
Among all regulatory elements, Transcription Factors (TFs) regulate cell processes by binding a specific DNA motif on promoter regions and affecting downstream gene expression.
By comparing the promoting regions of the genes considered in this study, the most interesting result was observed in the promoting region of GS-B2 gene, as a 132-bp indel was detected between the two parental lines. The analysis of this region outlined the presence of a 55-bp tandem repeat, as well as a number of TFBSs, such as EIN/EIL1, bZIP, AP2/ERF, SPL, ZF-HD, and NAC; NAM.
A recent review reported the role of EIN3/EIL1 transcription factors in Arabidospis, highlighting their key role as regulators of ethylene signaling [52]. Ethylene regulates many different aspects of plant development and stress responses, thus its signaling pathway needs proper modulation depending on the plant conditions. Salih et al. (2020) [53] reported a very recent study on the possible functions of EIL/EIN3 proteins in cotton. Their GO annotations and KEGG pathway analyses indicated that, besides being involved in the ethylene-activated signaling pathway and a number of cellular macromolecule metabolic process, a great number of EIL/EIN3 genes were also involved in nitrogen compound metabolic process.
In addition, a bZIP binding site was found in the polymorphic indel, which represents one of the largest and most variable families of TFs and are uniquely present in eukaryotes. A genome-wide analysis and gene ontology enrichment analysis of bZIP Transcription Factors performed on 191 bZIP TFs, identified in bread wheat, reported that some of them are involved in cellular metabolic processes related to nitrogen compounds [54]. More investigations will be needed to define and characterize the interaction between these TFs and GS2 gene in order to better explain the role of these TF families in nitrogen metabolism pathway.
AP2/ERF transcription factors were classified into five subfamilies [55], the most known of which are: AP2 (APETALA2), RAV (related to ABI3/VP1), DREB (dehydration-responsive element binding protein), and ERF (ethylene-responsive factor). The AP2/ERF transcription factors were shown to regulate diverse processes of plant development and stress responses, such as vegetative and reproductive development, cell proliferation, abiotic and biotic stress responses, and plant hormone responses [56,57,58].
SQUAMOSA Promoter-Binding Protein-Like (SPL) genes have been shown to play numerous important roles during plant growth and development [59]. SPLs are known to regulate several biological processes, including leaf development [60], phase transition [61], flower and fruit development [62], plant architecture [63], sporogenesis [64], GA signaling [65], and response to copper and fungal toxin [66,67].
Abu-Romman (2014) [68] reviewed ZF-HD TF family and reported that these classes of homeodomain proteins are involved in regulating intercellular trafficking [69], inflorescence stem growth [70], and hormone and stress signaling [71,72,73].
Among all TFBSs identified within the indel in the GS2 gene promoter in cv Svevo, the NAC; NAM TFBS was the most interesting and potentially involved in the different expression of GS2 gene and final GPC between the two analyzed cvs. Several studies have reported that wheat NAC TFs are involved in several biological processes such as senescence and nutrient remobilization [35,74], and stress response, both to biotic (such as stripe rust [75,76,77,78]) and abiotic stresses including drought and salt tolerance [79,80,81,82,83]. Specific studies have been carried out on NAM TFs in relation to GPG and nitrogen metabolism. Waters et al. (2009) [84] reported a TaNAM gene which increased protein content in the grain by increasing the remobilization of nitrogen from vegetative tissues. Interestingly, He et al. (2015) [85] described a TaNAC2 TF that positively regulated TaGS2 expression.
In conclusion, the data reported in the present work confirm that GS2 and Fd-GOGAT genes are involved in grain protein accumulation and suggest that the surrounding genomic regions and their promoters could affect gene expression. The identification of new useful superior alleles for both genes could be employed for marker-assisted selection and the constitution of wheat varieties with improved agronomic performance and N-use efficiency.
4. Materials and Methods
4.1. Plant Material
The Svevo × Ciccio mapping population, consisting of 120 F6:F7 RILs [28], was considered in previous studies to genetically dissect important agronomic traits, such as grain yield components, grain protein content, and yellow pigments [11,31]. We focused on data related to GPC, and, specifically, on RILs which have been shown to have residual heterozygosity at GS2 and Fd-GOGAT candidate genes. Near isogenic lines were indeed obtained starting from heterozygous lines at the genotype of the marker associated with the QTL of interest in the F6 RILs, SC45 and SC107, heterozygous at GS2 and Fd-GOGAT loci, respectively. The two set of heterogeneous inbred families (HIF) were generated by self-crossing to F8:F9 generation and subsequently used to validate individual effects of the putative QTLs in different environments.
4.2. Experimental Design and Phenotypic Evaluation
The two NILs sets, containing heterogeneous inbred families, respectively, three for Fd-GOGAT and six for GS2 genes, as well as the two parental lines Svevo and Ciccio, were grown for three growing seasons (2012–2013, 2013–2014, and 2014–2015) at the experimental field trials of Bari University, Valenzano (Bari), and Policoro (Potenza), both located in South Italy. Field trials were organized as a randomized complete blocked design (RCBD) for all entries, with ten total blocks, each including all lines, and each line was sown in a plot of one m2. In total, 14 genotypes were tested including parents as double check, for ten replicas (140 plots) each year. Standard agronomic practices were followed, and, during the growing season, 80 kg/ha of N were applied. At maturity, plots were hand harvested and phenotypic data and measurements were recorded for grain protein content (GPC), grain yield per spike (GYS) and thousand kernel weight (TKW). Grain protein content, expressed as a percentage of protein on a dry weight basis, was determined on 3 g sample of whole-meal flour using near-infrared reflectance spectroscopy (Spectra Alyzer Premium, Zeutec Büchi, Rendsburg, Germany). For each plot, the plants were harvested, seeds collected, five replicas analyzed, and a mean value given as line value. TKW was evaluated on a 15g seed sample per plot.
4.3. Statistical Analysis
The data were analyzed as a randomized design with three biological and five technical replicates and expressed as means ± SE. Statistical analysis was carried out using Sigma Plot software 12.0 (Systat Software, Inc., San Jose, CA, USA). One-way analysis of variance (ANOVA) and Tukey’s comparison test were used to calculate the difference between the genotypes and NILs families. Differences were considered statistically significant at a p-value of < 0.05.
4.4. Molecular Marker Analysis
Molecular analysis was carried out to confirm the segregation at loci of interest and to check for casual contaminations. DNA was extracted from leaves collected at tillering stage from both parental lines and NILs using a modified CTAB method. The chromosomal location of GS2 and Fd-GOGAT gene functional markers, along with their genetic distances on the durum wheat map, their sequences, and PCR conditions, are, respectively, reported in [26,27].
4.5. Promoter Region Sequencing
The promoter regions of both 2A and 2B homoeologous GS2 and Fd-GOGAT genes were retrieved from both bread and durum wheat genomes (cvs Chinese Spring and Svevo) (http://plants.ensembl.org/Triticum_aestivum/Info/Index, https://www.interomics.eu/durum-wheat-genome-intranet). Two sets of genome specific primer pairs were designed for each gene by using OligoExplorer software (http://www.genelink.com/tools/gl-oe.asp) and subsequently used to amplify both promoter regions in the durum wheat cvs Ciccio and Svevo (to exclude possible casual variety contamination) as well as in the two HIF-based NILs families, as previously reported [86]. Primer sequences are reported in Table S1. Single PCR fragments were cloned and sequenced as reported in [87].
The sequenced promoters of the two cvs were aligned and compared in order to detect possible polymorphisms potentially affecting transcription factors binding sites (TFBSs) by PlantPan3.0 database (http://plantpan.itps.ncku.edu.tw/index.html).
Supplementary Materials
Supplementary Materials can be found at https://www.mdpi.com/1422-0067/21/23/9253/s1. Table S1. Oligo name and sequences of primer combinations used to amplify promoter regions of GS2 and Fd-GOGAT homoeologous genes.
Author Contributions
Conceptualization, D.N. and A.G.; methodology, D.N., S.F., S.L.G., and E.M.; formal analysis, S.F. and S.L.G.; investigation, D.N., S.F., S.L.G., and E.M.; writing—original draft preparation, D.N.; writing—review and editing, D.N., E.M., and A.G.; supervision, D.N. and A.G.; project administration, D.N. and A.G.; and funding acquisition, D.N. and A.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by “Intervento cofinanziato dal Fondo di Sviluppo e Coesione 2007–2013—APQ Ricerca Regione Puglia “Programma regionale a sostegno della specializzazione intelligente e della sostenibilità sociale ed ambientale—FutureInResearch” (D.N.); financial support has been provided by PRIMA (CEREALMED), a programme supported by the European Union PRIMA 2019 (Italy) (A.G.); PSR-Puglia Misura 16.2 project “Iperdurum” (A.G.).
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| GPC | grain protein content |
| GS | glutamine synthetase |
| GOGAT | glutamate synthase |
| NUE | Nitrogen Use Efficiency |
| NILs | near-isogenic lines |
| HIF | heterogeneous inbred family |
References
- Fois, S.; Schlichting, L.; Marchylo, B.; Dexter, J.; Motzo, R.; Giunta, F. Environmental conditions affect semolina quality in durum wheat (Triticum turgidum ssp. durum L.) cultivars with different gluten strength and gluten protein composition. J. Sci. Food. Agric. 2011, 91, 2664–2673. [Google Scholar] [PubMed]
- Kaur, A.; Singh, N.; Kaur, S.; Katyal, M.; Virdi, A.S.; Kaur, D.; Ahlawat, A.K.; Singh, A.M. Relationship of various flour properties with noodle making characteristics among durum wheat varieties. Food Chem. 2015, 188, 517–526. [Google Scholar] [CrossRef] [PubMed]
- Chope, G.A.; Wan, Y.; Penson, S.P.; Bhandari, D.G.; Powers, S.J.; Shewry, P.R.; Hawkesford, M.J. Effects of genotype, season, and nitrogen nutrition on gene expression and protein accumulation in wheat grain. J. Agric. Food. Chem. 2014, 62, 4399–4407. [Google Scholar] [CrossRef] [PubMed]
- Lawlor, D.W. Carbon and nitrogen assimilation in relation to yield: Mechanisms are the key to understanding production systems. J. Exp. Bot. 2002, 53, 789–799. [Google Scholar] [CrossRef]
- Triboi, E.; Triboi-Blondel, A.M. Productivity and grain or seed composition: A new approach to an old problem. Eur. J. Agron. 2002, 16, 163–186. [Google Scholar] [CrossRef]
- Simmonds, N.W. The relation between yield and protein in cereal grain. J. Sci. Food. Agric. 1995, 67, 309–315. [Google Scholar] [CrossRef]
- Oury, F.X.; Berard, P.; Brancourt-Hulmel, M.; Depatureaux, C.; Doussignault, G.; Galic, N.; Giraud, A.; Heumez, E.; Lecompte, C.; Pluchard, P.; et al. Yield and grain protein concentration in bread wheat: A review and a study of multi-annual data from a French breeding program. J. Genet. Breed. 2003, 57, 59–68. [Google Scholar]
- Groos, C.; Robert, N.; Bervas, E.; Charmet, G. Genetic analysis of grain protein content, grain yield and thousand-kernel weight in bread wheat. Theor. Appl. Genet. 2003, 106, 1032–1040. [Google Scholar] [CrossRef]
- Habash, D.Z.; Bernard, S.; Schondelmaier, J.; Weyen, J.; Quarrie, S.A. The genetics of nitrogen use in hexaploid wheat: N utilisation, development and yield. Theor. Appl. Genet. 2007, 114, 403–419. [Google Scholar] [CrossRef]
- Bogard, M.; Jourdan, M.; Allard, V.; Martre, P.; Perretant, M.R.; Ravel, C.; Gaju, O. Anthesis date mainly explained correlations between post-anthesis leaf senescence, grain yield, and grain protein concentration in a winter wheat population segregating for flowering time QTLs. J. Exp. Bot. 2011, 62, 3621–3636. [Google Scholar] [CrossRef]
- Blanco, A.; Mangini, G.; Giancaspro, A.; Give, S.; Colasuonno, P.; Simeone, R.; Gadaleta, A. A relationships between grain protein content and grain yield components through quantitative trait locus analyses in a recombinant inbred line population derived from two elite durum wheat cultivars. Mol. Breed. 2012, 30, 79–92. [Google Scholar] [CrossRef]
- Cormier, F.; Le Gouis, J.; Dubreuil, P.; Lafarge, S.; Praud, S. A genome-wide identification of chromosomal regions determining nitrogen use efficiency components in wheat (Triticum aestivum L.). Theor. Appl. Genet. 2014, 127, 2679–2693. [Google Scholar] [CrossRef] [PubMed]
- Rapp, M.; Lein, V.; Lacoudre, F.; Lafferty, J.; Müller, E.; Vida, G.; Leiser, W.L. Simultaneous improvement of grain yield and protein content in durum wheat by different phenotypic indices and genomic selection. Theor. Appl. Genet. 2018, 131, 1315–1329. [Google Scholar] [CrossRef] [PubMed]
- Charmet, G.; Robert, N.; Branlard, G.; Linossier, L.; Martre, P.; Triboï, E. Genetic analysis of dry matter and nitrogen accumulation and protein composition in wheat kernels. Theor. Appl. Genet. 2005, 111, 540–550. [Google Scholar] [CrossRef]
- Suprayogi, Y.; Pozniak, C.J.; Clarke, F.R.; Clarke, J.M.; Knox, R.E.; Singh, A.K. Identification and validation of quantitative trait loci for grain protein concentration in adapted Canadian durum wheat populations. Theor. Appl. Genet. 2009, 119, 437–448. [Google Scholar] [CrossRef]
- Wang, L.I.N.; Cui, F.A.; Wang, J.; Jun, L.I.; Ding, A.; Zhao, C.; Wang, H. Conditional QTL mapping of protein content in wheat with respect to grain yield and its components. Thai. J. Genet. 2012, 91, 303–312. [Google Scholar] [CrossRef]
- Terasawa, Y.; Ito, M.; Tabiki, T.; Nagasawa, K.; Hatta, K.; Nishio, Z. Mapping of a major QTL associated with protein content on chromosome 2B in hard red winter wheat (Triticum aestivum L.). Breed. Sci. 2016, 66, 471–480. [Google Scholar] [CrossRef]
- Appels, R.; Eversole, K.; Feuillet, C.; Keller, B.; Rogers, J.; Stein, N.; Pozniak, C.J.; Choulet, F.; Distelfeld, A.; Poland, J.; et al. International Wheat Genome Sequencing Consortium (IWGSC) Shifting the limits in wheat research and breeding through a fully annotated and anchored reference genome sequence. Science 2018, 361, 6403. [Google Scholar]
- Maccaferri, M.; Harris, N.S.; Twardziok, S.O.; Pasam, R.K.; Gundlach, H.; Spannagl, M.; Ormanbekova, D.; Lux, T.; Prade, V.M.; Milner, S.G.; et al. Durum wheat genome highlights past domestication signatures and future improvement targets. Nat. Genet 2019, 51, 885. [Google Scholar] [CrossRef]
- Lea, P.J.; Azevedo, R.A. Nitrogen use efficiency. 2. Amino acid metabolism. Ann. Appl. Biol. 2007, 15, 1269–1275. [Google Scholar] [CrossRef]
- Zhou, R.; Zhu, Z.; Kong, X.; Huo, N.; Tian, Q.; Li, P.; Jin, C.; Dong, Y.; Jia, J. Development of wheat near-isogenic lines for powdery mildew resistance. Theor. Appl. Genet. 2005, 110, 640–648. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.; Hu, J.; Ao, Y.T.; Cheng, M.X.; Gao, G.J.; Zhang, Q.L.; He, G.C.; He, Y.Q. Development and evaluation of near-isogenic lines for brown planthopper resistance in rice cv. 9311. Sci. Rep. 2016, 6, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Darvasi, A.; Soller, M. Advanced intercross lines, an experimental population 390 for fine genetic mapping. Genetics 1995, 141, 1199–1207. [Google Scholar] [PubMed]
- Tuinstra, M.R.; Ejeta, G.; Goldsbrough, P.B. Heterogeneous inbred family (HIF) analysis: A method for developing near-isogenic lines that differ at quantitative trait loci. Theor. Appl. Genet. 1997, 95, 1005–1011. [Google Scholar] [CrossRef]
- Bai, X.; Luo, L.; Yan, W.; Kovi, M.R.; Zhan, W.; Xing, Y. Genetic dissection of rice grain shape using a recombinant inbred line population derived from two contrasting parents and fine mapping a pleiotropic quantitative trait locus qGL7. BMC. Genet. 2010, 11, 16. [Google Scholar] [CrossRef]
- Gadaleta, A.; Nigro, D.; Giancaspro, A.; Blanco, A. The glutamine synthetase (GS2) genes in relation to grain protein content of durum wheat. Funct. Integr. Genom. 2011, 11, 665–670. [Google Scholar] [CrossRef]
- Nigro, D.; Blanco, A.; Anderson, O.D.; Gadaleta, A. Characterization of Ferredoxin-Dependent Glutamine-Oxoglutarate Amidotransferase (Fd-GOGAT) Genes and Their Relationship with Grain Protein Content QTL in Wheat. PLoS ONE 2014, 9, e103869. [Google Scholar] [CrossRef]
- Gadaleta, A.; Giancaspro, A.; Giove, S.L.; Zacheo, S.; Mangini, G.; Simeone, R.; Signorile, A.; Blanco, A. Genetic and physical mapping of new EST-derived SSRs on the A and B genome chromosomes of wheat. Theor. Appl. Genet. 2009, 118, 1015–1025. [Google Scholar] [CrossRef]
- Nigro, D.; Fortunato, S.; Giove, S.L.; Mangini, G.; Yacoubi, I.; Simeone, R.; Blanco, A.; Gadaleta, A. Allelic Variants of Glutamine Synthetase and Glutamate Synthase Genes in a Collection of Durum Wheat and Association with Grain Protein Content. Diversity 2017, 9, 52. [Google Scholar] [CrossRef]
- Nigro, D.; Gadaleta, A.; Mangini, G.; Colasuonno, P.; Marcotuli, I.; Giancaspro, A.; Giove, S.L.; Simeone, R.; Blanco, A. Candidate genes and genome-wide association study of grain protein content and protein deviation in durum wheat. Planta 2019, 249, 1157–1175. [Google Scholar] [CrossRef]
- Nigro, D.; Fortunato, S.; Giove, S.L.; Paradiso, A.; Gu, Y.Q.; Blanco, A.; de Pinto, M.C.; Gadaleta, A. Glutamine synthetase in Durum Wheat: Genotypic Variation and Relationship with Grain Protein Content. Front. Plant. Sci. 2016, 7, 971. [Google Scholar] [CrossRef] [PubMed]
- Colasuonno, P.; Gadaleta, A.; Giancaspro, A.; Nigro, D.; Giove, S.; Incerti, O.; Mangini, G.; Signorile, A.; Simeone, R.; Blanco, A. Development of a high-density SNP-based linkage map and detection of yellow pigment content QTLs in durum wheat. Mol. Breed 2014, 34, 1563–1578. [Google Scholar] [CrossRef]
- Li, X.P.; Zhao, X.Q.; He, X.; Zhao, G.Y.; Li, B.; Liu, D.C.; Li, Z.S. Haplotype analysis of the genes encoding glutamine synthetase plastic isoforms and their association with nitrogen-use-and yield-related traits in bread wheat. New Phytol. 2011, 189, 449–458. [Google Scholar] [CrossRef] [PubMed]
- Hunt, A.G. Messenger RNA 3′ end formation in plants. In Nuclear Pre-mRNA Processing in Plants; Springer: Berlin/Heidelberg, Germany, 2008; pp. 151–177. [Google Scholar]
- Uauy, C.; Brevis, J.C.; Dubcovsky, J. The high grain protein content gene Gpc-B1 accelerates senescence and has pleiotropic effects on protein content in wheat. J. Exp. Bot 2006, 57, 2785–2794. [Google Scholar] [CrossRef] [PubMed]
- Uauy, C.; Distelfeld, A.; Fahima, T.; Blechl, A.; Dubcovsky, J. A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 2006, 314, 1298–1301. [Google Scholar] [CrossRef] [PubMed]
- Distelfeld, A.; Pearce, S.P.; Avni, R.; Scherer, B.; Uauy, C.; Piston, F.; Slade, A.; Zhao, R.; Dubcovsky, J. Divergent functions of orthologous NAC transcription factors in wheat and rice. Plant Mol. Biol. 2012, 78, 515–524. [Google Scholar] [CrossRef]
- Chen, X.Y.; Song, G.Q.; Zhang, S.J.; Li, Y.L.; Jie, G.A.O.; Shahidul, I.; Li, G.Y.; Ji, W.Q. The allelic distribution and variation analysis of the NAM-B1 gene in Chinese wheat cultivars. J. Integr. Agric. 2017, 16, 1294–1303. [Google Scholar] [CrossRef]
- Pumphrey, M.O.; Bernardo, R.; Anderson, J.A. Validating the Fhb1 QTL for Fusarium head blight resistance in near-isogenic wheat lines developed from breeding populations. Crop. Sci. 2007, 47, 200–206. [Google Scholar] [CrossRef]
- Barrero, J.M.; Cavanagh, C.; Verbyla, K.L.; Tibbits, J.F.; Verbyla, A.P.; Huang, B.E.; Rosewarne, G.M.; Stephen, S.; Wang, P.; Whan, A.; et al. Transcriptomic analysis of wheat near-isogenic lines identifies Pm19-A1 and A2 as candidates for a major dormancy QTL. Genome Biol. 2015, 16, 93. [Google Scholar] [CrossRef]
- Wang, X.; Liu, H.; Mia, M.S.; Siddique, K.H.; Yan, G. Development of near-isogenic lines targeting a major QTL on 3AL for pre-harvest sprouting resistance in bread wheat. Crop Pasture Sci. 2018, 69, 864–872. [Google Scholar] [CrossRef]
- Kuzay, S.; Xu, Y.; Zhang, J.; Katz, A.; Pearce, S.; Su, Z.; Fraser, M.; Anderson, J.A.; Brown-Guedira, G.; DeWitt, N.; et al. Identification of a candidate gene for a QTL for spikelet number per spike on wheat chromosome arm 7AL by high-resolution genetic mapping. Theor. Appl. Genet. 2019, 132, 2689–2705. [Google Scholar] [CrossRef] [PubMed]
- Masclaux-Daubresse, C.; Reisdorf-Cren, M.; Orsel, M. Leaf nitrogen remobilisation for plant development and grain filling. Plant. Biol. 2008, 10, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Criado, M.V.; Caputo, C.; Roberts, I.N.; Castro, M.A.; Barneix, A.J. Cytokinin-induced changes of nitrogen remobilization and chloroplast ultrastructure in wheat (Triticum aestivum). J. Plant. Physiol. 2009, 166, 1775–1785. [Google Scholar] [CrossRef] [PubMed]
- Masclaux-Daubresse, C.; Daniel-Vedele, F.; Dechorgnat, J.; Chardon, F.; Gaufichon, L.; Suzuki, A. Nitrogen uptake, assimilation and remobilization in plants: Challenges for sustainable and productive agriculture. Ann. Bot. 2010, 105, 1141–1157. [Google Scholar] [CrossRef] [PubMed]
- Joppa, L.R.; Cantrell, R.G. Chromosomal location of genes for grain protein content of wild tetraploid wheat. Crop Sci. 1990, 30, 1059–1064. [Google Scholar] [CrossRef]
- Prasad, M.; Kumar, N.; Kulwal, P.; Röder, M.; Balyan, H.; Dhaliwal, H.; Gupta, P. QTL analysis for grain protein content using SSR markers and validation studies using NILs in bread wheat. Theor. Appl. Genet. 2003, 106, 659–667. [Google Scholar] [CrossRef]
- Blanco, A.; Simeone, R.; Gadaleta, A. Detection of QTLs for grain protein content in durum wheat. Theor. Appl. Genet. 2006, 112, 1195–1204. [Google Scholar] [CrossRef]
- Kichey, T.; Le Gouis, J.; Sangwan, B.; Hirel, B.; Dubois, F. Changes in the cellular and subcellular localization of glutamine synthetase and glutamate dehydrogenase during flag leaf senescence in wheat (Triticum aestivum L.). Plant. Cell Physiol. 2005, 46, 964–974. [Google Scholar] [CrossRef]
- Fontaine, J.X.; Ravel, C.; Pageau, K.; Heumez, E.; Dubois, F.; Hirel, B.; Le Gouis, J. A quantitative genetic study for elucidating the contribution of glutamine synthetase, glutamate dehydrogenase and other nitrogen-related physiological traits to the agronomic performance of common wheat. Theor. Appl. Genet. 2009, 119, 645–662. [Google Scholar] [CrossRef]
- Coque, M.; Martin, A.; Veyrieras, J.; Hirel, B.; Gallais, A. Genetic variation for N-remobilization and postsilking N-uptake in a set of maize recombinant inbred lines. 3. QTL detection and coincidences. Theor. Appl. Genet. 2008, 117, 729–747. [Google Scholar] [CrossRef]
- Dolgikh, V.A.; Pukhovaya, E.M.; Zemlyanskaya, E.V. shaping ethylene response: The role of EIN3/EIL1 transcription factors. Front. Plant Sci. 2019, 10, 1030. [Google Scholar] [CrossRef] [PubMed]
- Salih, H.; He, S.; Li, H.; Peng, Z.; Du, X. Investigation of the EIL/EIN3 Transcription Factor Gene Family Members and Their Expression Levels in the Early Stage of Cotton Fiber Development. Plants 2020, 9, 128. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, P.; Baranwal, V.K.; Khurana, P. Genome-wide analysis of bZIP transcription factors in wheat and functional characterization of a TabZIP under abiotic stress. Sci. Rep. 2019, 9, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, Y.; Liu, Q.; Dubouzet, J.G.; Abe, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration -and cold- inducible gene expression. Biochem. Biophys. Res. Commun. 2002, 290, 998–1009. [Google Scholar] [CrossRef] [PubMed]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant. Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef] [PubMed]
- Licausi, F.; Giorgi, F.M.; Zenoni, S.; Osti, F.; Pezzotti, M.; Perata, P. Genomic and transcriptomic analysis of the AP2/ERF superfamily in Vitis vinifera. BMC Genom. 2010, 11, 719–734. [Google Scholar] [CrossRef]
- Sharoni, A.M.; Nuruzzaman, M.; Satoh, K.; Shimizu, T.; Kondoh, H.; Sasaya, T.; Choi, I.R.; Omura, T.; Kikuchi, S. Gene structures, classification and expression models of the AP2/EREBP transcription factor family in rice. Plant. Cell Physiol. 2011, 52, 344–360. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, Z.; Liu, D.; Zhang, K.; Li, A.; Mao, L. SQUAMOSA promoter-binding protein-like transcription factors: Star players for plant growth and development. J. Integr. Plant Biol. 2010, 52, 946–951. [Google Scholar] [CrossRef]
- Wu, G.; Poethig, R.S. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 2006, 133, 3539–3547. [Google Scholar] [CrossRef]
- Usami, T.; Horiguchi, G.; Yano, S.; Tsukaya, H. The more and smaller cells mutants of Arabidopsis thaliana identify novel roles for SQUAMOSA PROMOTER BINDING PROTEIN-LIKE genes in the control of heteroblasty. Development 2009, 136, 955–964. [Google Scholar] [CrossRef]
- Manning, K.; Tor, M.; Poole, M.; Hong, Y.; Thompson, A.J.; King, G.J.; Giovannoni, J.J.; Seymour, G.B. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 2006, 38, 948–952. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Wang, Y.; Xue, D.; Wang, J.; Yan, M.; Liu, G.; Dong, G.; Zeng, D.; Lu, Z.; Zhu, X.; et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 2010, 42, 541–544. [Google Scholar] [CrossRef]
- Unte, U.S.; Sorensen, A.M.; Pesaresi, P.; Gandikota, M.; Leister, D.; Saedler, H.; Huijser, P. SPL8, an SBP-box gene that affects pollen sac development in Arabidopsis. Plant Cell. 2003, 15, 1009–1019. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Schwarz, S.; Saedler, H.; Huijser, P. SPL8, a local regulator in a subset of gibberellin-mediated developmental processes in Arabidopsis. Plant Mol. Biol. 2007, 63, 429–439. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, M.; Moseley, J.L.; Tottey, S.; Del Campo, J.A.; Quinn, J.; Kim, Y.; Merchant, S. Genetic dissection of nutritional copper signaling in chlamydomonas distinguishes regulatory and target genes. Genetics 2004, 168, 795–807. [Google Scholar] [CrossRef] [PubMed]
- Stone, J.; Liang, X.; Nekl, E.; Stiers, J. Arabidopsis AtSPL14, a plant-specific SBP-domain transcription factor, participates in plant development and sensitivity to fumonisin B1. Plant J. 2005, 41, 744–754. [Google Scholar] [CrossRef]
- Abu-Romman, S. Molecular cloning and expression analysis of zinc finger-homeodomain transcription factor TaZFHD1 in wheat. S. Afr. J. Bot. 2014, 91, 32–36. [Google Scholar] [CrossRef]
- Kim, J.Y.; Rim, Y.; Wang, J.; Jackson, D. A novel cell-to-cell trafficking assay indicates that the KNOX homeodomain is necessary and sufficient for intercellular protein and mRNA trafficking. Genes Dev. 2005, 19, 788–793. [Google Scholar] [CrossRef]
- Smith, H.M.; Campbell, B.C.; Hake, S. Competence to respond to floral inductive signals requires the homeobox genes PENNYWISE and POUNDFOOLISH. Curr. Biol. 2004, 14, 812–817. [Google Scholar] [CrossRef]
- Himmelbach, A.; Hoffmann, T.; Leube, M.; Hohener, B.; Grill, E. Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. EMBO J. 2002, 21, 3029–3038. [Google Scholar] [CrossRef]
- Zhu, J.; Shi, H.; Lee, B.H.; Damsz, B.; Cheng, S.; Stirm, V.; Zhu, J.K.; Hasegawa, M.P.; Bressan, R.A. An Arabidopsis homeodomain transcription factor gene, HOS9, mediates cold tolerance through a CBF-independent pathway. Proc. Natl. Acad. Sci. USA 2004, 101, 9873–9878. [Google Scholar] [CrossRef] [PubMed]
- Coego, A.; Ramirez, V.; Gil, M.J.; Flors, V.; Mauch-Mani, B.; Vera, P. An Arabidposis homeodomain transcription factor, OVEREXPRESSOR OF CATIONIC PEROXIDASE 3, mediates resistance to infection by necrotrophic pathogens. Plant. Cell 2005, 17, 2123–2137. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Derkx, A.P.; Liu, D.C.; Buchner, P.; Hawkesford, M.J. Overexpression of a NAC transcription factor delays leaf senescence and increases grain nitrogen concentration in wheat. Plant Biol. 2015, 17, 904–913. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Duan, X.; Zhang, Q.; Li, X.; Wang, B.; Huang, L.; Wang, X.; Kang, Z. The target gene of tae-miR164, a novel NAC transcription factor from the NAM subfamily, negatively regulates resistance of wheat to stripe rust. Mol. Plant Pathol. 2014, 15, 284–296. [Google Scholar] [CrossRef] [PubMed]
- Xia, N.; Zhang, G.; Liu, X.-Y.; Deng, L.; Cai, G.-L.; Zhang, Y.; Wang, X.-J.; Zhao, J.; Huang, L.-L.; Kang, Z. Characterization of a novel wheat NAC transcription factor gene involved in defense response against stripe rust pathogen infection and abiotic stresses. Mol. Biol. Rep. 2010, 37, 3703–3712. [Google Scholar] [CrossRef]
- Xia, N.; Zhang, G.; Sun, Y.F.; Zhu, L.; Xu, L.S.; Chen, X.M.; Liu, B.; Yu, Y.T.; Wang, X.J.; Huang, L.L.; et al. TaNAC8, a novel NAC transcription factor gene in wheat, responds to stripe rust pathogen infection and abiotic stresses. Physiol. Mol. Plant. Pathol. 2010, 74, 394–402. [Google Scholar] [CrossRef]
- Wang, F.; Lin, R.; Feng, J.; Chen, W.; Qiu, D.; Xu, S. TaNAC1 acts as a negative regulator of stripe rust resistance in wheat, enhances susceptibility to Pseudomonas syringae, and promotes lateral root development in transgenic Arabidopsis thaliana. Front. Plant Sci. 2015, 6, 108. [Google Scholar] [CrossRef]
- Huang, Q.; Wang, Y.; Li, B.; Chang, J.; Chen, M.; Li, K.; Yang, G.; He, G. TaNAC29, a NAC transcription factor from wheat, enhances salt and drought tolerance in transgenic Arabidopsis. BMC Plant Biol. 2015, 15, 268. [Google Scholar] [CrossRef]
- Tang, Y.; Liu, M.; Gao, S.; Zhang, Z.; Zhao, C.; Zhang, F.; Chen, X. Molecular characterization of novel TaNAC genes in wheat and overexpression of TaNAC2a drought tolerance in tobacco. Physiol. Plant 2012, 144, 210–224. [Google Scholar] [CrossRef]
- Xue, G.P.; Bower, N.I.; McIntyre, C.L.; Riding, G.A.; Kazan, K.; Shorter, R. TaNAC69 from the NAC superfamily of transcription factors is up-regulated by abiotic stresses in wheat and recognises two consensus DNA-binding sequences. Funct. Plant Biol. 2006, 33, 43–57. [Google Scholar] [CrossRef]
- Mao, X.; Zhang, H.; Qian, X.; Li, A.; Zhao, G.; Jing, R. TaNAC2, a NAC type wheat transcription factor conferring enhanced multiple abiotic stress tolerances in Arabidopsis. J. Exp. Bot. 2012, 63, 2933–2946. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Chen, S.; Li, A.; Zhai, C.; Jing, R. Novel NAC transcription factor TaNAC67 confers enhanced multi-abiotic stress tolerances in Arabidopsis. PLoS ONE 2014, 9, e84359. [Google Scholar] [CrossRef] [PubMed]
- Waters, B.M.; Uauy, C.; Dubcovsky, J.; Grusak, M.A. Wheat (Triticum aestivum) NAM proteins regulate the translocation of iron, zinc, and nitrogen compounds from vegetative tissues to grain. J. Exp. Bot. 2009, 60, 4263–4274. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Qu, B.; Li, W.; Zhao, X.; Teng, W.; Ma, W.; Ren, Y.; Li, B.; Li, Z.; Tong, Y. The nitrate-inducible NAC transcription factor TaNAC2–5A controls nitrate response and increases wheat yield. Plant Physiol. 2015, 169, 1991–2005. [Google Scholar] [CrossRef] [PubMed]
- Gadaleta, A.; Nigro, D.; Marcotuli, I.; Giancaspro, A.; Giove, S.L.; Blanco, A. Isolation and characterisation of cytosolic glutamine synthetase (GSe) genes and association with grain protein content in durum wheat. Crop. Pasture Sci. 2014, 65, 38–45. [Google Scholar] [CrossRef]
- Nigro, D.; Gu, Y.Q.; Huo, N.; Marcotuli, I.; Blanco, A.; Gadaleta, A.; Anderson, O.D. Structural analysis of the wheat genes encoding NADH-dependent glutamine-2-oxoglutarate amidotransferases and correlation with grain protein content. PLoS ONE 2013, 8, e73751. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).