The Central Region of Testican-2 Forms a Compact Core and Promotes Cell Migration
Abstract
:1. Introduction
2. Results
2.1. EC Domain of Testican-2 Binds a Single Calcium Ion
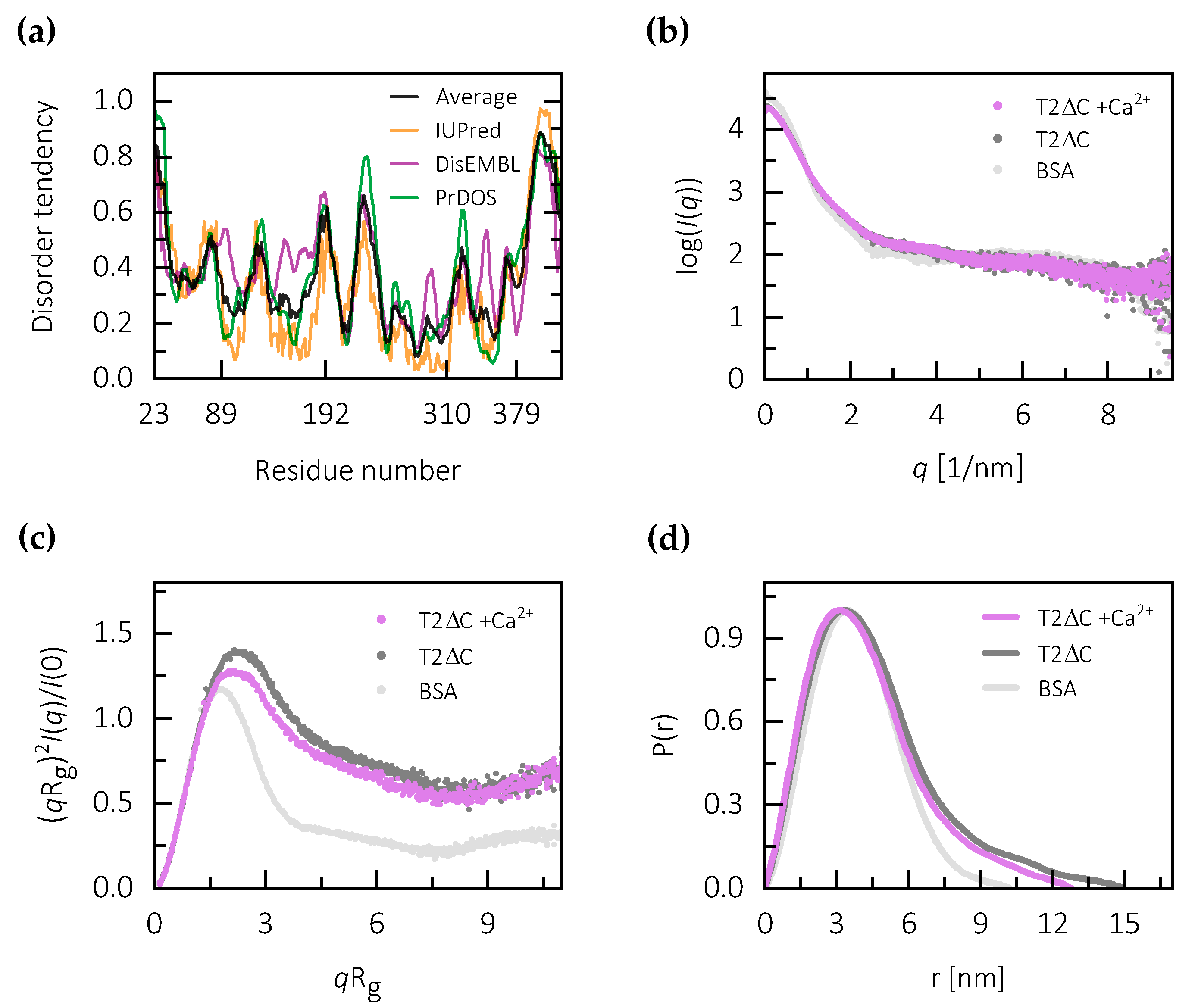
2.2. Calcium Binding Stabilizes the Central Core of Testican-2
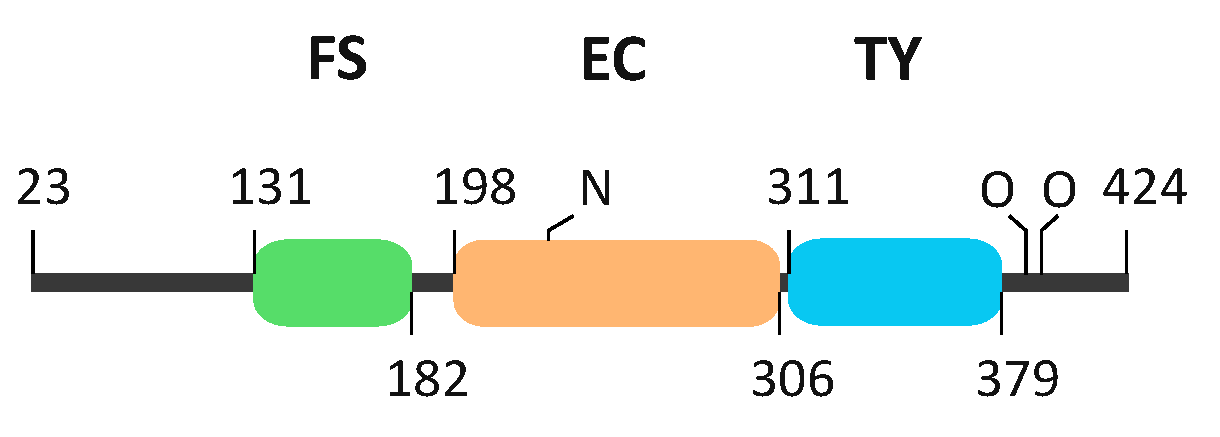
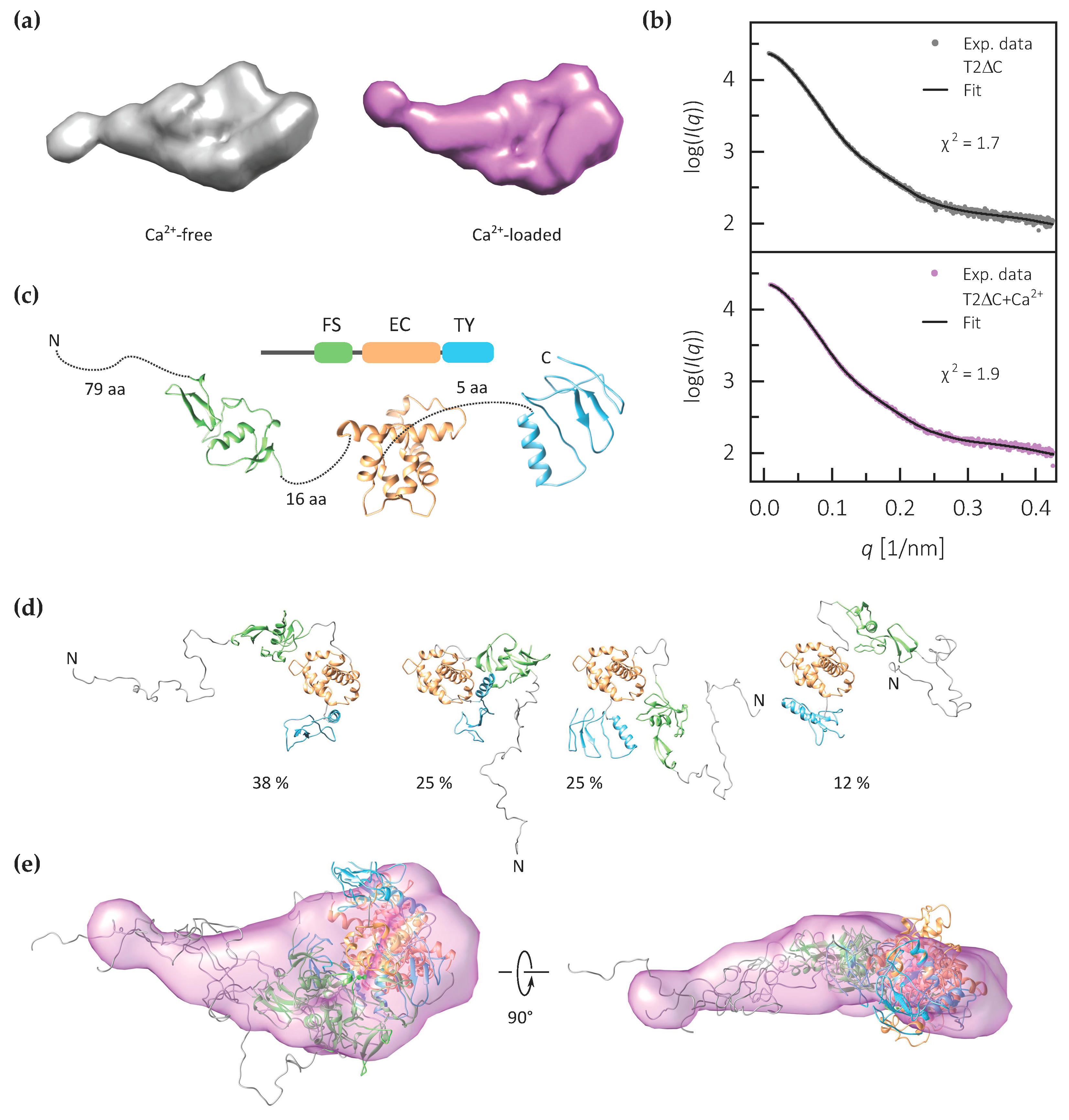
2.3. The FS–EC–TY Domain Triplet Forms a Compact Structural Core
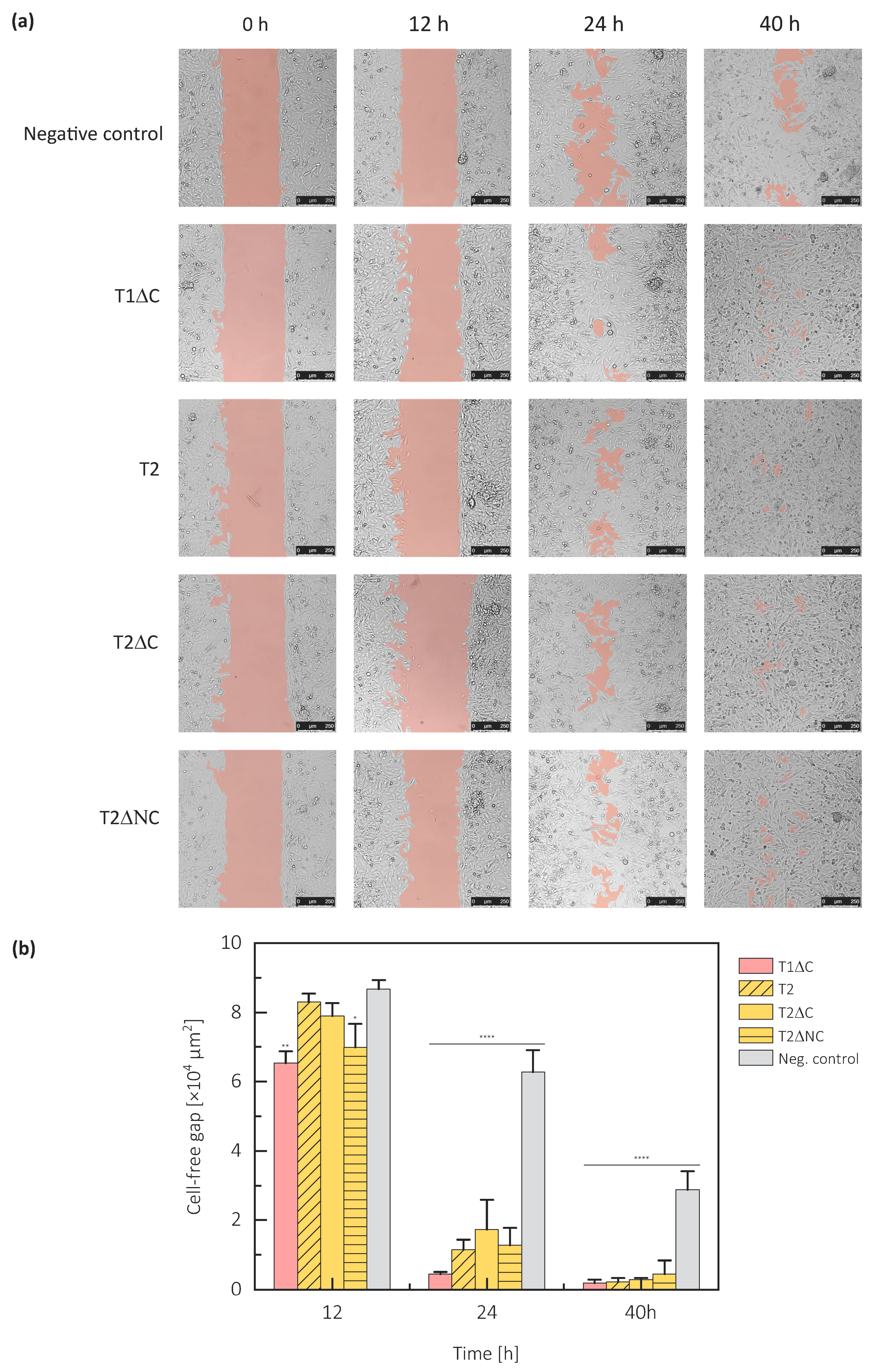
2.4. Protein Part of Testicans Promotes Cell Migration
3. Discussion
4. Materials and Methods
4.1. Cloning, Expression and Purification
4.2. Calcium-Binding Analysis
4.3. Sequence Analysis and Disorder Prediction
4.4. Small Angle X-ray Scattering and Modeling
4.5. Cell Exclusion Zone Assay
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BSA | bovine serum albumin |
| Dmax | maximum dimension |
| EC | extracellular calcium-binding (domain) |
| ECM | extracellular matrix |
| EOM | ensemble optimization method |
| FS | follistatin (domain) |
| G | Gibbs free energy |
| GAG | glycosaminoglycan |
| H | enthaply |
| I; I0 | scattered intensity; extrapolated scattering intensity at q = 0 |
| ITC | isothermal titration calorimetry |
| Kd | dissociation constant |
| MT-MMP | membrane-type matrix metaloprotease |
| Mw | molecular weight, in Da (daltons) |
| PDDF, P(r) | pair distance distribution function |
| q | momentum of transfer |
| Rg | radius of gyration |
| S | entropy |
| SAXS | small angle X-ray scattering |
| SEC | size-exclusion chromatography |
| T | temperature |
| TEV | tobbaco etch virus |
| TY | thyroglobulin type-1 (domain) |
References
- Frantz, C.; Stewart, K.M.; Weaver, V.M. The extracellular matrix at a glance. J. Cell Sci. 2010, 123, 4195–4200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mouw, J.K.; Ou, G.; Weaver, V.M. Extracellular matrix assembly: A multiscale deconstruction. Nat. Rev. Mol. Cell Biol. 2014, 15, 771–785. [Google Scholar] [CrossRef] [PubMed]
- Pickup, M.W.; Mouw, J.K.; Weaver, V.M. The extracellular matrix modulates the hallmarks of cancer. EMBO Rep. 2014, 15, 1243–1253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, P.; Guan, H.-T.; Dai, Z.-J.; Ma, Y.-G.; Liu, X.-X.; Wang, X.-J. Knockdown of SPOCK1 inhibits the proliferation and invasion in colorectal cancer cells by suppressing the PI3K/Akt pathway. Oncol. Res. 2016, 24, 437–445. [Google Scholar] [CrossRef]
- Yang, J.; Yang, Q.; Yu, J.; Li, X.; Yu, S.; Zhang, X. SPOCK1 promotes the proliferation, migration and invasion of glioma cells through PI3K/AKT and Wnt/β-catenin signaling pathways. Oncol. Rep. 2016. [Google Scholar] [CrossRef] [Green Version]
- Alliel, P.M.; Périn, J.P.; Jollès, P.; Bonnet, F.J. Testican, a multidomain testicular proteoglycan resembling modulators of cell social behaviour. Eur. J. Biochem. 1993, 214, 347–350. [Google Scholar] [CrossRef]
- Bonnet, F.; Périn, J.P.; Charbonnier, F.; Camuzat, A.; Roussel, G.; Nussbaum, J.L.; Alliel, P.M. Structure and cellular distribution of mouse brain testican. Association with the postsynaptic area of hippocampus pyramidal cells. J. Biol. Chem. 1996, 271, 4373–4380. [Google Scholar] [CrossRef] [Green Version]
- Vannahme, C.; Schübel, S.; Herud, M.; Gösling, S.; Hülsmann, H.; Paulsson, M.; Hartmann, U.; Maurer, P. Molecular cloning of testican-2: Defining a novel calcium-binding proteoglycan family expressed in brain. J. Neurochem. 1999, 73, 12–20. [Google Scholar] [CrossRef]
- Bradshaw, A.D. Diverse biological functions of the SPARC family of proteins. Int. J. Biochem. Cell Biol. 2012, 44, 480–488. [Google Scholar] [CrossRef] [Green Version]
- Hohenester, E.; Maurer, P.; Hohenadl, C.; Timpl, R.; Jansonius, J.N.; Engel, J. Structure of a novel extracellular Ca2+-binding module in BM-40. Nat. Struct. Mol. Biol. 1996, 3, 67–73. [Google Scholar] [CrossRef]
- Hartmann, U.; Hülsmann, H.; Seul, J.; Röll, S.; Midani, H.; Breloy, I.; Hechler, D.; Müller, R.; Paulsson, M. Testican-3: A brain-specific proteoglycan member of the BM-40/SPARC/osteonectin family. J. Neurochem. 2013, 125, 399–409. [Google Scholar] [CrossRef] [PubMed]
- Röll, S.; Seul, J.; Paulsson, M.; Hartmann, U. Testican-1 is dispensable for mouse development. Matrix Biol. 2006, 25, 373–381. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, A.; Uchiyama, K.; Nara, T.; Nishimura, N.; Hayasaka, M.; Hanaoka, K.; Yamamoto, T. Structural abnormalities of corpus callosum and cortical axonal tracts accompanied by decreased anxiety-like behavior and lowered sociability in spock3-mutant mice. Dev. Neurosci. 2014, 36, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Dhamija, R.; Graham, J.M.; Smaoui, N.; Thorland, E.; Kirmani, S. Novel de novo SPOCK1 mutation in a proband with developmental delay, microcephaly and agenesis of corpus callosum. Eur. J. Med. Genet. 2014, 57, 181–184. [Google Scholar] [CrossRef]
- Meh, P.; Pavšič, M.; Turk, V.; Baici, A.; Lenarčič, B. Dual concentration-dependent activity of thyroglobulin type-1 domain of testican: Specific inhibitor and substrate of cathepsin L. Biol. Chem. 2005, 386, 75–83. [Google Scholar] [CrossRef] [Green Version]
- Nakada, M.; Yamada, A.; Takino, T.; Miyamori, H.; Takahashi, T.; Yamashita, J.; Sato, H. Suppression of membrane-type 1 matrix metalloproteinase (MMP)-mediated MMP-2 activation and tumor invasion by testican 3 and its splicing variant gene product, N-Tes. Cancer Res. 2001, 61, 8896–8902. [Google Scholar]
- Mendes, S.R.; del Amo-Maestro, L.; Marino-Puertas, L.; de Diego, I.; Goulas, T.; Gomis-Rüth, F.X. Analysis of the inhibiting activity of reversion-inducing cysteine-rich protein with Kazal motifs (RECK) on matrix metalloproteinases. Sci. Rep. 2020, 10, 6317. [Google Scholar] [CrossRef] [Green Version]
- Nakada, M.; Miyamori, H.; Yamashita, J.; Sato, H. Testican 2 abrogates inhibition of membrane-type matrix metalloproteinases by other testican family proteins. Cancer Res. 2003, 63, 3364–3369. [Google Scholar]
- Marr, H.S.; Edgell, C.J.S. Testican-1 inhibits attachment of Neuro-2a cells. Matrix Biol. 2003, 22, 259–266. [Google Scholar] [CrossRef]
- Schnepp, A.; Lindgren, P.K.; Hülsmann, H.; Kröger, S.; Paulsson, M.; Hartmann, U. Mouse Testican-2. Expression, glycosylation, and effects on neurite outgrowth. J. Biol. Chem. 2005, 280, 11274–11280. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.-P.; Han, S.-W.; Song, S.-H.; Jeong, E.-G.; Lee, M.-Y.; Hwang, D.; Im, S.; Bang, Y.-J.; Kim, T.-Y. Testican-1-mediated epithelial-mesenchymal transition signaling confers acquired resistance to lapatinib in HER2-positive gastric cancer. Oncogene 2014, 33, 3334–3341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, C.; Fischer-Kešo, R.; Schlechter, T.; Ströbel, P.; Marx, A.; Hofmann, I. Plakophilin 1-deficient cells upregulate SPOCK1: Implications for prostate cancer progression. Tumour Biol. 2015, 36, 9567–9577. [Google Scholar] [CrossRef] [PubMed]
- Shu, Y.-J.; Weng, H.; Ye, Y.-Y.; Hu, Y.-P.; Bao, R.-F.; Cao, Y.; Wang, X.-A.; Zhang, F.; Xiang, S.-S.; Li, H.-F.; et al. SPOCK1 as a potential cancer prognostic marker promotes the proliferation and metastasis of gallbladder cancer cells by activating the PI3K/AKT pathway. Mol. Cancer 2015, 14, 12. [Google Scholar] [CrossRef] [Green Version]
- Alshargabi, R.; Sano, T.; Yamashita, A.; Takano, A.; Sanada, T.; Iwashita, M.; Shinjo, T.; Fukuda, T.; Sanui, T.; Kishida, S.; et al. SPOCK1 is a novel inducer of epithelial to mesenchymal transition in drug-induced gingival overgrowth. Sci. Rep. 2020, 10, 9785. [Google Scholar] [CrossRef] [PubMed]
- Takahata, T.; Shukla, R.; Yamamori, T.; Kaas, J.H. Differential expression patterns of striate cortex-enriched genes among Old World, New World, and prosimian primates. Cereb. Cortex 2012, 22, 2313–2321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barrera-Ocampo, A.; Arlt, S.; Matschke, J.; Hartmann, U.; Puig, B.; Ferrer, I.; Zürbig, P.; Glatzel, M.; Sepulveda-Falla, D.; Jahn, H. Amyloid-β precursor protein modulates the sorting of testican-1 and contributes to its accumulation in brain tissue and cerebrospinal fluid from patients with alzheimer disease. J. Neuropathol. Exp. Neurol. 2016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Castro, E.; Sigrist, C.J.A.; Gattiker, A.; Bulliard, V.; Langendijk-Genevaux, P.S.; Gasteiger, E.; Bairoch, A.; Hulo, N. ScanProsite: Detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006, 34, W362–W365. [Google Scholar] [CrossRef] [PubMed]
- Sigrist, C.J.A.; de Castro, E.; Cerutti, L.; Cuche, B.A.; Hulo, N.; Bridge, A.; Bougueleret, L.; Xenarios, I. New and continuing developments at PROSITE. Nucleic Acids Res. 2013, 41, D344–D347. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Ahmad, E.; Kumar, S.; Khan, R.; Gourinath, S. Flexibility of EF-hand motifs: Structural and thermodynamic studies of Calcium Binding Protein-1 from Entamoeba histolytica with Pb2+, Ba2+, and Sr2+. BMC Biophys. 2012, 5, 15. [Google Scholar] [CrossRef] [Green Version]
- Denessiouk, K.; Permyakov, S.; Denesyuk, A.; Permyakov, E.; Johnson, M.S. Two Structural Motifs within Canonical EF-Hand Calcium-Binding Domains Identify Five Different Classes of Calcium Buffers and Sensors. PLoS ONE 2014, 9, e109287. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Harrison, P.M. fLPS: Fast discovery of compositional biases for the protein universe. BMC Bioinform. 2017, 18, 476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radivojac, P.; Obradovic, Z.; Smith, D.K.; Zhu, G.; Vucetic, S.; Brown, C.J.; Lawson, J.D.; Dunker, A.K. Protein flexibility and intrinsic disorder. Protein Sci. 2004, 13, 71–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, A.H.; Crick, S.L.; Vitalis, A.; Chicoine, C.L.; Pappu, R.V. Net charge per residue modulates conformational ensembles of intrinsically disordered proteins. Proc. Natl. Acad. Sci. USA 2010, 107, 8183–8188. [Google Scholar] [CrossRef] [Green Version]
- Mészáros, B.; Erdős, G.; Dosztányi, Z. IUPred2A: Context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res. 2018, 46, W329–W337. [Google Scholar] [CrossRef]
- Linding, R.; Jensen, L.J.; Diella, F.; Bork, P.; Gibson, T.J.; Russell, R.B. Protein Disorder Prediction. Structure 2003, 11, 1453–1459. [Google Scholar] [CrossRef] [Green Version]
- Ishida, T.; Kinoshita, K. PrDOS: Prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 2007, 35, W460–W464. [Google Scholar] [CrossRef]
- Lord, M.S.; Whitelock, J.M. Recombinant production of proteoglycans and their bioactive domains. FEBS J. 2013, 280, 2490–2510. [Google Scholar] [CrossRef]
- Pavšič, M.; Vito, T.; Lenarčič, B. Purification and characterization of a recombinant human testican-2 expressed in baculovirus-infected Sf9 insect cells. Protein Exp. Purif. 2008, 58, 132–139. [Google Scholar] [CrossRef]
- Kohfeldt, E.; Maurer, P.; Vannahme, C.; Timpl, R. Properties of the extracellular calcium binding module of the proteoglycan testican. FEBS Lett. 1997, 414, 557–561. [Google Scholar] [CrossRef]
- Maurer, P.; Hohenadl, C.; Hohenester, E.; Göhring, W.; Timpl, R.; Engel, J. The C-terminal portion of BM-40 (SPARC/osteonectin) is an autonomously folding and crystallisable domain that binds calcium and collagen IV. J. Mol. Biol. 1995, 253, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Drmota Prebil, S.; Slapšak, U.; Pavšič, M.; Ilc, G.; Puž, V.; de Almeida Ribeiro, E.; Anrather, D.; Hartl, M.; Backman, L.; Plavec, J.; et al. Structure and calcium-binding studies of calmodulin-like domain of human non-muscle alpha-actinin-1. Sci. Rep. 2016, 6, 27383. [Google Scholar] [CrossRef] [PubMed]
- Hurwitz, S. Homeostatic control of plasma calcium concentration. Crit. Rev. Biochem. Mol. Biol. 1996, 31, 41–100. [Google Scholar] [CrossRef] [PubMed]
- Bernadó, P.; Blanchard, L.; Timmins, P.; Marion, D.; Ruigrok, R.W.H.; Blackledge, M. A structural model for unfolded proteins from residual dipolar couplings and small-angle x-ray scattering. Proc. Natl. Acad. Sci. USA 2005, 102, 17002–17007. [Google Scholar] [CrossRef] [Green Version]
- Kikhney, A.G.; Svergun, D.I. A practical guide to small angle X-ray scattering (SAXS) of flexible and intrinsically disordered proteins. FEBS Lett. 2015, 589, 2570–2577. [Google Scholar] [CrossRef] [Green Version]
- Hung, J.-Y.; Yen, M.-C.; Jian, S.-F.; Wu, C.-Y.; Chang, W.-A.; Liu, K.-T.; Hsu, Y.-L.; Chong, I.-W.; Kuo, P.-L. Secreted protein acidic and rich in cysteine (SPARC) induces cell migration and epithelial mesenchymal transition through WNK1/snail in non-small cell lung cancer. Oncotarget 2017, 8, 63691–63702. [Google Scholar] [CrossRef]
- Sullivan, M.M.; Puolakkainen, P.A.; Barker, T.H.; Funk, S.E.; Sage, E.H. Altered tissue repair in hevin-null mice: Inhibition of fibroblast migration by a matricellular SPARC homolog. Wound Repair Regen. 2008, 16, 310–319. [Google Scholar] [CrossRef]
- Fahmi, M.; Ito, M. Evolutionary Approach of Intrinsically Disordered CIP/KIP Proteins. Sci. Rep. 2019, 9, 1575. [Google Scholar] [CrossRef] [Green Version]
- Santofimia-Castaño, P.; Rizzuti, B.; Xia, Y.; Abian, O.; Peng, L.; Velázquez-Campoy, A.; Neira, J.L.; Iovanna, J. Targeting intrinsically disordered proteins involved in cancer. Cell Mol. Life Sci. 2020, 77, 1695–1707. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Zhang, Y.; Da, J.; Jia, Z.; Wu, H.; Gu, K. Downregulation of SPARC Expression Decreases Cell Migration and Invasion Involving Epithelial-Mesenchymal Transition through the p-FAK/p-ERK Pathway in Esophageal Squamous Cell Carcinoma. J. Cancer 2020, 11, 414–420. [Google Scholar] [CrossRef]
- Nagase, T.; Seki, N.; Ishikawa, K.; Ohira, M.; Kawarabayasi, Y.; Ohara, O.; Tanaka, A.; Kotani, H.; Miyajima, N.; Nomura, N. Prediction of the coding sequences of unidentified human genes. VI. The coding sequences of 80 new genes (KIAA0201-KIAA0280) deduced by analysis of cDNA clones from cell line KG-1 and brain. DNA Res. 1996, 3, 321–329, 341–354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berger, I.; Fitzgerald, D.J.; Richmond, T.J. Baculovirus expression system for heterologous multiprotein complexes. Nat. Biotechnol. 2004, 22, 1583–1587. [Google Scholar] [CrossRef] [PubMed]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franke, D.; Petoukhov, M.V.; Konarev, P.V.; Panjkovich, A.; Tuukkanen, A.; Mertens, H.D.T.; Kikhney, A.G.; Hajizadeh, N.R.; Franklin, J.M.; Jeffries, C.M.; et al. ATSAS 2.8: A comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J. Appl. Crystallogr. 2017, 50, 1212–1225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Svergun, D.I. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 1992, 25, 495–503. [Google Scholar] [CrossRef]
- Franke, D.; Svergun, D.I. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2009, 42, 342–346. [Google Scholar] [CrossRef] [Green Version]
- Volkov, V.V.; Svergun, D.I. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2003, 36, 860–864. [Google Scholar] [CrossRef] [Green Version]
- Tria, G.; Mertens, H.D.T.; Kachala, M.; Svergun, D.I. Advanced ensemble modelling of flexible macromolecules using X-ray solution scattering. IUCrJ 2015, 2, 207–217. [Google Scholar] [CrossRef] [Green Version]
- Kikhney, A.G.; Borges, C.R.; Molodenskiy, D.S.; Jeffries, C.M.; Svergun, D.I. SASBDB: Towards an automatically curated and validated repository for biological scattering data. Protein Sci. 2020, 29, 66–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Sato, H.; Ebisawa, K.; Takanari, K.; Yagi, S.; Toriyama, K.; Yamawaki-Ogata, A.; Kamei, Y. Skin-Derived Precursor Cells Promote Wound Healing in Diabetic Mice. Ann. Plast. Surg. 2015, 74, 114–120. [Google Scholar] [CrossRef] [PubMed]

| Kd [µM] | n | ΔH [kcal/mol] | TΔS [kcal/mol] | ΔG [kcal/mol] |
|---|---|---|---|---|
| 17.1 ± 3.4 | 1.3 ± 0.2 | −17.64 ± 2.4 | 11.25 ± 0.22 | −6.39 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krajnc, A.; Gaber, A.; Lenarčič, B.; Pavšič, M. The Central Region of Testican-2 Forms a Compact Core and Promotes Cell Migration. Int. J. Mol. Sci. 2020, 21, 9413. https://doi.org/10.3390/ijms21249413
Krajnc A, Gaber A, Lenarčič B, Pavšič M. The Central Region of Testican-2 Forms a Compact Core and Promotes Cell Migration. International Journal of Molecular Sciences. 2020; 21(24):9413. https://doi.org/10.3390/ijms21249413
Chicago/Turabian StyleKrajnc, Anja, Aljaž Gaber, Brigita Lenarčič, and Miha Pavšič. 2020. "The Central Region of Testican-2 Forms a Compact Core and Promotes Cell Migration" International Journal of Molecular Sciences 21, no. 24: 9413. https://doi.org/10.3390/ijms21249413





