Inhibitory Monoclonal Antibodies and Their Recombinant Derivatives Targeting Surface-Exposed Carbonic Anhydrase XII on Cancer Cells
Abstract
1. Introduction
2. Results
2.1. Generation and Characterization of MAbs Specific to Human CA XII
2.2. Inhibition of Recombinant CA XII by the MAbs
2.3. MAb 14D6 Effect on A549 and A498 Cell Migration
2.4. Inhibitory Activity of MAb 14D6 in Three-Dimensional Cultures of A549 and A498 Cells
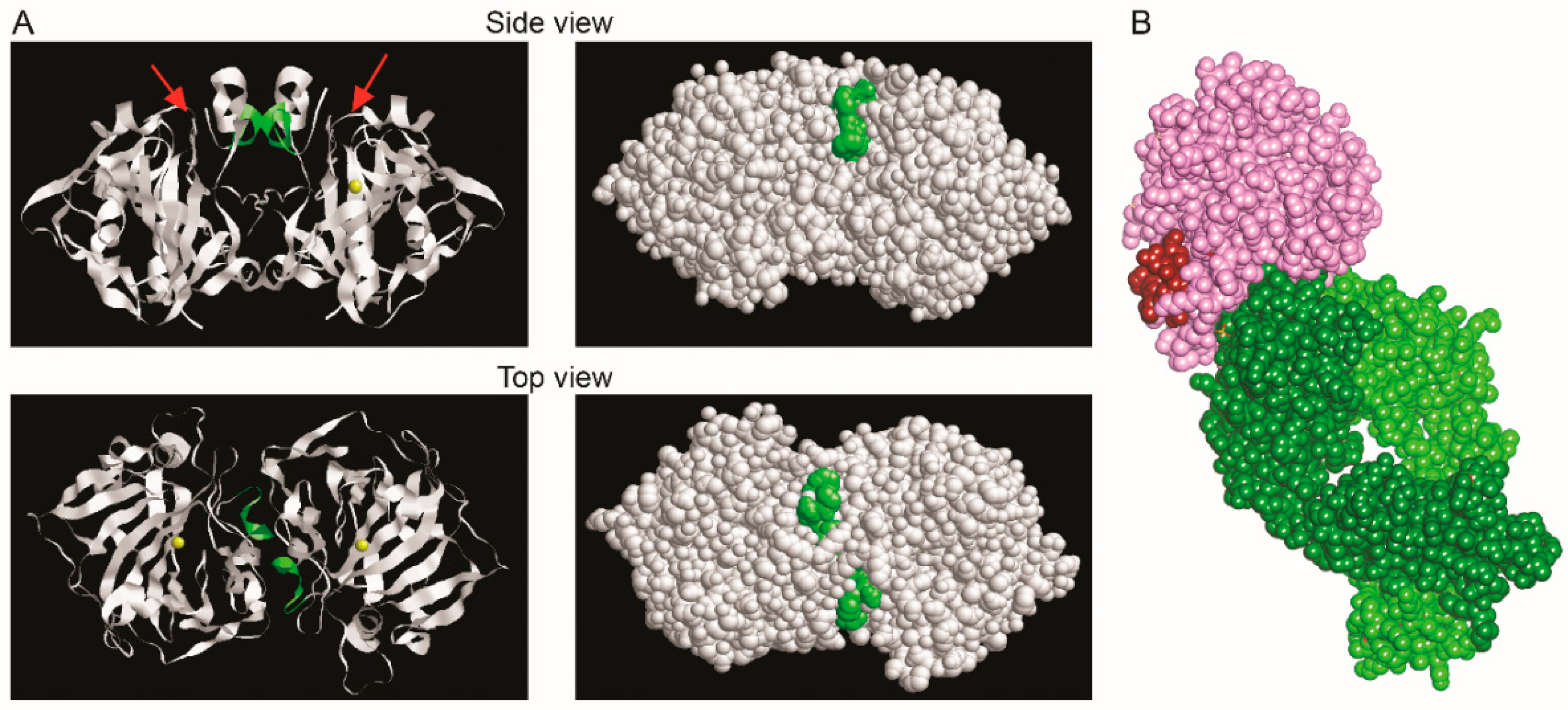
2.5. Epitope Mapping and Competitive Properties of the MAbs
2.6. Sequencing of the Variable Region of MAb 14D6
2.7. Production of 14D6-Derived scFv in E. coli
2.8. Production of 14D6–Derived scFv-Fc in CHO Cell Line
2.9. Activity Testing of 14D6-Derived scFv and scFv-Fc
3. Discussion
4. Materials and Methods
4.1. Recombinant CA Antigens for Immunization, ELISA, and Western Blot
4.2. Generation of Hybridomas
4.3. Enzyme-Linked ImmunoSorbent Assay (ELISA)
4.3.1. Indirect ELISA
4.3.2. Competitive ELISA
4.3.3. Direct ELISA
4.4. Protein Electrophoresis and Western Blot
4.5. Flow Cytometry
4.6. Purification of MAbs and scFv-Fc
4.7. Conjugation of MAbs with Horseradish Peroxidase
4.8. CA XII Inhibition Assay
4.9. Sequencing of Antibody Variable Regions
4.10. Construction of Expression Vectors for 14D6-Derived scFv and scFv-Fc
4.11. Generation of Recombinant scFv and scFv-Fc
4.12. Refolding and Purification of scFv
4.13. Cell Culturing for Functional Assays
4.14. Testing the Inhibitory Activity of the MAb 14D6 on A549 and A498 Cell Migration by ‘Wound Healing’ Assay
4.15. Testing the Inhibitory Activity of the MAb 14D6 in Cancer Spheroids
4.16. Production of His-Tagged CA XII Fragments for Epitope Mapping
5. Patents
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| aa | amino acids |
| ADCC | Antibody-Dependent Cellular Cytotoxicity |
| AZM | acetazolamide |
| BSA | Bovine Serum Albumin |
| CA | carbonic anhydrase |
| CA IX | carbonic anhydrase IX |
| CA XII | carbonic anhydrase XII |
| CA XIIDE3 | recombinant extracellular domain of human CA XII produced in E. coli DE3 strain |
| CA XIIHEK | recombinant extracellular domain of human CA XII expressed in human cell line HEK–293 |
| CDR | Complementarity–Determining Regions |
| ELISA | Enzyme-Linked ImmunoSorbent Assay |
| FC | flow cytometry |
| FR | framework region |
| H | Ig heavy chain |
| HAMA | human anti-mouse antibody |
| HRP | horseradish peroxidase |
| Ig | immunoglobulins |
| IgG | immunoglobulin G |
| Kd | apparent dissociation constant |
| L | Ig light chain |
| MAbs | monoclonal antibodies |
| OD | optical density |
| PBS-T | 0.1% Tween-20 in PBS |
| PDB | Protein Data Bank |
| PET | positron emission tomography |
| PVDF | polyvinylidene difluoride |
| RAbs | recombinant antibodies |
| RIT | radioimmunotherapy |
| RT | room temperature |
| scFv | single-chain fragment variable |
| scFv-Fc | scFv fused to human IgG1 Fc fragment |
| SDS–PAG | sodium dodecyl sulfate-polyacrylamide gel |
| SDS–PAGE | SDS–PAG electrophoresis |
| TSPyV | trichodysplasia spinulosa-associated polyomavirus |
| V | variable region of Ig |
| VH | variable region of Ig heavy chain |
| VL | variable region of Ig light chain |
| VLY | vaginolysin |
| WB | Western blot |
References
- Chaplin, D.D. Overview of the immune response. J. Allergy Clin. Immunol. 2010, 125, S3–S23. [Google Scholar] [CrossRef] [PubMed]
- Wootla, B.; Denic, A.; Rodriguez, M. Polyclonal and Monoclonal Antibodies in Clinic; Humana Press: Totowa, NJ, USA, 2014; pp. 79–110. [Google Scholar]
- Ahmad, Z.A.; Yeap, S.K.; Ali, A.M.; Ho, W.Y.; Alitheen, N.B.M.; Hamid, M. scFv Antibody: Principles and Clinical Application. Clin. Dev. Immunol. 2012, 2012, 980250. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.Y.; Pop, L.M.; Vitetta, E.S. Engineering therapeutic monoclonal antibodies. Immunol. Rev. 2008, 222, 9–27. [Google Scholar] [CrossRef] [PubMed]
- Krauss, J. Recombinant Antibodies for the Diagnosis and Treatment of Cancer. Mol. Biotechnol. 2003, 25, 1–18. [Google Scholar] [CrossRef]
- Petricevic, B.; Laengle, J.; Singer, J.; Sachet, M.; Fazekas, J.; Steger, G.G.; Bartsch, R.; Jensen-Jarolim, E.; Bergmann, M. Trastuzumab mediates antibody-dependent cell-mediated cytotoxicity and phagocytosis to the same extent in both adjuvant and metastatic HER2/neu breast cancer patients. J. Transl. Med. 2013, 11, 307. [Google Scholar] [CrossRef]
- Vanneman, M.; Dranoff, G. Combining immunotherapy and targeted therapies in cancer treatment. Nat. Rev. Cancer 2012, 12, 237–251. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Belov, L.; Solomon, M.J.; Chan, C.; Clarke, S.J.; Christopherson, R.I. Colorectal cancer cell surface protein profiling using an antibody microarray and fluorescence multiplexing. J. Vis. Exp. 2011, e3322. [Google Scholar] [CrossRef]
- Scott, A.M.; Wolchok, J.D.; Old, L.J. Antibody therapy of cancer. Nat. Rev. Cancer 2012, 12, 278–287. [Google Scholar] [CrossRef]
- Chien, M.-H.; Ying, T.-H.; Hsieh, Y.-H.; Lin, C.-H.; Shih, C.-H.; Wei, L.-H.; Yang, S.-F. Tumor-associated carbonic anhydrase XII is linked to the growth of primary oral squamous cell carcinoma and its poor prognosis. Oral Oncol. 2012, 48, 417–423. [Google Scholar] [CrossRef]
- Doyen, J.; Parks, S.K.; Marcié, S.; Pouysségur, J.; Chiche, J. Knock-down of hypoxia-induced carbonic anhydrases IX and XII radiosensitizes tumor cells by increasing intracellular acidosis. Front. Oncol. 2012, 2, 199. [Google Scholar] [CrossRef]
- Nordfors, K.; Haapasalo, J.; Korja, M.; Niemelä, A.; Laine, J.; Parkkila, A.-K.; Pastorekova, S.; Pastorek, J.; Waheed, A.; Sly, W.S.; et al. The tumour-associated carbonic anhydrases CA II, CA IX and CA XII in a group of medulloblastomas and supratentorial primitive neuroectodermal tumours: An association of CA IX with poor prognosis. BMC Cancer 2010, 10, 148. [Google Scholar] [CrossRef] [PubMed]
- Carradori, S.; De Monte, C.; D’Ascenzio, M.; Secci, D.; Celik, G.; Ceruso, M.; Vullo, D.; Scozzafava, A.; Supuran, C.T. Salen and tetrahydrosalen derivatives act as effective inhibitors of the tumor-associated carbonic anhydrase XII—A new scaffold for designing isoform-selective inhibitors. Bioorganic Med. Chem. Lett. 2013, 23, 6759–6763. [Google Scholar] [CrossRef] [PubMed]
- Lounnas, N.; Rosilio, C.; Nebout, M.; Mary, D.; Griessinger, E.; Neffati, Z.; Chiche, J.; Spits, H.; Hagenbeek, T.J.; Asnafi, V.; et al. Pharmacological inhibition of carbonic anhydrase XII interferes with cell proliferation and induces cell apoptosis in T-cell lymphomas. Cancer Lett. 2013, 333, 76–88. [Google Scholar] [CrossRef] [PubMed]
- Imai, K.; Takaoka, A. Comparing antibody and small-molecule therapies for cancer. Nat. Rev. Cancer 2006, 6, 714–727. [Google Scholar] [CrossRef] [PubMed]
- Thiry, A.; Dogné, J.M.; Masereel, B.; Supuran, C.T. Targeting tumor-associated carbonic anhydrase IX in cancer therapy. Trends Pharmacol. Sci. 2006, 27, 566–573. [Google Scholar] [CrossRef] [PubMed]
- Ahlskog, J.K.J.; Dumelin, C.E.; Trüssel, S.; Mårlind, J.; Neri, D. In vivo targeting of tumor-associated carbonic anhydrases using acetazolamide derivatives. Bioorganic Med. Chem. Lett. 2009, 19, 4851–4856. [Google Scholar] [CrossRef]
- Morris, J.C.; Chiche, J.; Grellier, C.; Lopez, M.; Bornaghi, L.F.; Maresca, A.; Supuran, C.T.; Pouysségur, J.; Poulsen, S.-A. Targeting Hypoxic Tumor Cell Viability with Carbohydrate-Based Carbonic Anhydrase IX and XII Inhibitors. J. Med. Chem. 2011, 54, 6905–6918. [Google Scholar] [CrossRef]
- Touisni, N.; Maresca, A.; McDonald, P.C.; Lou, Y.; Scozzafava, A.; Dedhar, S.; Winum, J.-Y.; Supuran, C.T. Glycosyl Coumarin Carbonic Anhydrase IX and XII Inhibitors Strongly Attenuate the Growth of Primary Breast Tumors. J. Med. Chem. 2011, 54, 8271–8277. [Google Scholar] [CrossRef]
- Tauro, M.; Loiodice, F.; Ceruso, M.; Supuran, C.T.; Tortorella, P. Arylamino bisphosphonates: Potent and selective inhibitors of the tumor-associated carbonic anhydrase XII. Bioorganic Med. Chem. Lett. 2014, 24, 1941–1943. [Google Scholar] [CrossRef]
- Oosterwdk, E.; Ruiter, D.J.; Hoedemaeker, P.J.; Pauwels, E.K.J.; Jonas, U.; Zwartendijk, I.; Warnaar, S.O. Monoclonal antibody G 250 recognizes a determinant present in renal-cell carcinoma and absent from normal kidney. Int. J. Cancer 1986, 38, 489–494. [Google Scholar] [CrossRef]
- Divgi, C.R.; Bander, N.H.; Scott, A.M.; O’Donoghue, J.A.; Sgouros, G.; Welt, S.; Finn, R.D.; Morrissey, F.; Capitelli, P.; Williams, J.M.; et al. Phase I/II radioimmunotherapy trial with iodine-131-labeled monoclonal antibody G250 in metastatic renal cell carcinoma. Clin. Cancer Res. 1998, 4, 2729–2739. [Google Scholar] [PubMed]
- Oosterwijk-Wakka, J.C.; Boerman, O.C.; Mulders, P.F.; Oosterwijk, E. Application of Monoclonal Antibody G250 Recognizing Carbonic Anhydrase IX in Renal Cell Carcinoma. Int. J. Mol. Sci. 2013, 14, 11402–11423. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Lo, A.; Yammanuru, A.; Tallarico, A.S.C.; Brady, K.; Murakami, A.; Barteneva, N.; Zhu, Q.; Marasco, W.A. Unique Biological Properties of Catalytic Domain Directed Human Anti-CAIX Antibodies Discovered through Phage-Display Technology. PLoS ONE 2010, 5, e9625. [Google Scholar] [CrossRef] [PubMed]
- Murri-Plesko, M.T.; Hulikova, A.; Oosterwijk, E.; Scott, A.M.; Zortea, A.; Harris, A.L.; Ritter, G.; Old, L.; Bauer, S.; Swietach, P.; et al. Antibody inhibiting enzymatic activity of tumour-associated carbonic anhydrase isoform IX. Eur. J. Pharmacol. 2011, 657, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Chiche, J.; Ilc, K.; Laferrière, J.; Trottier, E.; Dayan, F.; Mazure, N.M.; Brahimi-Horn, M.C.; Pouysségur, J. Hypoxia-Inducible Carbonic Anhydrase IX and XII Promote Tumor Cell Growth by Counteracting Acidosis through the Regulation of the Intracellular pH. Cancer Res. 2009, 69, 358–368. [Google Scholar] [CrossRef]
- Battke, C.; Kremmer, E.; Mysliwietz, J.; Gondi, G.; Dumitru, C.; Brandau, S.; Lang, S.; Vullo, D.; Supuran, C.; Zeidler, R. Generation and characterization of the first inhibitory antibody targeting tumour-associated carbonic anhydrase XII. Cancer Immunol. Immunother. 2011, 60, 649–658. [Google Scholar] [CrossRef]
- Gondi, G.; Mysliwietz, J.; Hulikova, A.; Jen, J.P.; Swietach, P.; Kremmer, E.; Zeidler, R. Antitumor Efficacy of a Monoclonal Antibody That Inhibits the Activity of Cancer-Associated Carbonic Anhydrase XII. Cancer Res. 2013, 73, 6494–6503. [Google Scholar] [CrossRef]
- Dekaminaviciute, D.; Kairys, V.; Zilnyte, M.; Petrikaite, V.; Jogaite, V.; Matuliene, J.; Gudleviciene, Z.; Vullo, D.; Supuran, C.T.; Zvirbliene, A. Monoclonal antibodies raised against 167–180 aa sequence of human carbonic anhydrase XII inhibit its enzymatic activity. J. Enzym. Inhib. Med. Chem. 2014, 29, 804–810. [Google Scholar] [CrossRef]
- Fiedler, L.; Kellner, M.; Gosewisch, A.; Oos, R.; Böning, G.; Lindner, S.; Albert, N.; Bartenstein, P.; Reulen, H.-J.; Zeidler, R.; et al. Evaluation of 177Lu[Lu]-CHX-A″-DTPA-6A10 Fab as a radioimmunotherapy agent targeting carbonic anhydrase XII. Nucl. Med. Biol. 2018, 60, 55–62. [Google Scholar] [CrossRef]
- Fiedler, L.; Kellner, M.; Oos, R.; Böning, G.; Ziegler, S.; Bartenstein, P.; Zeidler, R.; Gildehaus, F.J.; Lindner, S. Fully Automated Production and Characterization of64Cu and Proof-of-Principle Small-Animal PET Imaging Using64Cu-Labelled CA XII Targeting 6A10 Fab. ChemMedChem 2018, 13, 1230–1237. [Google Scholar] [CrossRef]
- Tamošiunas, P.L.; Petraitytė-Burneikienė, R.; Lasickienė, R.; Akatov, A.; Kundrotas, G.; Sereika, V.; Lelešius, R.; Žvirblienė, A.; Sasnauskas, K. Generation of Recombinant Porcine Parvovirus Virus-Like Particles inSaccharomyces cerevisiaeand Development of Virus-Specific Monoclonal Antibodies. J. Immunol. Res. 2014, 2014, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Mickevičiūtė, A.; Juozapaitienė, V.; Michailovienė, V.; Jachno, J.; Matulienė, J.; Matulis, D. Recombinant Production of 12 Catalytically Active Human CA Isoforms. In Carbonic Anhydrase as Drug Target; Springer International Publishing: Basel, Switzerland, 2019; pp. 15–37. [Google Scholar]
- Liang, C.-C.; Park, A.Y.; Guan, J.-L. In vitro scratch assay: A convenient and inexpensive method for analysis of cell migration in vitro. Nat. Protoc. 2007, 2, 329–333. [Google Scholar] [CrossRef] [PubMed]
- Gedvilaite, A.; Kucinskaite-Kodze, I.; Lasickiene, R.; Timinskas, A.; Vaitiekaite, A.; Ziogiene, D.; Žvirblienė, A. Evaluation of Trichodysplasia Spinulosa-Associated Polyomavirus Capsid Protein as a New Carrier for Construction of Chimeric Virus-Like Particles Harboring Foreign Epitopes. Viruses 2015, 7, 4204–4229. [Google Scholar] [CrossRef] [PubMed]
- Whittington, D.A.; Waheed, A.; Ulmasov, B.; Shah, G.N.; Grubb, J.H.; Sly, W.S.; Christianson, D.W. Crystal structure of the dimeric extracellular domain of human carbonic anhydrase XII, a bitopic membrane protein overexpressed in certain cancer tumor cells. Proc. Natl. Acad. Sci. USA 2001, 98, 9545–9550. [Google Scholar] [CrossRef] [PubMed]
- Alterio, V.; Kellner, M.; Esposito, D.; Liesche-Starnecker, F.; Bua, S.; Supuran, C.T.; Monti, S.M.; Zeidler, R.; De Simone, G. Biochemical and Structural Insights into Carbonic Anhydrase XII/Fab6A10 Complex. J. Mol. Biol. 2019, 431, 4910–4921. [Google Scholar] [CrossRef]
- Wang, Z.; Raifu, M.; Howard, M.; Smith, L.; Hansen, D.; Goldsby, R.; Ratner, D.I. Universal PCR amplification of mouse immunoglobulin gene variable regions: The design of degenerate primers and an assessment of the effect of DNA polymerase 3′ to 5′ exonuclease activity. J. Immunol. Methods 2000, 233, 167–177. [Google Scholar] [CrossRef]
- Orlandi, R.; Gussow, D.H.; Jones, P.T.; Winter, G. Cloning immunoglobulin variable domains for expression by the polymerase chain reaction. Proc. Natl. Acad. Sci. USA 1989, 86, 3833–3837. [Google Scholar] [CrossRef]
- Krebber, A.; Bornhauser, S.; Burmester, J.; Honegger, A.; Willuda, J.; Bosshard, H.R.; Plückthun, A. Reliable cloning of functional antibody variable domains from hybridomas and spleen cell repertoires employing a reengineered phage display system. J. Immunol. Methods 1997, 201, 35–55. [Google Scholar] [CrossRef]
- Ye, J.; Ma, N.; Madden, T.L.; Ostell, J.M. IgBLAST: An immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 2013, 41, W34–W40. [Google Scholar] [CrossRef]
- Brochet, X.; Lefranc, M.-P.; Giudicelli, V. IMGT/V-QUEST: The highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008, 36, W503–W508. [Google Scholar] [CrossRef]
- Giudicelli, V.; Brochet, X.; Lefranc, M.-P. IMGT/V-QUEST: IMGT Standardized Analysis of the Immunoglobulin (IG) and T Cell Receptor (TR) Nucleotide Sequences. Cold Spring Harb. Protoc. 2011, 2011, 695–715. [Google Scholar] [CrossRef] [PubMed]
- Ehrenmann, F.; Lefranc, M.-P. IMGT/DomainGapAlign: IMGT Standardized Analysis of Amino Acid Sequences of Variable, Constant, and Groove Domains (IG, TR, MH, IgSF, MhSF). Cold Spring Harb. Protoc. 2011, 2011, 737–749. [Google Scholar] [CrossRef] [PubMed]
- Ehrenmann, F.; Kaas, Q.; Lefranc, M.-P. IMGT/3Dstructure-DB and IMGT/DomainGapAlign: A database and a tool for immunoglobulins or antibodies, T cell receptors, MHC, IgSF and MhcSF. Nucleic Acids Res. 2010, 38, D301–D307. [Google Scholar] [CrossRef]
- Kaas, Q.; Lefranc, M.-P. IMGT Colliers de Perles: Standardized Sequence-Structure Representations of the IgSF and MhcSF Superfamily Domains. Curr. Bioinform. 2007, 2, 21–30. [Google Scholar] [CrossRef]
- Kaas, Q.; Ehrenmann, F.; Lefranc, M.-P. IG, TR and IgSF, MHC and MhcSF: What do we learn from the IMGT Colliers de Perles? Briefings Funct. Genom. Proteom. 2008, 6, 253–264. [Google Scholar] [CrossRef]
- Žvirblienė, A.; Pleckaityte, M.; Lasickiene, R.; Kucinskaite-Kodze, I.; Zvirblis, G. Production and characterization of monoclonal antibodies against vaginolysin: Mapping of a region critical for its cytolytic activity. Toxicon 2010, 56, 19–28. [Google Scholar] [CrossRef]
- Dekaminaviciute, D.; Lasickiene, R.; Parkkila, S.; Jogaite, V.; Matuliene, J.; Matulis, D.; Žvirblienė, A. Development and Characterization of New Monoclonal Antibodies against Human Recombinant CA XII. BioMed Res. Int. 2014, 2014, 1–11. [Google Scholar] [CrossRef]
- Uda, N.R.; Stenner, F.; Seibert, V.; Herzig, P.; Markuly, N.; Van Dijk, M.; Zippelius, A.; Renner, C. Humanized Monoclonal Antibody Blocking Carbonic Anhydrase 12 Enzymatic Activity Leads to Reduced Tumor Growth In Vitro. Anticancer Res. 2019, 39, 4117–4128. [Google Scholar] [CrossRef]
- Bandaranayake, A.D.; Almo, S.C. Recent advances in mammalian protein production. FEBS Lett. 2014, 588, 253–260. [Google Scholar] [CrossRef]
- Kallio, H.; Martinez, A.R.; Hilvo, M.; Hyrskyluoto, A. Cancer-Associated Carbonic Anhydrases IX and XII: Effect of Growth Factors on Gene Expression in Human Cancer Cell Lines. J. Cancer Mol. 2010, 5, 73–78. [Google Scholar]
- Wykoff, C.C.; Beasley, N.J.; Watson, P.H.; Turner, K.J.; Pastorek, J.; Sibtain, A.; Wilson, G.D.; Turley, H.; Talks, K.L.; Maxwell, P.H.; et al. Hypoxia-inducible expression of tumor-associated carbonic anhydrases. Cancer Res. 2000, 60, 7075–7083. [Google Scholar] [PubMed]
- Hsieh, M.-J.; Chen, K.-S.; Chiou, H.-L.; Hsieh, Y.-S. Carbonic anhydrase XII promotes invasion and migration ability of MDA-MB-231 breast cancer cells through the p38 MAPK signaling pathway. Eur. J. Cell Biol. 2010, 89, 598–606. [Google Scholar] [CrossRef] [PubMed]
- Daunys, S.; Petrikaitė, V. The roles of carbonic anhydrases IX and XII in cancer cell adhesion, migration, invasion and metastasis. Biol. Cell 2020. [Google Scholar] [CrossRef] [PubMed]
- Kopecka, J.; Campia, I.; Jacobs, A.; Frei, A.P.; Ghigo, D.; Wollscheid, B.; Riganti, C. Carbonic anhydrase XII is a new therapeutic target to overcome chemoresistance in cancer cells. Oncotarget 2015, 6, 6776–6793. [Google Scholar] [CrossRef]
- Frenzel, A.; Schirrmann, T.; Hust, M. Phage display-derived human antibodies in clinical development and therapy. mAbs 2016, 8, 1177–1194. [Google Scholar] [CrossRef]
- Wan, H. An Overall Comparison of Small Molecules and Large Biologics in ADME Testing. ADMET DMPK 2016, 4, 1–22. [Google Scholar] [CrossRef]
- Almåsbak, H.; Aarvak, T.; Vemuri, M.C. CAR T Cell Therapy: A Game Changer in Cancer Treatment. J. Immunol. Res. 2016, 2016, 1–10. [Google Scholar] [CrossRef]
- Mullard, A. FDA approves first CAR T therapy. Nat. Rev. Drug Discov. 2017, 16, 669. [Google Scholar] [CrossRef] [PubMed]
- Unverdorben, F.; Richter, F.; Hutt, M.; Seifert, O.; Malinge, P.; Fischer, N.; Kontermann, R.E. Pharmacokinetic properties of IgG and various Fc fusion proteins in mice. mAbs 2015, 8, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Bujotzek, A.; Fuchs, A.; Qu, C.; Benz, J.O.; Klostermann, S.; Antes, I.; Georges, G. MoFvAb: Modeling the Fv region of antibodies. mAbs 2015, 7, 838–852. [Google Scholar] [CrossRef] [PubMed]
- Meyer, L.; López, T.; Espinosa, R.; Arias, C.F.; Vollmers, C.; Dubois, R.M. A simplified workflow for monoclonal antibody sequencing. PLoS ONE 2019, 14, e0218717. [Google Scholar] [CrossRef]
- Kopecka, J.; Rankin, G.M.; Salaroglio, I.C.; Poulsen, S.-A.; Riganti, C. P-glycoprotein-mediated chemoresistance is reversed by carbonic anhydrase XII inhibitors. Oncotarget 2016, 7, 85861–85875. [Google Scholar] [CrossRef] [PubMed]
- Čapkauskaitė, E.; Zubrienė, A.; Smirnov, A.; Torresan, J.; Kisonaite, M.; Kazokaitė, J.; Gylytė, J.; Michailoviene, V.; Jogaitė, V.; Manakova, E.; et al. Benzenesulfonamides with pyrimidine moiety as inhibitors of human carbonic anhydrases I, II, VI, VII, XII, and XIII. Bioorganic Med. Chem. 2013, 21, 6937–6947. [Google Scholar] [CrossRef] [PubMed]
- Cimmperman, P.; Baranauskiene, L.; Jachimovičiūtė, S.; Jachno, J.; Torresan, J.; Michailoviene, V.; Matuliene, J.; Sereikaitė, J.; Bumelis, V.; Matulis, D. A Quantitative Model of Thermal Stabilization and Destabilization of Proteins by Ligands. Biophys. J. 2008, 95, 3222–3231. [Google Scholar] [CrossRef] [PubMed]
- Dudutienė, V.; Matulienė, J.; Smirnov, A.; Timm, D.D.; Zubrienė, A.; Baranauskienė, L.; Morkunaitė, V.; Smirnovienė, J.; Michailovienė, V.; Juozapaitienė, V.; et al. Discovery and Characterization of Novel Selective Inhibitors of Carbonic Anhydrase IX. J. Med. Chem. 2014, 57, 9435–9446. [Google Scholar] [CrossRef] [PubMed]
- Sudzius, J.; Baranauskiene, L.; Golovenko, D.; Matuliene, J.; Michailoviene, V.; Torresan, J.; Jachno, J.; Sukackaitė, R.; Manakova, E.; Gražulis, S.; et al. 4-[N-(Substituted 4-pyrimidinyl)amino]benzenesulfonamides as inhibitors of carbonic anhydrase isozymes I, II, VII, and XIII. Bioorganic Med. Chem. 2010, 18, 7413–7421. [Google Scholar] [CrossRef] [PubMed]
- Jogaitė, V.; Zubrienė, A.; Michailoviene, V.; Gylytė, J.; Morkūnaitė, V.; Matulis, D. Characterization of human carbonic anhydrase XII stability and inhibitor binding. Bioorganic Med. Chem. 2013, 21, 1431–1436. [Google Scholar] [CrossRef] [PubMed]
- Köhler, G.; Milstein, C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nat. Cell Biol. 1975, 256, 495–497. [Google Scholar] [CrossRef]
- Wilson, M.B. Nakane Recent developments in the periodate method of conjugating horseradish peroxidase (HRPO) to antibodies. In Immunofluorescence and Related Staining Techniques; Knapp, W., Holubar, K., Wick, G., Eds.; Elsevier North-Holland: Amsterdam, The Netherlands, 1978; p. 363. [Google Scholar]
- Khalifah, R.G. The carbon dioxide hydration activity of carbonic anhydrase. I. Stop-flow kinetic studies on the native human isoenzymes B and C. J. Biol. Chem. 1971, 246, 2561–2573. [Google Scholar]
- Kuzmič, P. History, Variants and Usage of the “Morrison Equation” in Enzyme Inhibition Kinetics; BioKin Ltd.: Watertown, MA, USA, 2015. [Google Scholar]
- Morrison, J. Kinetics of the reversible inhibition of enzyme-catalysed reactions by tight-binding inhibitors. Biochim. Biophys. Acta Enzymol. 1969, 185, 269–286. [Google Scholar] [CrossRef]
- Pleckaityte, M.; Mistiniene, E.; Lasickiene, R.; Zvirblis, G.; Žvirblienė, A. Generation of recombinant single-chain antibodies neutralizing the cytolytic activity of vaginolysin, the main virulence factor of Gardnerella vaginalis. BMC Biotechnol. 2011, 11, 100. [Google Scholar] [CrossRef] [PubMed]
- Kucinskaite-Kodze, I.; Pleckaityte, M.; Bremer, C.M.; Seiz, P.L.; Zilnyte, M.; Bulavaite, A.; Mickiene, G.; Zvirblis, G.; Sasnauskas, K.; Glebe, D.; et al. New broadly reactive neutralizing antibodies against hepatitis B virus surface antigen. Virus Res. 2016, 211, 209–221. [Google Scholar] [CrossRef] [PubMed]
- Čeponytė, U.; Paškevičiūtė, M.; Petrikaitė, V. Comparison of NSAIDs activity in COX-2 expressing and non-expressing 2D and 3D pancreatic cancer cell cultures. Cancer Manag. Res. 2018, 10, 1543–1551. [Google Scholar] [CrossRef] [PubMed]










| MAb Clone | Isotype | ELISA, WB | Cross-Reactivities (ELISA, Wb) | FC | Kd, M | |||
|---|---|---|---|---|---|---|---|---|
| CA XIIHEK | CA XIIDE3 | A498 | A549 | Jurkat | ||||
| 1B10 | IgG1 | + | + | – | + | + | – | 2.4 × 10−10 |
| 1E7 | IgG2b | + | + | CA XIII | + | + | – | 1.4 × 10−9 |
| 2G10 | IgG2b | + | + | CA XIII | + | + | – | 5.3 × 10−10 |
| 6D12 | IgG1 | + | + | – | + | + | – | 1.1 × 10−9 |
| 6G5 | IgG2a | + | + | CA I, CA II | – | – | – | ND * |
| 6G10 | IgG1 | + | + | – | + | + | – | 1.2 × 10−9 |
| 6H2 | IgG1 | + | + | – | + | + | – | 1.7 × 10−8 |
| 9B9 | IgG1 | + | – | CA IV, CA IX | – | – | – | ND * |
| 9D6 | IgG1 | + | + | – | + | + | – | 2.3 × 10−10 |
| 10C7 | IgG1 | + | + | – | + | + | – | 2.4 × 10−9 |
| 11A12 | IgG2a | + | + | – | + | + | – | 1.4 × 10−8 |
| 11C5 | IgG1 | + | + | – | + | + | – | 2.5 × 10−10 |
| 12H2 | IgG1 | + | + | – | – | – | – | ND * |
| 13G2 | IgG1 | + | – | CA IV, CA IX | – | – | – | ND * |
| 14D6 | IgG1 | + | + | CA II | + | + | – | 2.0 × 10−9 |
| 15B4 | IgG1 | + | + | – | + | + | – | 7.3 × 10−9 |
| 15H1 | IgG1 | + | + | – | + | + | – | 3.3 × 10−9 |
| 16A8 | IgG1 | + | + | – | + | + | – | 7.7 × 10−10 |
| 17E9 | IgG1 | + | + | – | + | + | – | 1.5 × 10−9 |
| 17A3 | IgG1 | + | + | – | + | + | – | 1.8 × 10−9 |
| 20C3 | IgG1 | + | + | – | + | + | – | 1.6 × 10−9 |
| 20C4 | IgG1 | + | – | CA IV, CA IX | – | – | – | ND * |
| 20G7 | IgG2b | + | + | CA II, CA VI | – | – | – | ND * |
| 22A2 | IgG2b | + | + | – | + | + | – | 1.2×10−9 |
| Primer Name | Primer Sequence | Amplified Chain | Ref. |
|---|---|---|---|
| IgG1 | 5′–TTA ATA GAC AGA TGG GGG TGT CGT TTT GGC | H | [38] |
| MH1 | 5′–CAT ATG SAR GTN MAG CTG SAG SAG TC | H | |
| Kc | 5′–TTA GGA TAC AGT TGG TGC AGC ATC | Lκ | |
| Mk | 5′–CAT ATG GAY ATT GTG MTS ACM CAR WCT MCA | Lκ | |
| VH1FOR | 5′–TGA GGA GAC GGT GAC CGT GGT CCC TTG GCC CCA G | H | [39] |
| VHlBACK | 5′–AGG TSM ARC TGC AGS AGT CWG G | H | |
| VK2FOR | 5′–GTT ATT TGA TCT CCA GCT TGG TCC C | Lκ | |
| VK1BACK | 5′–GAC ATT CAG CTG ACC CAG TCT CCA | Lκ | |
| LB6 | 5′–CAT ATG ATT MAG ATR AMC CAG TC | Lκ | [40] |
| LB10 | 5′–CAT ATG ATT GWG CTS ACC CAA TC | Lκ | |
| LB11 | 5′–CAT ATG ATT STR ATG ACC CAR TC | Lκ | |
| LB12 | 5′–CAT ATG RTT KTG ATG ACC CAR AC | Lκ | |
| LB16 | 5′–CAT ATG ATT GTG ATG ACA CAA CC | Lκ | |
| LB17 | 5′–CAT ATG ATT TTG CTG ACT CAG TC | Lκ |
| Fragment Number | Amino Acid Sequence | PCR Primers | Fragments Size, kDa |
|---|---|---|---|
| #1 | V27–G123 | 5′– ATA CATATG TTA CCCCCAGTGCAGGTG 5′– ATG CATATG GTGAACGGTTCCAAGTG | 13.5 |
| #2 | Y67–F211 | 5′– ATT CATATG TTA GAATGCTTCCTGGCCTTTGT 5′– TTT CATATG TATGACGCCAGCCTCACG | 17.7 |
| #3 | H192–S290 | 5′– GCC CATATG TTA GGAGAAGGAGGTGTAT 5′– GAC CATATG CATGTAAAGTACAAAGGCCA | 14.3 |
| #4 | P101–S290 | 5′– TTT CATATG TTA GGAGAAGGAGGTGTAT 5′– ATT CATATG CCCTCGGACATGCACA | 24.3 |
| #5 | F35–R220 | 5′– AGT CATATG TTTGGTCCTGATGGGGAGAA 5′– A GGATCC TTA GCGGTA ATATTCAGCG GTCC | 23.4 |
| #6 | W43–E209 | 5′– GGA CATATG TGGTCCAAGAAGTACCCGTC 5′– A GGATCC TTA CT CTTCAATGTT GAATCCCG | 21.2 |
| #7 | G51–N231 | 5′– A CATATG GGGGGCCTGC TGCAGT 5′– A GGATCC TTA GGGTTGCA AGGGGGTG | 23.1 |
| #8 | D59–Y221 | 5′– ATT CATATG GACCTGCACAGTGACATCCT 5′– TT GGATCC TTA GTAGCGGTAATATTCAGCGG | 21.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stravinskiene, D.; Sliziene, A.; Baranauskiene, L.; Petrikaite, V.; Zvirbliene, A. Inhibitory Monoclonal Antibodies and Their Recombinant Derivatives Targeting Surface-Exposed Carbonic Anhydrase XII on Cancer Cells. Int. J. Mol. Sci. 2020, 21, 9411. https://doi.org/10.3390/ijms21249411
Stravinskiene D, Sliziene A, Baranauskiene L, Petrikaite V, Zvirbliene A. Inhibitory Monoclonal Antibodies and Their Recombinant Derivatives Targeting Surface-Exposed Carbonic Anhydrase XII on Cancer Cells. International Journal of Molecular Sciences. 2020; 21(24):9411. https://doi.org/10.3390/ijms21249411
Chicago/Turabian StyleStravinskiene, Dovile, Aiste Sliziene, Lina Baranauskiene, Vilma Petrikaite, and Aurelija Zvirbliene. 2020. "Inhibitory Monoclonal Antibodies and Their Recombinant Derivatives Targeting Surface-Exposed Carbonic Anhydrase XII on Cancer Cells" International Journal of Molecular Sciences 21, no. 24: 9411. https://doi.org/10.3390/ijms21249411
APA StyleStravinskiene, D., Sliziene, A., Baranauskiene, L., Petrikaite, V., & Zvirbliene, A. (2020). Inhibitory Monoclonal Antibodies and Their Recombinant Derivatives Targeting Surface-Exposed Carbonic Anhydrase XII on Cancer Cells. International Journal of Molecular Sciences, 21(24), 9411. https://doi.org/10.3390/ijms21249411






