NO Scavenging through Reductive Nitrosylation of Ferric Mycobacterium tuberculosis and Homo sapiens Nitrobindins
Abstract
:1. Introduction
2. Results
3. Discussion
- (i)
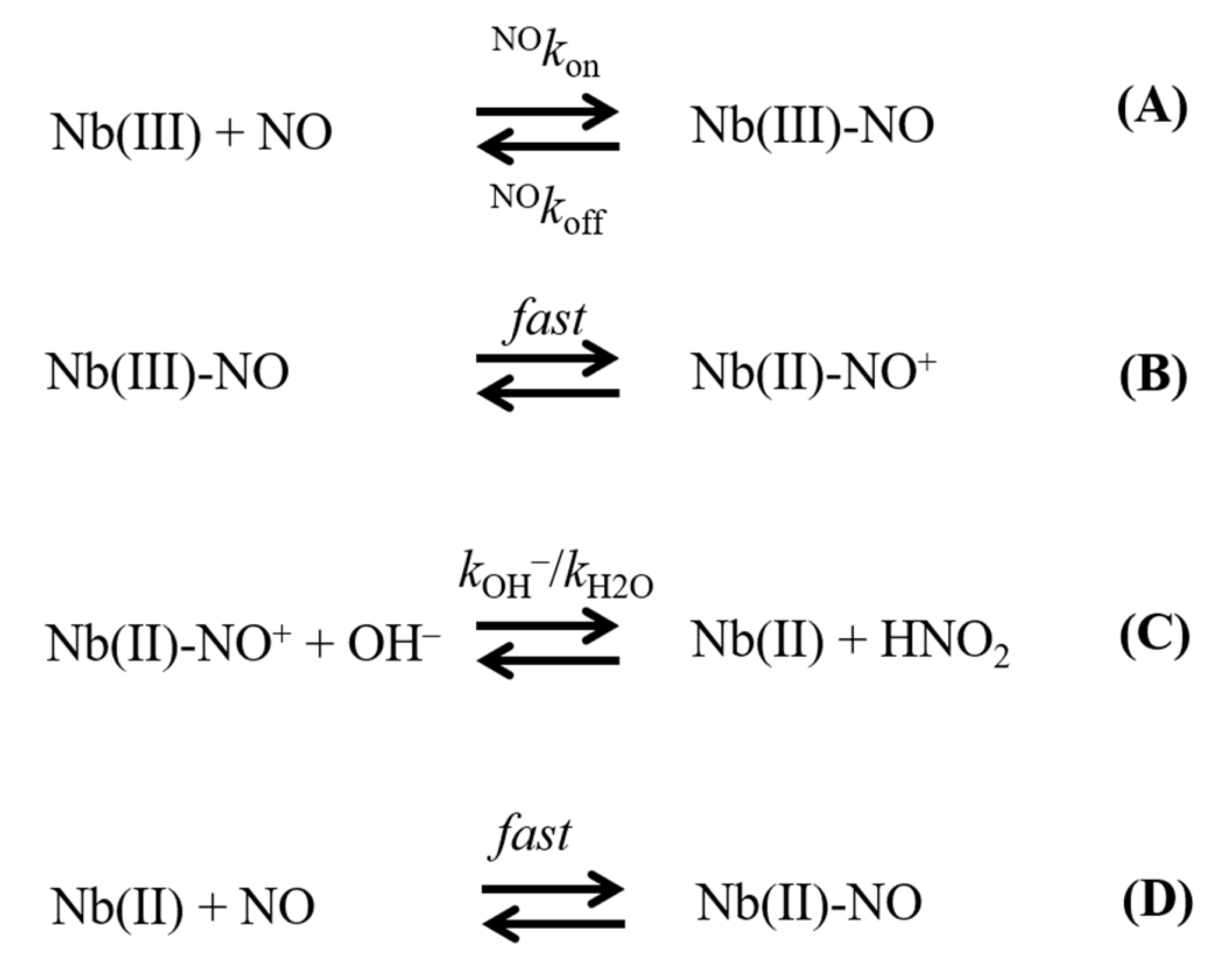
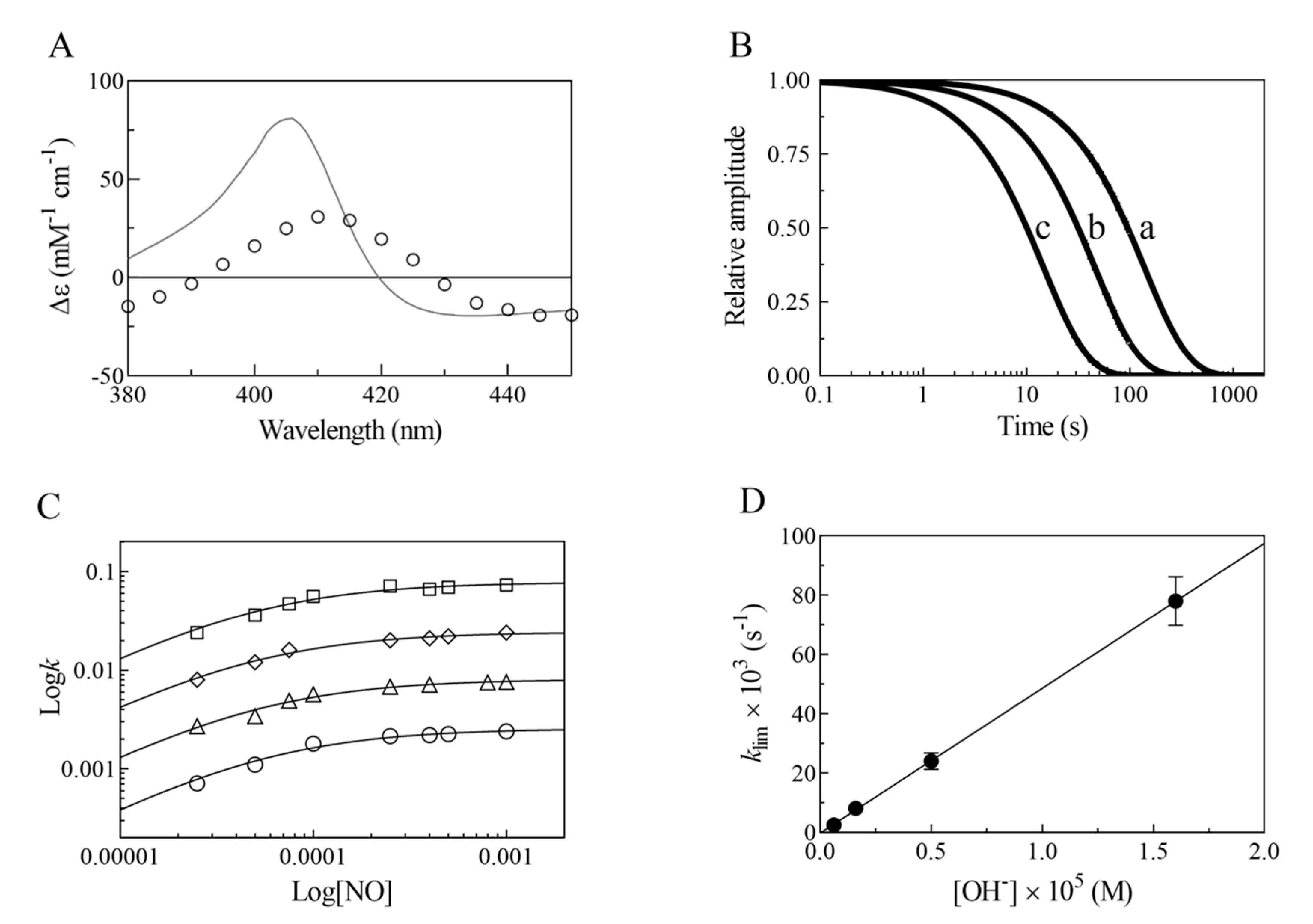
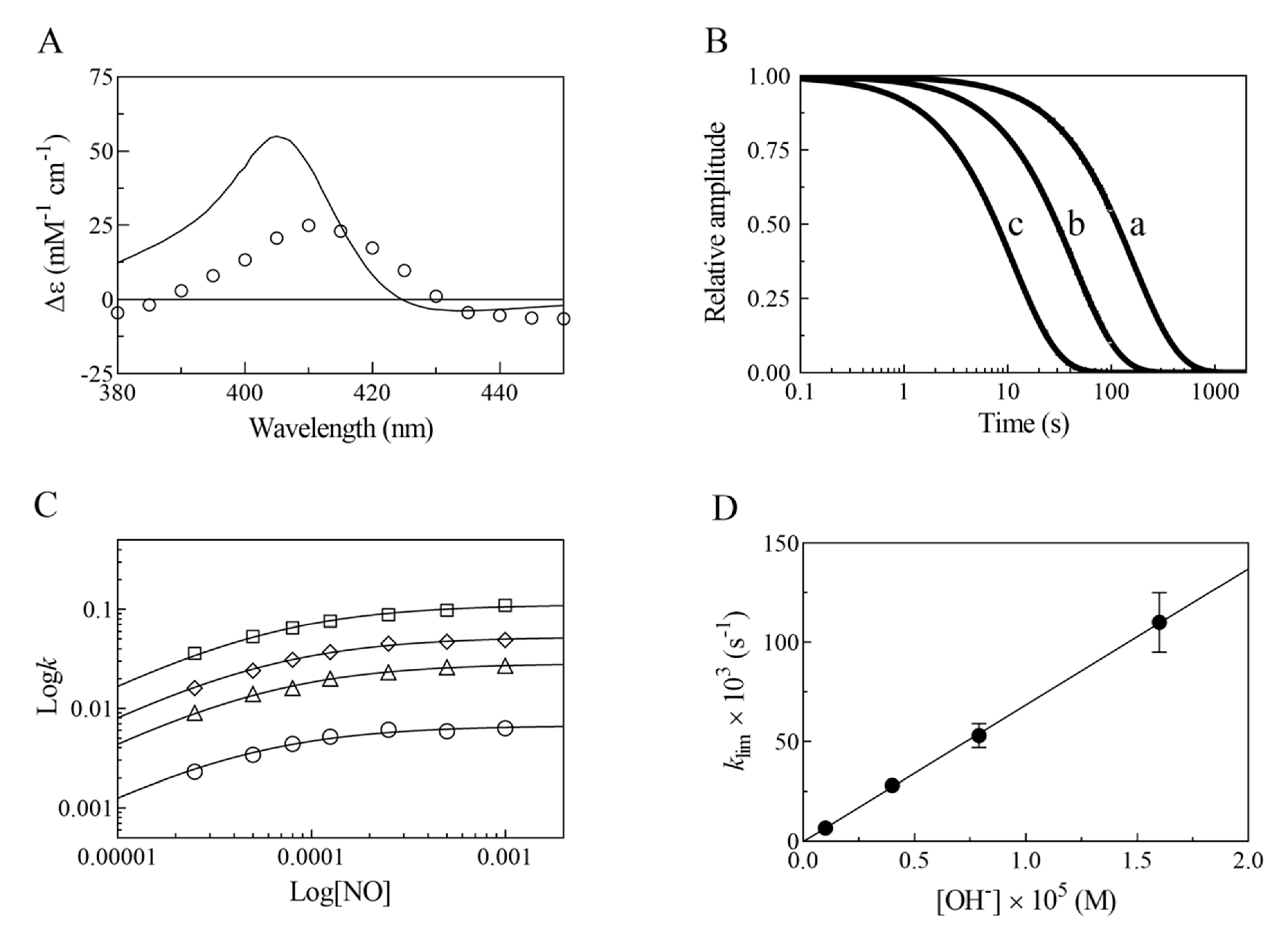
- The very fast NO binding kinetics of Mt-Nb(III) and Hs-Nb(III) [13], so as to be lost in the dead-time of the rapid-mixing stopped-flow apparatus at the employed NO concentrations (ranging between 2.5 × 10−5 and 1.0 × 10−3 M), reflect a specific feature of Nbs, which is shared with NPs [9,31], since all other heme-proteins show an about 10-fold slower reactivity (Table 1).
- (ii)
- Despite the very different structural organization of the heme-proteins examined (all-β-barrel versus all-α-helix structures) [4,5,6,11,13,32,33,34,35,36], the values of K for NO binding to ferric heme-proteins are closely similar (ranging between 1.4 × 10−5 and 2.1 × 10−4 M) (present study and [13,22,23,24,25,26,27,30]) (Table 1). This suggests a balance between the NO association and dissociation rate constants among most heme-proteins, such that the energy barriers for ligand entry and exit are affected to the same extent by structural constraints.
- (iii)
- The linear dependence of k on [OH–] indicates that no additional features are involved in the irreversible reductive nitrosylation of Mt-Nb(III) and Hs-Nb(III). Moreover, the values of kOH– for ferric heme-protein reductive nitrosylation range between 1.7 × 102 M−1 s−1 and 6.9 × 103 M−1 s−1 [13,22,23,24,25,26,27,30] (Table 1), possibly reflecting the different anion accessibility to the heme pocket and/or the heme-Fe(III) protein reduction potentials [6,22,23,34,36,37,38,39]. The close similarity of kOH- between all-β-barrel Nbs and all-α-helical Hb indicates that the grossly different folding [4,6,10,11,13,36,40,41] does not significantly affect anion accessibility to the heme pocket.
- (iv)
- The rate of the conversion of heme-Fe(II)-NO+ to heme-Fe(II) + HNO2 (Scheme 1C) represents the rate-limiting step of heme-protein reductive nitrosylation (present study and [22,23,24,25,26,27,28,30]). Of note, the OH– species catalyzes the conversion of Mt-Nb(II)-NO+ to Mt-Nb(II) + HNO2 and of Hs-Nb(II)-NO+ to Hs-Nb(II) + HNO2 much more efficiently than H2O (kH2O ~ 0 s−1). This may reflect either the higher affinity of negatively charged ligands for ferric heme-proteins with respect to uncharged compounds and/or the deprotonation rate of the incoming ligand [5,37,42].
- (v)
4. Materials
5. Methods
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Brunori, M. Nitric oxide, cytochrome-c oxidase and myoglobin. Trends Biochem. Sci. 2001, 26, 21–23. [Google Scholar] [CrossRef]
- Smith, L.J.; Kahraman, A.; Thornton, J.M. Heme proteins diversity in structural characteristics, function, and folding. Proteins 2010, 78, 2349–2368. [Google Scholar] [CrossRef]
- Ascenzi, P.; Brunori, M. A molecule for all seasons: The heme. J. Porphyr. Phthalocyanines 2016, 20, 134–149. [Google Scholar] [CrossRef]
- Perutz, M.F. Regulation of oxygen affinity of hemoglobin: Influence of structure of the globin on the heme iron. Annu. Rev. Biochem. 1979, 48, 327–386. [Google Scholar] [CrossRef] [PubMed]
- Bolognesi, M.; Bordo, D.; Rizzi, M.; Tarricone, C.; Ascenzi, P. Non-vertebrate hemoglobins: Structural bases for reactivity. Prog. Biophys. Mol. Biol. 1997, 68, 29–68. [Google Scholar] [CrossRef]
- Pesce, A.; Bolognesi, M.; Nardini, M. The diversity of 2/2 (truncated) globins. Adv. Microb. Physiol. 2013, 63, 49–78. [Google Scholar] [PubMed]
- Andersen, J.F.; Weichsel, A.; Balfour, C.A.; Champagne, D.E.; Montfort, W.R. The crystal structure of nitrophorin 4 at 1.5 Å resolution: Transport of nitric oxide by a lipocalin-based heme protein. Structure 1998, 6, 1315–1327. [Google Scholar] [CrossRef] [Green Version]
- Weichsel, A.; Andersen, J.F.; Champagne, D.E.; Walker, F.A.; Montfort, W.R. Crystal structures of a nitric oxide transport protein from a blood-sucking insect. Nat. Struct. Biol. 1998, 4, 304–309. [Google Scholar] [CrossRef]
- Andersen, J.F.; Ding, X.D.; Balfour, C.; Shokhireva, T.K.; Champagne, D.E.; Walker, F.A.; Montfort, W.R. Kinetics and equilibria in ligand binding by nitrophorins 1–4: Evidence for stabilization of a nitric oxide-ferriheme complex through a ligand-induced conformational trap. Biochemistry 2000, 39, 10118–10131. [Google Scholar] [CrossRef]
- Bianchetti, C.M.; Blouin, G.C.; Bitto, E.; Olson, J.S.; Phillips, G.N., Jr. The structure and NO binding properties of the nitrophorin-like heme-binding protein from Arabidopsis thaliana gene locus At1g79260.1. Proteins 2010, 78, 917–931. [Google Scholar] [CrossRef] [Green Version]
- Bianchetti, C.M.; Bingman, C.A.; Phillips, G.N., Jr. Structure of the C-terminal heme-binding domain of THAP domain containing protein 4 from Homo sapiens. Proteins 2011, 79, 1337–1341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Simone, G.; Ascenzi, P.; Polticelli, F. Nitrobindin: An ubiquitous family of all β-barrel heme-proteins. IUBMB Life 2016, 68, 423–428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Simone, G.; di Masi, A.; Vita, G.M.; Polticelli, F.; Pesce, A.; Nardini, M.; Bolognesi, M.; Ciaccio, C.; Coletta, M.; Turilli, E.S.; et al. Mycobacterial and human nitrobindins: Structure and function. Antioxid. Redox Signal. 2020, 33, 229–246. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, J.M.; Hazzard, J.M.; Nussenzveig, R.H.; Champagne, D.E.; Walker, F.A. Reversible binding of nitric oxide by a salivary heme protein from a bloodsucking insect. Science 1993, 260, 539–541. [Google Scholar] [CrossRef]
- Montfort, W.R.; Weichsel, A.; Andersen, J.F. Nitrophorins and related antihemostatic lipocalins from Rhodnius prolixus and other blood-sucking arthropods. Biochim. Biophys. Acta 2000, 1482, 110–118. [Google Scholar] [CrossRef]
- Kondrashov, D.A.; Roberts, S.A.; Weichsel, A.; Montfort, W.R. Protein functional cycle viewed at atomic resolution: Conformational change and mobility in nitrophorin 4 as a function of pH and NO binding. Biochemistry 2004, 43, 13637–13647. [Google Scholar] [CrossRef]
- Andersen, J.F. Structure and mechanism in salivary proteins from blood-feeding arthropods. Toxicon 2010, 56, 1120–1129. [Google Scholar] [CrossRef] [Green Version]
- Knipp, M.; Ogata, H.; Soavi, G.; Cerullo, G.; Allegri, A.; Abbruzzetti, S.; Bruno, S.; Viappiani, C.; Bidon-Chanal, A.; Luque, F.J. Structure and dynamics of the membrane attaching nitric oxide transporter nitrophorin 7. F1000Research 2015, 4, 45. [Google Scholar] [CrossRef] [Green Version]
- De Simone, G.; Ascenzi, P.; di Masi, A.; Polticelli, F. Nitrophorins and nitrobindins: Structure and function. Biomol. Concepts 2017, 8, 105–118. [Google Scholar] [CrossRef]
- Weichsel, A.; Maes, E.M.; Andersen, J.F.; Valenzuela, J.G.; Shokhireva, T.K.; Walker, F.A.; Montfort, W.R. Heme-assisted S-nitrosation of a proximal thiolate in a nitric oxide transport protein. Proc. Natl. Acad. Sci. USA 2005, 102, 594–599. [Google Scholar] [CrossRef] [Green Version]
- De Simone, G.; di Masi, A.; Polticelli, F.; Ascenzi, P. Human nitrobindin: The first example of an all-β-barrel ferric heme-protein that catalyzes peroxynitrite detoxification. FEBS Open Bio. 2018, 8, 2002–2010. [Google Scholar] [CrossRef]
- Hoshino, M.; Ozawa, K.; Seki, H.; Ford, P.C. Photochemistry of nitric oxide adducts of water-soluble iron(III) porphyrin and ferrihemoproteins studied by nanosecond laser photolysis. J. Am. Chem. Soc. 1993, 115, 9568–9575. [Google Scholar] [CrossRef] [Green Version]
- Hoshino, M.; Maeda, M.; Konishi, R.; Seki, H.; Ford, P.C. Studies on the reaction mechanism for reductive nitrosylation of ferrihemoproteins in buffer solutions. J. Am. Chem. Soc. 1996, 118, 5702–5707. [Google Scholar] [CrossRef]
- Herold, S.; Puppo, A. Kinetics and mechanistic studies of the reactions of metleghemoglobin, ferrylleghemoglobin, and nitrosylleghemoglobin with reactive nitrogen species. J. Biol. Inorg. Chem. 2005, 10, 946–957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ascenzi, P.; Yu, C.; di Masi, A.; Gullotta, F.; De Sanctis, G.; Fanali, G.; Fasano, M.; Coletta, M. Reductive nitrosylation of ferric human serum heme-albumin. FEBS J. 2010, 277, 2474–2485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ascenzi, P.; Pesce, A.; Nardini, M.; Bolognesi, M.; Ciaccio, C.; Coletta, M.; Dewilde, S. Reductive nitrosylation of Methanosarcina acetivorans protoglobin: A comparative study. Biochem. Biophys. Res. Commun. 2013, 430, 3001–3005. [Google Scholar] [CrossRef]
- Ascenzi, P.; di Masi, A.; Tundo, G.R.; Pesce, A.; Visca, P.; Coletta, M. Nitrosylation mechanisms of Mycobacterium tuberculosis and Campylobacter jejuni truncated hemoglobins N, O, and P. PLoS ONE 2014, 9, e102811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ascenzi, P.; De Simone, G.; Polticelli, F.; Gioia, M.; Coletta, M. Reductive nitrosylation of ferric human hemoglobin bound to human haptoglobin 1-1 and 2-2. J. Biol. Inorg. Chem. 2018, 23, 437–445. [Google Scholar] [CrossRef]
- Ascenzi, P.; De Simone, G.; Sbardella, D.; Coletta, M. Reductive nitrosylation of ferric microperoxidase-11. J. Biol. Inorg. Chem. 2019, 24, 21–29. [Google Scholar] [CrossRef]
- Ascenzi, P.; di Masi, A.; Gullotta, F.; Mattu, M.; Ciaccio, C.; Coletta, M. Reductive nitrosylation of ferric cyanide horse heart myoglobin is limited by cyanide dissociation. Biochem. Biophys. Res. Commun. 2010, 393, 196–200. [Google Scholar] [CrossRef] [Green Version]
- Maes, E.M.; Weichsel, A.; Andersen, J.F.; Shepley, D.; Montfort, W.R. Role of binding site loops in controlling nitric oxide release: Structure and kinetics of mutant forms of nitrophorin 4. Biochemistry 2004, 43, 6679–6690. [Google Scholar] [CrossRef]
- Bushnell, G.W.; Louie, G.V.; Brayer, G.D. High-resolution three-dimensional structure of horse heart cytochrome c. J. Mol. Biol. 1990, 214, 585–595. [Google Scholar] [CrossRef]
- Ellis, P.J.; Appleby, C.A.; Guss, J.M.; Hunter, W.N.; Ollis, D.L.; Freeman, H.C. Structure of ferric soybean leghemoglobin a nicotinate at 2.3 Å resolution. Acta Crystallogr. D Biol. Crystallogr. 1997, 53, 302–310. [Google Scholar] [CrossRef] [PubMed]
- Brucker, E.A.; Olson, J.S.; Ikeda-Saito, M.; Phillips, G.N., Jr. Nitric oxide myoglobin: Crystal structure and analysis of ligand geometry. Proteins 1998, 30, 352–356. [Google Scholar] [CrossRef]
- Wardell, M.; Wang, Z.; Ho, J.X.; Robert, J.; Ruker, F.; Ruble, J.; Carter, D.C. The atomic structure of human methemalbumin at 1.9 Å. Biochem. Biophys. Res. Commun. 2002, 291, 813–819. [Google Scholar] [CrossRef] [PubMed]
- Pesce, A.; Bolognesi, M.; Nardini, M. Protoglobin: Structure and ligand-binding properties. Adv. Microb. Physiol. 2013, 63, 79–96. [Google Scholar] [PubMed]
- Beetlestone, J.G.; Adeosun, O.S.; Goddard, J.E.; Kushimo, J.B.; Ogunlesi, M.M.; Ogunmola, G.B.; Okonjo, K.O.; Seamonds, B. Reactivity difference between haemoglobins. Part XIX. J. Chem. Soc. Dalton Trans. 1976, 1251–1278. [Google Scholar] [CrossRef]
- Perutz, M.F. Myoglobin and haemoglobin: Role of distal residues in reactions with haem ligands. Trends Biochem. Sci. 1989, 14, 42–44. [Google Scholar] [CrossRef]
- Miele, A.E.; Santanché, S.; Travaglini-Allocatelli, C.; Vallone, B.; Brunori, M.; Bellelli, A. Modulation of ligand binding in engineered human hemoglobin distal pocket. J. Mol. Biol. 1999, 290, 515–524. [Google Scholar] [CrossRef]
- Hargrove, M.S.; Barry, J.K.; Brucker, E.A.; Berry, M.B.; Phillips, G.N., Jr.; Olson, J.S.; Arredondo-Peter, R.; Dean, J.M.; Klucas, R.V.; Sarath, G. Characterization of recombinant soybean leghemoglobin a and apolar distal histidine mutants. J. Mol. Biol. 1997, 266, 1032–1042. [Google Scholar] [CrossRef]
- di Masi, A.; De Simone, G.; Ciaccio, C.; D’Orso, S.; Coletta, M.; Ascenzi, P. Haptoglobin: From hemoglobin scavenging to human health. Mol. Asp. Med. 2020, 73, 100851. [Google Scholar] [CrossRef] [PubMed]
- Giacometti, G.M.; Ascenzi, P.; Bolognesi, M.; Brunori, M. Reactivity of ferric Aplysia myoglobin towards anionic ligands in the acidic region. Proposal for a structural model. J. Mol. Biol. 1981, 146, 363–374. [Google Scholar] [CrossRef]
- Barbosa, R.M.; Lopes Jesus, A.J.; Santos, R.M.; Pereira, C.L.; Marques, C.F.; Rocha, B.S.; Ferreira, N.R.; Ledo, A.; Laranjin, J. Preparation, standardization and measuremet of nitric oxide solutions. Glob. J. Anal. Chem. 2011, 2, 272–284. [Google Scholar]
- Antonini, E.; Brunori, M. Hemoglobin and Myoglobin in Their Reactions with Ligands; North-Holland Publishing Co.: Amsterdam, The Netherland, 1971. [Google Scholar]
- Ascenzi, P.; Brunori, M.; Pennesi, G.; Ercolani, C.; Monacelli, F. Equilibrium and kinetic study of nitric oxide binding to phthalocyaninatoiron(II) in dimethyl sulphoxide. J. Chem. Soc. Dalton Trans. 1987, 369–371. [Google Scholar] [CrossRef]
| Heme-Protein | NOkon (M−1 s−1) | K (M) | kOH− (M−1 s−1) |
|---|---|---|---|
| Mt-Nb | 1.8 × 106 a | 5.2 × 10−5 b | 4.9 × 103 c |
| Hs-Nb | 1.1 × 106 a | 5.3 × 10−5 d | 6.9 × 103 c |
| Mt-trHbN | 1.4 × 105 e | 1.4 × 10−5 e | 1.7 × 102 f |
| Mt-trHbO | 9.2 × 103 e | 1.5 × 10−5 e | 2.4 × 102 f |
| Cj-trHbP | 9.8 × 104 e | 5.4 × 10−5 e | 9.1 × 102 f |
| Methanosarcina acetivorans Pgb | 4.8 × 104 g | 6.1 × 10−5 g | 2.9 × 103 h |
| Glycine max legHb | 1.4 × 105 i | 2.1 × 10−5 i | 3.3 × 103 j |
| Horse heart Mb | 6.8 × 104 k | 1.2 × 10−4 k | 3.9 × 102 l |
| Sperm whale Mb | 1.9 × 105 m | 7.7 × 10−5 m | 3.2 × 102 n |
| Human Hb | --- o | 8.3 × 10−5 p | 3.2 × 103 p |
| Human Hb:Hp1-1 | --- o | 9.1 × 10−5 q | 4.9 × 103 r |
| Human Hb:Hp2-2 | --- o | 1.7 × 10−4 q | 6.7 × 103 r |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Simone, G.; di Masi, A.; Ciaccio, C.; Coletta, M.; Ascenzi, P. NO Scavenging through Reductive Nitrosylation of Ferric Mycobacterium tuberculosis and Homo sapiens Nitrobindins. Int. J. Mol. Sci. 2020, 21, 9395. https://doi.org/10.3390/ijms21249395
De Simone G, di Masi A, Ciaccio C, Coletta M, Ascenzi P. NO Scavenging through Reductive Nitrosylation of Ferric Mycobacterium tuberculosis and Homo sapiens Nitrobindins. International Journal of Molecular Sciences. 2020; 21(24):9395. https://doi.org/10.3390/ijms21249395
Chicago/Turabian StyleDe Simone, Giovanna, Alessandra di Masi, Chiara Ciaccio, Massimo Coletta, and Paolo Ascenzi. 2020. "NO Scavenging through Reductive Nitrosylation of Ferric Mycobacterium tuberculosis and Homo sapiens Nitrobindins" International Journal of Molecular Sciences 21, no. 24: 9395. https://doi.org/10.3390/ijms21249395
APA StyleDe Simone, G., di Masi, A., Ciaccio, C., Coletta, M., & Ascenzi, P. (2020). NO Scavenging through Reductive Nitrosylation of Ferric Mycobacterium tuberculosis and Homo sapiens Nitrobindins. International Journal of Molecular Sciences, 21(24), 9395. https://doi.org/10.3390/ijms21249395








