Potential of the Electronic Nose for the Detection of Respiratory Diseases with and without Infection
Abstract
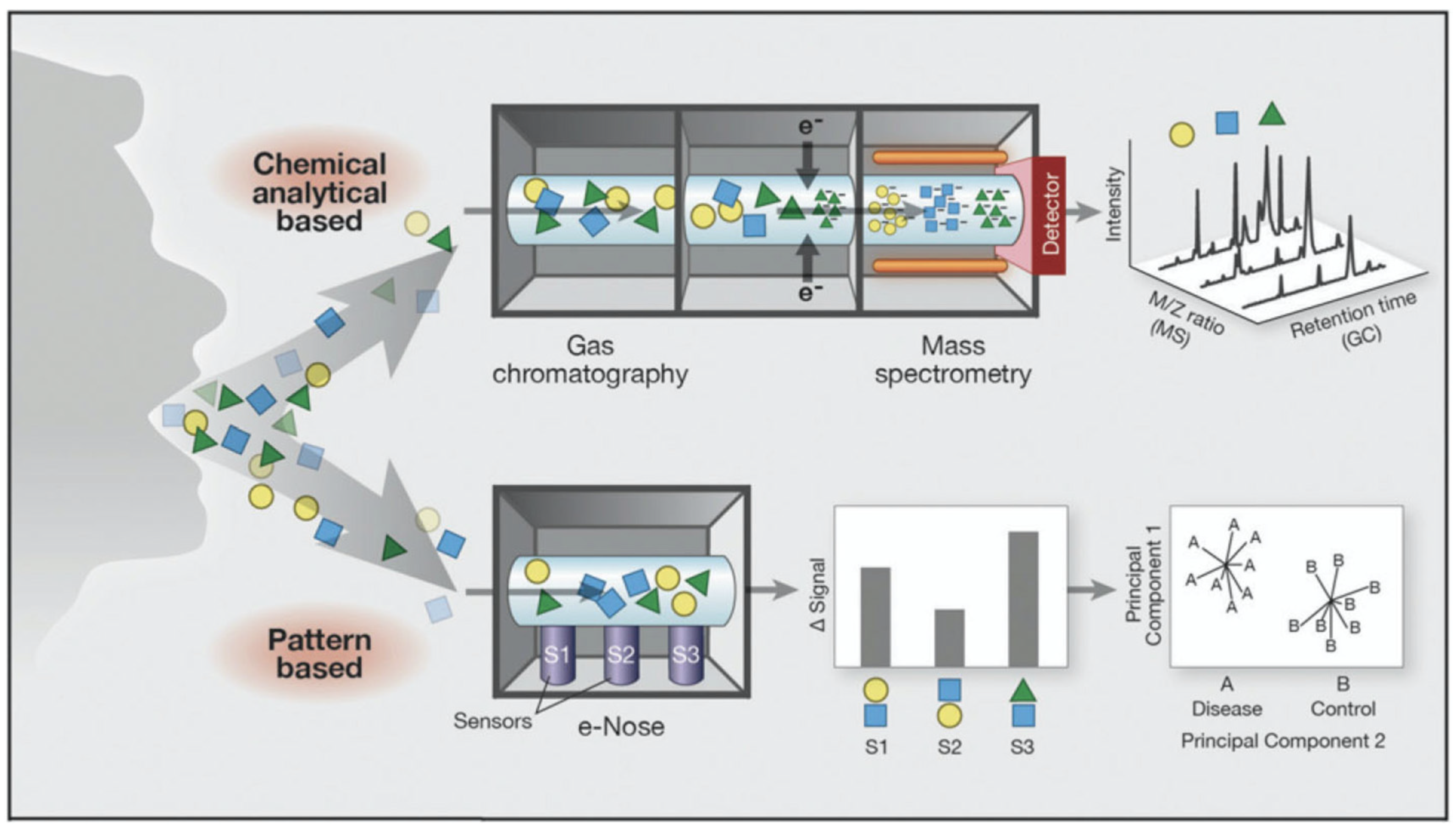
:1. Introduction
2. eNose Technology in Respiratory Disease and Infection
2.1. Lung Cancer
2.2. Asthma
2.3. Chronic Obstructive Pulmonary Disease
2.4. Cystic Fibrosis, Bronchiectasis and Primary Ciliary Dyskinesia
3. Future Directions and Need for Future VOC-based Studies
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AEx | Asymptomatic former asbestos-exposed subjects |
| ARD | Benign asbestos-related diseases |
| AUROC | Area under the receiver operator characteristic |
| BAL | Broncho-alveolar lavage |
| BODE | Body mass index, obstruction, dyspnea, and exercise |
| CD | Crohn’s disease |
| CF | Cystic fibrosis |
| CFTR | Cystic fibrosis transmembrane conductance regulator |
| COPD | Chronic obstructive pulmonary disorder |
| CVV | Cross-validation value |
| ECOPD | Exacerbations of COPD |
| eNose | Electronic nose |
| GC-MS | Gas chromatography-mass spectrometry |
| IA | Invasive pulmonary aspergillosis |
| IBD | Inflammatory bowel disease |
| LCDA | Linear canonical discriminant analysis |
| LRA | Logistic regression analysis |
| MPM | Malignant pleural mesothelioma |
| PA | Pseudomonas aeruginosa |
| PCA | Principal component analysis |
| PCD | Primary ciliary dyskinesia |
| PCIN | Prolonged chemotherapy-induced neutropenia |
| PLS-DA | Partial least square discriminant analysis |
| POC | Point of care |
| PPMs | Potential pathogenic micro-organisms |
| SA | Staphylococcus aureus |
| TB | Mycobacterium tuberculosis |
| UC | Ulcerative colitis |
| VOCs | Volatile organic compounds |
| 6MWD | Six-minute walk test distance |
References
- Dragonieri, S.; Pennazza, G.; Carratu, P.; Resta, O. Electronic Nose Technology in Respiratory Diseases. Lung 2017, 195, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Gordon, S.M.; Szidon, J.P.; Krotoszynski, B.K.; Gibbons, R.D.; O’Neill, H.J. Volatile organic compounds in exhaled air from patients with lung cancer. Clin. Chem. 1985, 31, 1278–1282. [Google Scholar] [CrossRef] [PubMed]
- Behera, B.; Joshi, R.; Anil Vishnu, G.K.; Bhalerao, S.; Pandya, H.J. Electronic nose: A non-invasive technology for breath analysis of diabetes and lung cancer patients. J. Breath Res. 2019, 13, 024001. [Google Scholar] [CrossRef] [PubMed]
- Tiele, A.; Wicaksono, A.; Kansara, J.; Arasaradnam, R.P.; Covington, J.A. Breath Analysis Using eNose and Ion Mobility Technology to Diagnose Inflammatory Bowel Disease-A Pilot Study. Biosensors 2019, 9, 55. [Google Scholar] [CrossRef] [Green Version]
- Monasta, L.; Pierobon, C.; Princivalle, A.; Martelossi, S.; Marcuzzi, A.; Pasini, F.; Perbellini, L. Inflammatory bowel disease and patterns of volatile organic compounds in the exhaled breath of children: A case-control study using Ion Molecule Reaction-Mass Spectrometry. PLoS ONE 2017, 12, e0184118. [Google Scholar] [CrossRef]
- Arasaradnam, R.P.; McFarlane, M.; Daulton, E.; Skinner, J.; O’Connell, N.; Wurie, S.; Chambers, S.; Nwokolo, C.; Bardhan, K.; Savage, R.; et al. Non-invasive exhaled volatile organic biomarker analysis to detect inflammatory bowel disease (IBD). Dig. Liver Dis. 2016, 48, 148–153. [Google Scholar] [CrossRef] [Green Version]
- Gardner, J.W.; Bartlett, P.N. A brief history of electronic noses. Sens. Actuators B Chem. 1994, 18, 210–211. [Google Scholar] [CrossRef]
- Persaud, K.; Dodd, G. Analysis of discrimination mechanisms in the mammalian olfactory system using a model nose. Nature 1982, 299, 352–355. [Google Scholar] [CrossRef]
- van de Goor, R.; van Hooren, M.; Dingemans, A.M.; Kremer, B.; Kross, K. Training and Validating a Portable Electronic Nose for Lung Cancer Screening. J. Thorac. Oncol. 2018, 13, 676–681. [Google Scholar] [CrossRef] [Green Version]
- Santonico, M.; Pennazza, G.; Grasso, S.; D’Amico, A.; Bizzarri, M. Design and test of a Biosensor-based multisensorial system: A proof of concept study. Sensors 2013, 13, 16625–16640. [Google Scholar] [CrossRef]
- Rocco, R.; Incalzi, R.A.; Pennazza, G.; Santonico, M.; Pedone, C.; Bartoli, I.R.; Vernile, C.; Mangiameli, G.; Rocca, A.L.; De Luca, G.; et al. BIONOTE e-nose technology may reduce false positives in lung cancer screening programmes. Eur. J. Cardio Thorac. Surg. 2016, 49, 1112–1117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewis, N.S. Comparisons between mammalian and artificial olfaction based on arrays of carbon black-polymer composite vapor detectors. Acc. Chem. Res. 2004, 37, 663–672. [Google Scholar] [CrossRef] [PubMed]
- Di Natale, C.; Paolesse, R.; D’Amico, A. Metalloporphyrins based artificial olfactory receptors. Sens. Actuators B Chem. 2007, 121, 238–246. [Google Scholar] [CrossRef]
- Bos, L.D.J.; Van Walree, I.C.; Kolk, A.H.J.; Janssen, H.G.; Sterk, P.J.; Schultz, M.J. Alterations in exhaled breath metabolite-mixtures in two rat models of lipopolysaccharide-induced lung injury. J. Appl. Physiol. 2013, 115, 1487–1495. [Google Scholar] [CrossRef] [Green Version]
- Arasaradnam, R.P.; Mcfarlane, M.J.; Ryan-Fisher, C.; Westenbrink, E.; Hodges, P.; Thomas, M.G.; Chambers, S.; O’Connell, N.; Bailey, C.; Harmston, C.; et al. Detection of colorectal cancer (CRC) by urinary volatile organic compound analysis. PLoS ONE 2014, 9, e108750. [Google Scholar] [CrossRef]
- De Vries, R.; Muller, M.; Van Der Noort, V.; Theelen, W.S.M.E.; Schouten, R.D.; Hummelink, K.; Muller, S.H.; Wolf-Lansdorf, M.; Dagelet, J.W.F.; Monkhorst, K.; et al. Prediction of response to anti-PD-1 therapy in patients with non-small-cell lung cancer by electronic nose analysis of exhaled breath. Ann. Oncol. 2019, 30, 1660–1666. [Google Scholar] [CrossRef] [Green Version]
- International Agency for Research on Cancer (IARC). Latest Global Cancer Data, 2018; World Health Organization: Geneva, Switzerland, 2018. [Google Scholar]
- Machado, R.F.; Laskowski, D.; Deffenderfer, O.; Burch, T.; Zheng, S.; Mazzone, P.J.; Mekhail, T.; Jennings, C.; Stoller, J.K.; Pyle, J.; et al. Detection of lung cancer by sensor array analyses of exhaled breath. Am. J. Respir. Crit. Care Med. 2005, 171, 1286–1291. [Google Scholar] [CrossRef] [Green Version]
- Phillips, M.; Altorki, N.; Austin, J.H.M.; Cameron, R.B.; Cataneo, R.N.; Kloss, R.; Maxfield, R.A.; Munawar, M.I.; Pass, H.I.; Rashid, A.; et al. Detection of lung cancer using weighted digital analysis of breath biomarkers. Clin. Chim. Acta 2008, 393, 76–84. [Google Scholar] [CrossRef] [Green Version]
- D’Amico, A.; Pennazza, G.; Santonico, M.; Martinelli, E.; Roscioni, C.; Galluccio, G.; Paolesse, R.; Di Natale, C. An investigation on electronic nose diagnosis of lung cancer. Lung Cancer 2010, 68, 170–176. [Google Scholar] [CrossRef]
- Dragonieri, S.; Annema, J.T.; Schot, R.; van der Schee, M.P.C.; Spanevello, A.; Carratú, P.; Resta, O.; Rabe, K.F.; Sterk, P.J. An electronic nose in the discrimination of patients with non-small cell lung cancer and COPD. Lung Cancer 2009, 64, 166–170. [Google Scholar] [CrossRef]
- Mazzone, P.J.; Hammel, J.; Dweik, R.; Na, J.; Czich, C.; Laskowski, D.; Mekhail, T. Diagnosis of lung cancer by the analysis of exhaled breath with a colorimetric sensor array. Thorax 2007, 62, 565–568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hakim, M.; Billan, S.; Tisch, U.; Peng, G.; Dvrokind, I.; Marom, O.; Abdah-Bortnyak, R.; Kuten, A.; Haick, H. Diagnosis of head-and-neck cancer from exhaled breath. Br. J. Cancer 2011, 104, 1649–1655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Natale, C.; Macagnano, A.; Martinelli, E.; Paolesse, R.; D’Arcangelo, G.; Roscioni, C.; Finazzi-Agrò, A.; D’Amico, A. Lung cancer identification by the analysis of breath by means of an array of non-selective gas sensors. Biosens. Bioelectron. 2003, 18, 1209–1218. [Google Scholar] [CrossRef]
- Tran, V.H.; Chan, H.P.; Thurston, M.; Jackson, P.; Lewis, C.; Yates, D.; Bell, G.; Thomas, P.S. Breath analysis of lung cancer patients using an electronic nose detection system. IEEE Sens. J. 2010, 10, 1514–1518. [Google Scholar] [CrossRef]
- McWilliams, A.; Beigi, P.; Srinidhi, A.; Lam, S.; MacAulay, C.E. Sex and smoking status effects on the early detection of early lung cancer in high-risk smokers using an electronic nose. IEEE Trans. Biomed. Eng. 2015, 62, 2044–2054. [Google Scholar] [CrossRef] [PubMed]
- Dung, T.T.; Oh, Y.; Choi, S.J.; Kim, I.D.; Oh, M.K.; Kim, M. Applications and advances in bioelectronic noses for odour sensing. Sensors 2018, 18, 103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, A.D. Advances in electronic-nose technologies for the detection of volatile biomarker metabolites in the human breath. Metabolites 2015, 5, 140–163. [Google Scholar] [CrossRef] [PubMed]
- Gibson, T.D.; Prosser, O.; Hulbert, J.N.; Marshall, R.W.; Corcoran, P.; Lowery, P.; Ruck-Keene, E.A.; Heron, S. Detection and simultaneous identification of microorganisms from headspace samples using an electronic nose. Sens. Actuators B Chem. 1997, 44, 413–422. [Google Scholar] [CrossRef]
- Pavlou, A.K.; Magan, N.; Jones, J.M.; Brown, J.; Klatser, P.; Turner, A.P.F. Detection of Mycobacterium tuberculosis (TB) in vitro and in situ using an electronic nose in combination with a neural network system. Biosens. Bioelectron. 2004, 20, 538–544. [Google Scholar] [CrossRef]
- Bruins, M.; Rahim, Z.; Bos, A.; Van De Sande, W.W.J.; Endtz, H.P.; Van Belkum, A. Diagnosis of active tuberculosis by e-nose analysis of exhaled air. Tuberculosis 2013, 93, 232–238. [Google Scholar] [CrossRef]
- Thaler, E.R.; Hanson, C.W. Use of an electronic nose to diagnose bacterial sinusitis. Am. J. Rhinol. 2006, 20, 170–172. [Google Scholar] [CrossRef] [PubMed]
- Shafiek, H.; Fiorentino, F.; Merino, J.L.; López, C.; Oliver, A.; Segura, J.; De Paul, I.; Sibila, O.; Agustí, A.; Cosío, B.G. Using the electronic nose to identify airway infection during COPD exacerbations. PLoS ONE 2015, 10, e0135199. [Google Scholar] [CrossRef] [Green Version]
- Suarez-Cuartin, G.; Giner, J.; Merino, J.L.; Rodrigo-Troyano, A.; Feliu, A.; Perea, L.; Sanchez-Reus, F.; Castillo, D.; Plaza, V.; Chalmers, J.D.; et al. Identification of Pseudomonas aeruginosa and airway bacterial colonization by an electronic nose in bronchiectasis. Respir. Med. 2018, 136, 111–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robroeks, C.M.H.H.T.; Van Berkel, J.J.B.N.; Dallinga, J.W.; Jöbsis, Q.; Zimmermann, L.J.I.; Hendriks, H.J.E.; Wouters, M.F.M.; Van Der Grinten, C.P.M.; Van De Kant, K.D.G.; Van Schooten, F.J.; et al. Metabolomics of volatile organic compounds in cystic fibrosis patients and controls. Pediatr. Res. 2010, 68, 75–80. [Google Scholar] [CrossRef] [Green Version]
- Montuschi, P.; Santonico, M.; Mondino, C.; Pennazza, G.; Maritini, G.; Martinelli, E.; Capuano, R.; Ciabattoni, G.; Paolesse, R.; Di Natale, C.; et al. Diagnostic performance of an electronic nose, fractional exhaled nitric oxide, and lung function testing in asthma. Chest 2010, 137, 790–796. [Google Scholar] [CrossRef] [PubMed]
- American Cancer Society. Facts & Figures 2019. Am. Cancer Soc. 2019, 69, 1–71. [Google Scholar]
- Tirzïte, M.; Bukovskis, M.; Strazda, G.; Jurka, N.; Taivans, I. Detection of lung cancer with electronic nose and logistic regression analysis. J. Breath Res. 2019, 13, 016006. [Google Scholar] [CrossRef] [Green Version]
- Brereton, R.G.; Lloyd, G.R. Partial least squares discriminant analysis: Taking the magic away. J. Chemom. 2014, 28, 213–225. [Google Scholar] [CrossRef]
- Lamote, K.; Brinkman, P.; Vandermeersch, L.; Vynck, M.; Sterk, P.J.; Van Langenhove, H.; Thas, O.; Van Cleemput, J.; Nackaerts, K.; van Meerbeeck, J.P. Breath analysis by gas chromatography-mass spectrometry and electronic nose to screen for pleural mesothelioma: A crosssectional case-control study. Oncotarget 2017, 8, 91593–91602. [Google Scholar] [CrossRef] [Green Version]
- Dragonieri, S.; Schot, R.; Mertens, B.J.A.; Le Cessie, S.; Gauw, S.A.; Spanevello, A.; Resta, O.; Willard, N.P.; Vink, T.J.; Rabe, K.F.; et al. An electronic nose in the discrimination of patients with asthma and controls. J. Allergy Clin. Immunol. 2007, 120, 856–862. [Google Scholar] [CrossRef] [Green Version]
- Plaza, V.; Crespo, A.; Giner, J.; Merino, J.L.; Ramos-Barbón, D.; Mateus, E.F.; Torrego, A.; Cosio, B.G.; Agustí, A.; Sibila, O. Inflammatory asthma phenotype discrimination using an electronic nose breath analyzer. J. Investig. Allergol. Clin. Immunol. 2015, 25, 431–437. [Google Scholar]
- Ibrahim, B.; Basanta, M.; Cadden, P.; Singh, D.; Douce, D.; Woodcock, A.; Fowler, S.J. Non-invasive phenotyping using exhaled volatile organic compounds in asthma. Thorax 2011, 66, 804–809. [Google Scholar] [CrossRef] [Green Version]
- Wagener, A.; Brinkman, P.; Zwinderman, A.; D’Amico, A.; Pennazza, G.S.M. Exhaled Breath Profiling and Eosinophilic Airway Inflammation in Asthma—Results of A Pilot Study. Am. J. Respir. Crit. Care Med. 2013, 201, A2392. [Google Scholar]
- Van der Schee, M.P.; Palmay, R.; Cowan, J.O.; Taylor, D.R. Predicting steroid responsiveness in patients with asthma using exhaled breath profiling. Clin. Exp. Allergy 2013, 43, 1217–1225. [Google Scholar] [CrossRef]
- Brinkman, P.; Wagener, A.H.; Hekking, P.P.; Bansal, A.T.; van der Zee, A.H.M.; Wang, Y.; Weda, H.; Knobel, H.H.; Vink, T.J.; Rattray, N.J.; et al. Identification and prospective stability of electronic nose (eNose)–derived inflammatory phenotypes in patients with severe asthma. J. Allergy Clin. Immunol. 2019, 143, 1811–1820. [Google Scholar] [CrossRef] [Green Version]
- Tenero, L.; Sandri, M.; Piazza, M.; Paiola, G.; Zaffanello, M.; Piacentini, G. Electronic nose in discrimination of children with uncontrolled asthma. J. Breath Res. 2020, 14, 46003. [Google Scholar] [CrossRef]
- Bannier, M.A.G.E.; Van De Kant, K.D.G.; Jöbsis, Q.; Dompeling, E. Feasibility and diagnostic accuracy of an electronic nose in children with asthma and cystic fibrosis. J. Breath Res. 2019, 13, 036009. [Google Scholar] [CrossRef]
- Global Initiative for Asthma. GINA Report: Global Strategy for Asthma Management and Prevention 2019; Global Initiative for Asthma: Fontana, WI, USA, 2019. [Google Scholar]
- Fens, N.; Roldaan, A.C.; van der Schee, M.P.; Boksem, R.J.; Zwinderman, A.H.; Bel, E.H.; Sterk, P.J. External validation of exhaled breath profiling using an electronic nose in the discrimination of asthma with fixed airways obstruction and chronic obstructive pulmonary disease. Clin. Exp. Allergy 2011, 41, 1371–1378. [Google Scholar] [CrossRef]
- Brinkman, P.; van de Pol, M.A.; Gerritsen, M.G.; Bos, L.D.; Dekker, T.; Smids, B.S.; Sinha, A.; Majoor, C.J.; Sneeboer, M.M.; Knobel, H.H.; et al. Exhaled breath profiles in the monitoring of loss of control and clinical recovery in asthma. Clin. Exp. Allergy 2017, 47, 1159. [Google Scholar] [CrossRef]
- Papi, A.; Bellettato, C.M.; Braccioni, F.; Romagnoli, M.; Casolari, P.; Caramori, G.; Fabbri, L.M.; Johnston, S.L. Infections and airway inflammation in chronic obstructive pulmonary disease severe exacerbations. Am. J. Respir. Crit. Care Med. 2006, 173, 1114–1121. [Google Scholar] [CrossRef]
- Smith, C.B.; Golden, C.; Klauber, M.R.; Kanner, R.; Renzetti, A. Interactions between viruses and bacteria in patients with chronic bronchitis. J. Infect. Dis. 1976, 134, 552–561. [Google Scholar] [CrossRef] [PubMed]
- Donaldson, G.C.; Seemungal, T.A.R.; Bhowmik, A.; Wedzicha, J.A. Relationship between exacerbation frequency and lung function decline in chronic obstructive pulmonary disease. Thorax 2002, 57, 847–852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spencer, S.; Jones, P.W. Time course of recovery of health status following an infective exacerbation of chronic bronchitis. Thorax 2003, 58, 589–593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monsó, E.; Ruiz, J.; Rosell, A.; Manterola, J.; Fiz, J.; Morera, J.; Ausina, V. Bacterial infection in chronic obstructive pulmonary disease: A study of stable and exacerbated outpatients using the protected specimen brush. Am. J. Respir. Crit. Care Med. 1995, 152, 1316–1320. [Google Scholar] [CrossRef]
- Fagon, J.Y.; Chastre, J.; Trouillet, J.L.; Domart, Y.; Dombret, M.C.; Bornet, M.; Gibert, C. Characterization of distal bronchial microflora during acute exacerbation of chronic bronchitis: Use of the protected specimen brush technique in 54 mechanically ventilated patients. Am. Rev. Respir. Dis. 1990, 142, 1004–1008. [Google Scholar] [CrossRef]
- Finamore, P.; Pedone, C.; Scarlata, S.; Di Paolo, A.; Grasso, S.; Santonico, M.; Pennazza, G.; Incalzi, R.A. Validation of exhaled volatile organic compounds analysis using electronic nose as index of COPD severity. Int. J. COPD 2018, 13, 1441–1448. [Google Scholar] [CrossRef] [Green Version]
- van Velzen, P.; Brinkman, P.; Knobel, H.H.; van den Berg, J.W.K.; Jonkers, R.E.; Loijmans, R.J.; Prins, J.M.; Sterk, P.J. Exhaled Breath Profiles Before, During and After Exacerbation of COPD: A Prospective Follow-Up Study. COPD J. Chronic Obstr. Pulm. Dis. 2019, 16, 330–337. [Google Scholar] [CrossRef] [Green Version]
- Cohen-Cymberknoh, M.; Simanovsky, N.; Hiller, N.; Gileles Hillel, A.; Shoseyov, D.; Kerem, E. Differences in disease expression between primary ciliary dyskinesia and cystic fibrosis with and without pancreatic insufficiency. Chest 2014, 145, 738–744. [Google Scholar] [CrossRef]
- Ratjen, F.; Waters, V.; Klingel, M.; McDonald, N.; Dell, S.; Leahy, T.R.; Yau, Y.; Grasemann, H. Changes in airway inflammation during pulmonary exacerbations in patients with cystic fibrosis and primary ciliary dyskinesia. Eur. Respir. J. 2016, 47, 829–836. [Google Scholar] [CrossRef] [Green Version]
- Butterfield, R. Primary ciliary dyskinesia. Pediatr. Rev. 2017, 38, 145–146. [Google Scholar] [CrossRef]
- Walker, W.T.; Jackson, C.L.; Lackie, P.M.; Hogg, C.; Lucas, J.S. Nitric oxide in primary ciliary dyskinesia. Eur. Respir. J. 2012, 40, 1024–1032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paff, T.; van der Schee, M.P.; Daniels, J.M.A.; Pals, G.; Postmus, P.E.; Sterk, P.J.; Haarman, E.G. Exhaled molecular profiles in the assessment of cystic fibrosis and primary ciliary dyskinesia. J. Cyst. Fibros. 2013, 12, 454–460. [Google Scholar] [CrossRef] [Green Version]
- Bush, A.; Payne, D.; Pike, S.; Jenkins, G.; Henke, M.O.; Rubin, B.K. Mucus properties in children with primary ciliary dyskinesia: Comparison with cystic fibrosis. Chest 2006, 129, 118–123. [Google Scholar] [CrossRef] [PubMed]
- MacKerness, K.J.; Jose, P.J.; Bush, A. Differences in airway inflammation in cystic fibrosis and primary ciliary dyskinesia. Pediatr. Asthma Allergy Immunol. 2009, 22, 163–168. [Google Scholar] [CrossRef]
- Tramper-Stranders, G.A.; van der Ent, C.K.; Wolfs, T.F.W. Detection of Pseudomonas aeruginosa in patients with cystic fibrosis. J. Cyst. Fibros. 2005, 4, 37–43. [Google Scholar] [CrossRef] [Green Version]
- Middleton, P.G.; Mall, M.A.; Dřevínek, P.; Lands, L.C.; McKone, E.F.; Polineni, D.; Ramsey, B.W.; Taylor-Cousar, J.L.; Tullis, E.; Vermeulen, F.; et al. Elexacaftor–Tezacaftor–Ivacaftor for Cystic Fibrosis with a Single Phe508del Allele. N. Engl. J. Med. 2019, 381, 1809–1819. [Google Scholar] [CrossRef] [PubMed]
- De Heer, K.; Van Der Schee, M.P.; Zwinderman, K.; Van Den Berk, I.A.H.; Visser, C.E.; Van Oers, R.; Sterk, P.J. Electronic nose technology for detection of invasive pulmonary aspergillosis in prolonged chemotherapy-induced neutropenia: A proof-of-principle study. J. Clin. Microbiol. 2013, 51, 1490–1495. [Google Scholar] [CrossRef] [Green Version]
- De Heer, K.; Kok, M.G.M.; Fens, N.; Weersink, E.J.M.; Zwinderman, A.H.; Van Der Schee, M.P.C.; Visser, C.E.; Van Oers, M.H.J.; Sterk, P.J. Detection of Airway Colonization by Aspergillus fumigatus by Use of Electronic Nose Technology in Patients with Cystic Fibrosis. J. Clin. Microbiol. 2016, 54, 569–575. [Google Scholar] [CrossRef] [Green Version]
- Chambers, S.T.; Syhre, M.; Murdoch, D.R.; McCartin, F.; Epton, M.J. Detection of 2-Pentylfuran in the breath of patients with Aspergillus fumigatus. Med. Mycol. 2009, 47, 468–476. [Google Scholar] [CrossRef] [Green Version]
- Pamukcu, A.; Bush, A.; Buchdahl, R. Effects of Pseudomonas aeruginosa colonization on lung function and anthropometric variables in children with cystic fibrosis. Pediatr. Pulmonol. 1995, 19, 10–15. [Google Scholar] [CrossRef]
- Joensen, O.; Paff, T.; Haarman, E.G.; Skovgaard, I.M.; Jensen, P.; Bjarnsholt, T.; Nielsen, K.G. Exhaled breath analysis using electronic nose in cystic fibrosis and primary ciliary dyskinesia patients with chronic pulmonary infections. PLoS ONE 2014, 9, e115584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nasir, M.; Bean, H.D.; Smolinska, A.; Rees, C.A.; Zemanick, E.T.; Hill, J.E. Volatile molecules from bronchoalveolar lavage fluid can “rule-in” Pseudomonas aeruginosa and “rule-out” Staphylococcus aureus infections in cystic fibrosis patients. Sci. Rep. 2018, 8, 826. [Google Scholar] [CrossRef] [PubMed]
- Ringshausen, F.C.; De Roux, A.; Diel, R.; Hohmann, D.; Welte, T.; Rademacher, J. Bronchiectasis in Germany: A population-based estimation of disease prevalence. Eur. Respir. J. 2015, 46, 1805–1807. [Google Scholar] [CrossRef] [Green Version]
- Seitz, A.E.; Olivier, K.N.; Steiner, C.A.; De Oca, R.M.; Holland, S.M.; Prevots, D.R. Trends and burden of bronchiectasis-associated hospitalizations in the United States, 1993–2006. Chest 2010, 138, 944–949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pasteur, M.C.; Bilton, D.; Hill, A.T. British thoracic society guideline for non-CF bronchiectasis. Thorax 2010, 65, 1–58. [Google Scholar] [CrossRef] [Green Version]
- Finch, S.; McDonnell, M.J.; Abo-Leyah, H.; Aliberti, S.; Chalmers, J.D. A comprehensive analysis of the impact of pseudomonas aeruginosa colonization on prognosis in adult bronchiectasis. Ann. Am. Thorac. Soc. 2015, 12, 1602–1611. [Google Scholar]
- Krauss, E.; Haberer, J.; Maurer, O.; Barreto, G.; Drakopanagiotakis, F.; Degen, M.; Seeger, W.; Guenther, A. Exploring the Ability of Electronic Nose Technology to Recognize Interstitial Lung Diseases (ILD) by Non-Invasive Breath Screening of Exhaled Volatile Compounds (VOC): A Pilot Study from the European IPF Registry (eurIPFreg) and Biobank. J. Clin. Med. 2019, 8, 1698. [Google Scholar] [CrossRef]
- Moor, C.C.; Oppenheimer, J.C.; Nakshbandi, G.; Aerts, J.G.J.V.; Brinkman, P.; van der Zee, A.-H.M.; Wijsenbeek, M.S. Exhaled breath analysis by use of eNose technology: A novel diagnostic tool for interstitial lung disease. Eur. Respir. J. 2020, 56, 2002042. [Google Scholar] [CrossRef]
- Peters, Y.; Schrauwen, R.W.M.; Tan, A.C.; Bogers, S.K.; De Jong, B.; Siersema, P.D. Detection of Barrett’s oesophagus through exhaled breath using an electronic nose device. Gut 2020, 69, 1169–1172. [Google Scholar] [CrossRef]
- van Keulen, K.E.; Jansen, M.E.; Schrauwen, R.W.M.; Kolkman, J.J.; Siersema, P.D. Volatile organic compounds in breath can serve as a non-invasive diagnostic biomarker for the detection of advanced adenomas and colorectal cancer. Aliment. Pharmacol. Ther. 2020, 51, 334–346. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-Aguilar, M.; Ramírez-García, S.; Ilizaliturri-Hernández, C.; Gómez-Gómez, A.; Van-Brussel, E.; Díaz-Barriga, F.; Medellín-Garibay, S.; Flores-Ramírez, R. Ultrafast gas chromatography coupled to electronic nose to identify volatile biomarkers in exhaled breath from chronic obstructive pulmonary disease patients: A pilot study. Biomed. Chromatogr. 2019, 33, 4684. [Google Scholar] [CrossRef] [PubMed]
- Wingelaar, T.T.; Brinkman, P.; de Vries, R.; van Ooij, P.J.A.M.; Hoencamp, R.; Maitland-Van der Zee, A.H.; Hollmann, M.W.; van Hulst, R.A. Detecting pulmonary oxygen toxicity using enose technology and associations between electronic nose and gas chromatography–mass spectrometry data. Metabolites 2019, 9, 286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neerincx, A.H.; Geurts, B.P.; Van Loon, J.; Tiemes, V.; Jansen, J.J.; Harren, F.J.M.; Kluijtmans, L.A.J.; Merkus, P.J.F.M.; Cristescu, S.M.; Buydens, L.M.C.; et al. Detection of Staphylococcus aureus in cystic fibrosis patients using breath VOC profiles. J. Breath Res. 2016, 10, 046014. [Google Scholar] [CrossRef]
- Bos, L.D.J.; Sterk, P.J.; Schultz, M.J. Volatile Metabolites of Pathogens: A Systematic Review. PLoS Pathog. 2013, 9, e1003311. [Google Scholar] [CrossRef] [Green Version]
- Filipiak, W.; Sponring, A.; Baur, M.M.; Filipiak, A.; Ager, C.; Wiesenhofer, H.; Nagl, M.; Troppmair, J.; Amann, A. Molecular analysis of volatile metabolites released specifically by staphylococcus aureus and pseudomonas aeruginosa. BMC Microbiol. 2012, 12, 113. [Google Scholar] [CrossRef] [Green Version]
- Koo, S.; Thomas, H.R.; Daniels, S.D.; Lynch, R.C.; Fortier, S.M.; Shea, M.M.; Rearden, P.; Comolli, J.C.; Baden, L.R.; Marty, F.M. A breath fungal secondary metabolite signature to diagnose invasive aspergillosis. Clin. Infect. Dis. 2014, 59, 1733–1740. [Google Scholar] [CrossRef] [Green Version]
- Van Der Schee, M.P.; Paff, T.; Brinkman, P.; Van Aalderen, W.M.C.; Haarman, E.G.; Sterk, P.J. Breathomics in lung disease. Chest 2015, 147, 224–231. [Google Scholar] [CrossRef]
| Publication [Ref.] | Disease | Number of Patients/Total Study Participants | Type of VOC Detection Device | Main Findings |
|---|---|---|---|---|
| Tirzīte et al. [38] | Lung cancer | 252/475 | Cyranose 320 | High-risk controls vs. cancer Sensitivity: 96%, specificity: 92% Non-smokers vs. cancer Sensitivity: 96%, specificity: 91% |
| Van de Goor et al. [9] | Lung cancer | 52/144 | Aeonose | High-risk controls vs. cancer Sensitivity: 83%, specificity: 84% |
| McWilliams et al. [26] | Lung cancer | 25/191 | Cyranose 320 | High-risk controls vs. cancer Sensitivity: 81.3%, specificity: 88% |
| Rocco et al. [11] | Lung cancer | 23/100 | BIONOTE eNose | High-risk controls vs. cancer Sensitivity: 86%, specificity: 95% |
| Lamote et al. [40] | Lung cancer (malignant pleural mesothelioma, [MPM]) | 35/64 (19 asymptomatic asbestos-exposed subjects [AEx] + 16 control, 14 MPM, 15 benign disease [ARD]) | Common Invent, Owlstone Lonestar, Cyranose 320, Tor Vergata eNoses + GC-MS | AEx subjects vs. MPM eNose: 97% accuracy GC-MS: 97% accuracy MPM vs. AEx + ARD eNose: 74% accuracy GC-MS: 94% |
| De Vries et al. [16] | Lung cancer | 143/143 | SpiroNose | Responders vs. Non-responders to anti-PD-1 therapy Sensitivity: 81%, specificity: 50% |
| Dragonieri et al. [41] | Asthma | 20/40 | Cyranose 320 | Mild asthma vs. young controls Cross-validation: 100% Severe asthma vs. old controls Cross-validation: 90% |
| Plaza et al. [42] | Asthma | 52/52 | Cyranose 320 | Eosinophilic vs. neutrophilic Accuracy: 73%, AUROC: 0.92 Eosinophilic vs. paucigranulocytic Accuracy: 74%, AUROC: 0.79 Neutrophilic vs. paucigranulocytic Accuracy: 89%, AUROC: 0.88 |
| Brinkman et al. [46] | Asthma | 78/78 | Common Invent, Owlstone Lonestar, Cyranose 320, Tor Vergata eNoses | Inflammatory phenotypes in severe asthma Three distinct clusters (n = 26, n = 33, n = 19) |
| Van der Schee et al. [45] | Asthma | 25/45 | Cyranose 320 | Asthma vs. controls Sensitivity: 80%, specificity: 65% |
| Tenero et al. [47] | Asthma | 28/38 | Cyranose 320 | Non-symptomatic asthma (control + controlled asthma) vs. symptomatic asthma (partially controlled + uncontrolled asthma) Sensitivity: 0.79, specificity: 0.84 |
| Montuschi et al. [36] | Asthma | 27/51 | Tor Vergata eNose + GC-MS | Asthma vs. controls eNose: 87.5% accuracy GC-MS: “significantly different” |
| Brinkman et al. [51] | Asthma | 23/23 | Common Invent, Owlstone Lonestar, Cyranose 320, Tor Vergata eNoses + GC-MS | eNose: baseline vs. loss of control: 95% accuracy loss of control vs. recovery: 86% GC-MS: baseline vs. loss of control: 68% accuracy loss of control vs. recovery: 77% |
| Bannier et al. [48] | Asthma & CF | 33/55 (20 asthma, 13 CF) | Aeonose | Asthma vs. CF Sensitivity: 0.89, specificity: 0.77 CF vs. controls Sensitivity: 0.85, specificity: 0.77 Asthma vs. controls Sensitivity: 0.84, specificity: 0.91 |
| Fens et al. [50] | Asthma & COPD | 60 asthma, 40 COPD | Cyranose 320 | COPD vs. fixed asthma Sensitivity: 85%, specificity: 90% COPD vs. classical asthma Sensitivity: 91%, specificity: 90% |
| Shafiek et al. [33] | COPD | 143/173 (90 Exacerbated COPD [ECOPD], 50 stable COPD [SCOPD]) | Cyranose 320 | SCOPD vs. controls Sensitivity: 72%, specificity: 70% ECOPD vs. controls Sensitivity: 66%, specificity: 80% ECOPD vs. SCOPD Sensitivity: 89%, specificity: 48% |
| Finnamore et al. [58] | COPD | 63/63 | BIONOTE eNose | BODE functional status predicted via eNose Sensitivity: 0.71, specificity: 0.93 |
| Van Velzen et al. [59] | COPD | 31/68 (31 ECOPD, 37 COPD) | Common Invent, Owlstone Lonestar, Cyranose 320, Tor Vergata eNoses + GC-MS | ECOPD vs. COPD eNose: Accuracy: 75% GC-MS: Accuracy: 71% |
| Paff et al. [64] | CF & PCD | 50/73 (25 CF, 25 PCD) | Cyranose 320 | CF vs. controls Sensitivity: 84%, specificity: 65% PCD vs. controls Sensitivity: 88%, specificity: 52% CF vs. PCD Sensitivity: 84%, specificity: 60% |
| Joensen et al. [73] | CF & PCD | 85/106 (64 CF, 21 PCD) | Cyranose 320 | CF with P. aeruginosa (PA) vs. CF without PA Sensitivity: 71.4%, specificity: 63.3% No sig. difference between: CF with non-PA infection vs. CF without infection & PCD with PA/other infection vs. PCD without infection |
| De Heer et al. [70] | CF | 27/27 | Cyranose 320 | CF with (n = 9) and without (n = 18) A. Fumigatus Sensitivity: 78%, specificity: 94% |
| Suarez-Cuartin et al. [34] | Bronchiecta-sis | 73/73 | Cyranose 320 | Bronchiectasis with PA vs. Bronchiectasis without PA Sensitivity: 92%, specificity: 85% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Licht, J.-C.; Grasemann, H. Potential of the Electronic Nose for the Detection of Respiratory Diseases with and without Infection. Int. J. Mol. Sci. 2020, 21, 9416. https://doi.org/10.3390/ijms21249416
Licht J-C, Grasemann H. Potential of the Electronic Nose for the Detection of Respiratory Diseases with and without Infection. International Journal of Molecular Sciences. 2020; 21(24):9416. https://doi.org/10.3390/ijms21249416
Chicago/Turabian StyleLicht, Johann-Christoph, and Hartmut Grasemann. 2020. "Potential of the Electronic Nose for the Detection of Respiratory Diseases with and without Infection" International Journal of Molecular Sciences 21, no. 24: 9416. https://doi.org/10.3390/ijms21249416




