Putative Role of Protein Palmitoylation in Cardiac Lipid-Induced Insulin Resistance
Abstract
1. Introduction
2. Cardiac Insulin Signaling and Lipid-Induced Insulin Resistance
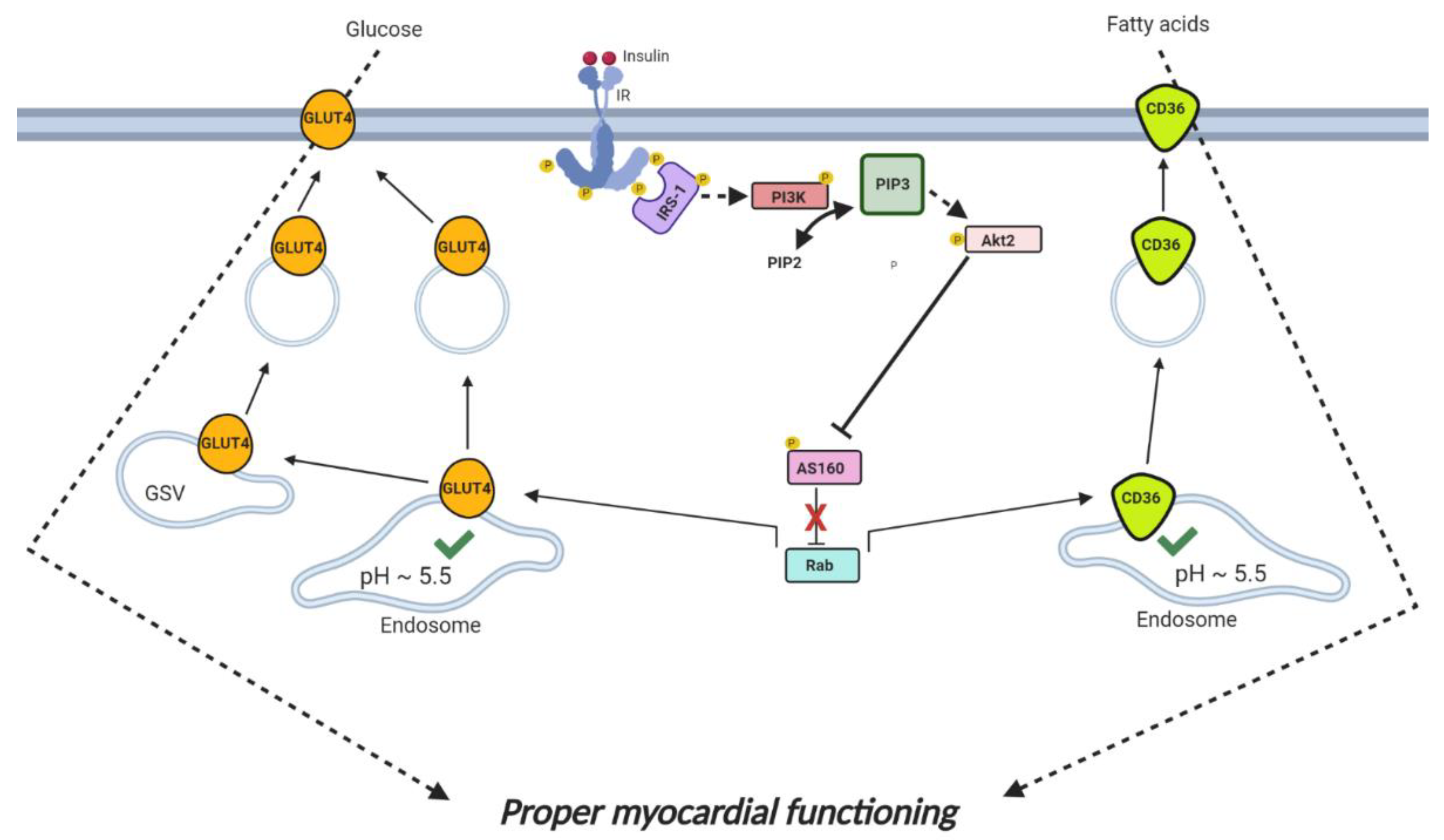
2.1. Cardiac Insulin Signaling
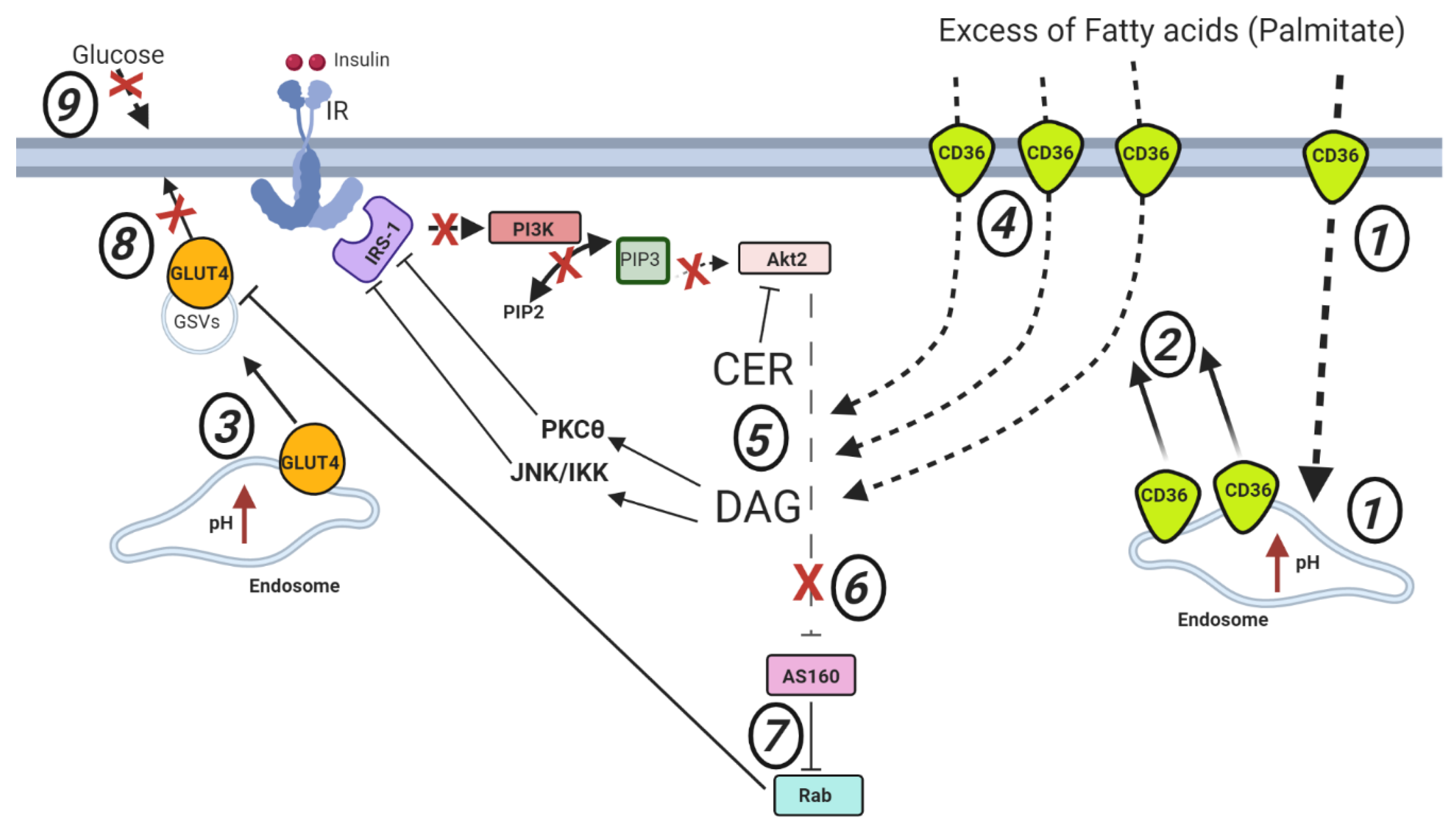
2.2. Cardiac Lipid-Induced Insulin Resistance
3. Palmitoylation and Its Enzymatic Regulation
3.1. Palmitoylation Function
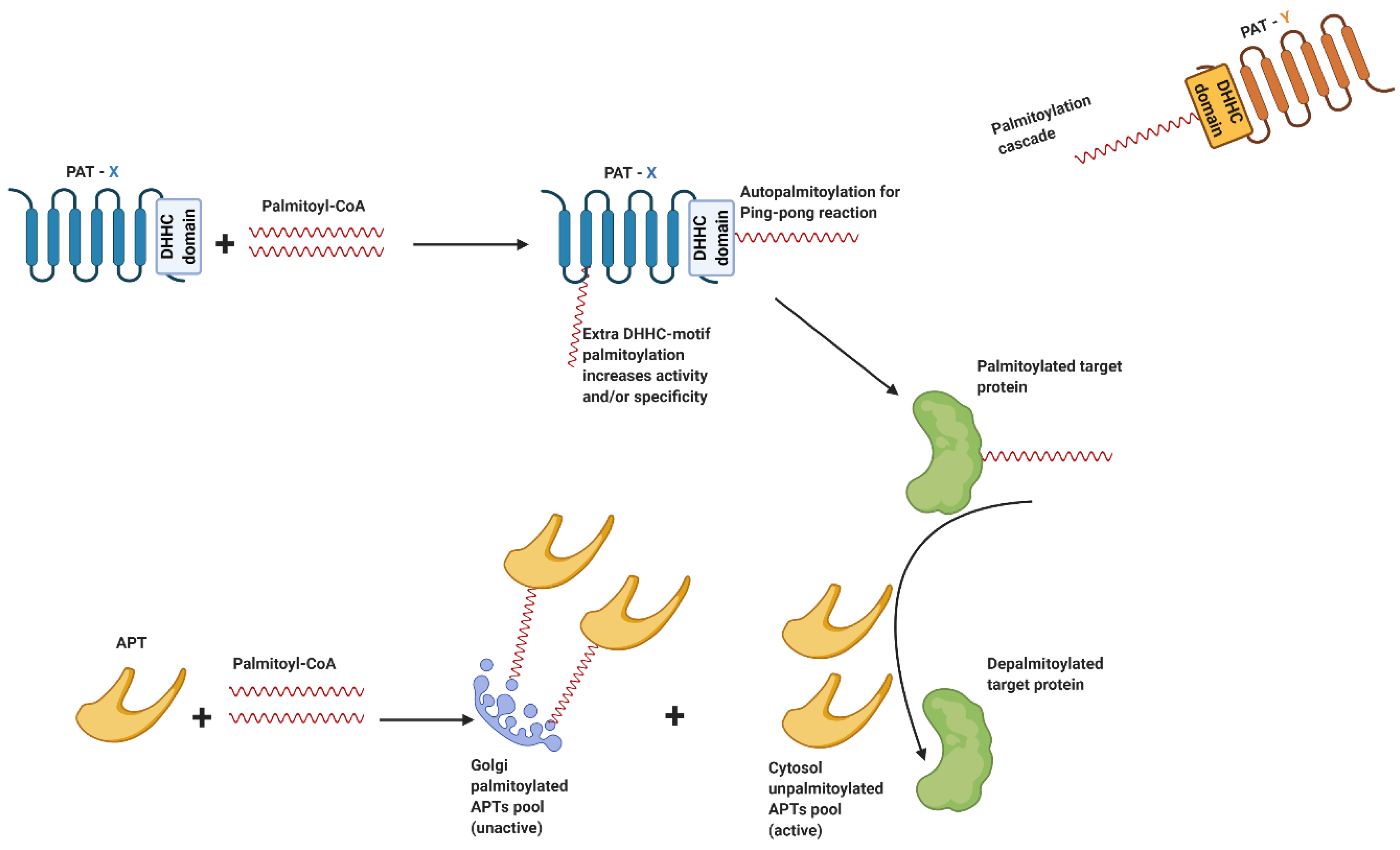
3.2. Palmitoylation Enzymatic Machinery
3.3. Regulation of PATs and APTs
4. Role of Palmitoylation of Signaling and Trafficking Proteins in Insulin-Stimulated Substrate Transporter Translocation
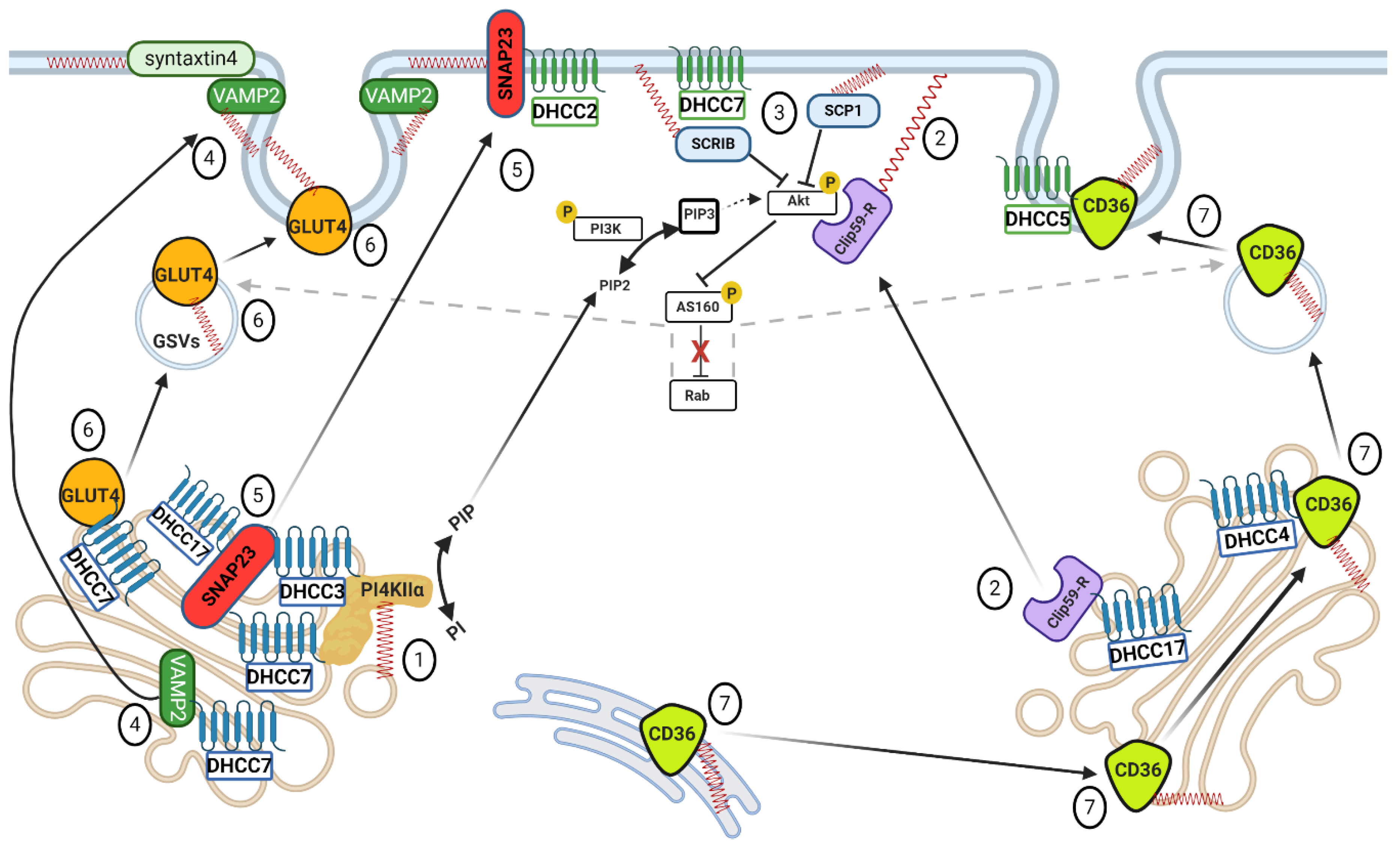
4.1. Caveolins
4.2. Phosphatidylinositol 4-Kinase II-α
4.3. ClipR-59
4.4. Tumor Suppressor SCRIB and Phosphatase SCP1
4.5. SNARE Proteins
4.6. GLUT4
4.7. CD36
4.8. Corollary for Insulin-Stimulated Substrate Transporter Translocation
5. Aberrant Protein Palmitoylation Driving Insulin Resistance
5.1. PKCε
5.2. GAPDH
5.3. GLUT4
5.4. CD36
5.5. Corollary for the Role of Palmitoylation in the Development of Insulin Resistance
6. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| CD36 (SR-B2) | Fatty acid transporter (scavenger receptor B2) |
| GLUT4 | Glucose transporter |
| GLUT1 | Glucose transporter |
| Cav-1 | Caveolin-1 |
| Cav-2 | Caveolin-2 |
| Cav-3 | Caveolin 3 |
| IR | Insulin receptor |
| IRS-1 | Insulin receptor substrate-1 |
| PI3K | Phosphatidylinositol 3-kinase |
| Akt2 | RAC-beta serine/threonine–protein kinase |
| Rab | Ras-associated binding proteins |
| Rab GAP AS160 | Rab GTPase-activating protein AS160 |
| PI4P | Phosphatidylinositol-4-phosphate |
| PIP2 | Phosphatidylinositol-4, 5-bisphosphate |
| PIP3 | Phosphatidylinositol-3,4,5-trisphosphate |
| PI4KIIα | Phosphatidylinositol 4-kinase II-alpha |
| PI4KIIβ | Phosphatidylinositol 4-kinase II-beta |
| VAMP | Vesicle-associated membrane protein |
| v-SNAREs | Vesicle-associated soluble N-ethylmaleimide-sensitive factor attachment protein receptors |
| t-SNAREs | Target membrane-associated soluble N-ethylmaleimide-sensitive factor attachment protein receptors |
| DAG | Diacylglycerol |
| CER | Ceramide |
| JNK | c-Junk N-terminal Kinase |
| IKK | IκB kinase |
| (n)PKC | (novel) protein kinase-C |
| PKCε | protein kinase-C epsilon |
| PKCδ | protein kinase-C delta |
| PKCθ | protein kinase-C theta |
| DHHC | Asp-His-His-Cys cysteine-rich domain |
| PAT | Palmitoyl-acyltransferase |
| APT | Acyl-protein thioesterase |
| PPT1 | Palmitoyl protein thioesterase 1 |
| Csk | C-terminal Src kinase |
| EGF | Epidermal growth factor |
| PDGF | Platelet-derived growth factor |
| ClipR-59 | Cytoplasmic linker protein 170-related 59 kDa protein |
| SCRIB | Scribble |
| SCP1 | Synaptonemal complex protein 1 |
| IRAP | Insulin responsive aminopeptidase |
| SNAP23 | Synaptosomal-associated protein 23 |
References
- Ouwens, D.M.; Diamant, M.; Fodor, M.; Habets, D.D.J.; Pelsers, M.M.A.L.; Hasnaoui, M.E.; Dang, Z.C.; Vlasblom, R.; Rietdijk, A.; Boer, C.; et al. Cardiac contractile dysfunction in insulin-resistant rats fed a high-fat diet is associated with elevated CD36-mediated fatty acid uptake and esterification. Diabetologia 2007, 1938–1948. [Google Scholar] [CrossRef] [PubMed]
- Coort, S.L.M.; Bonen, A.; van der Vusse, G.J.; Glatz, J.F.C.; Luiken, J.J.F.P. Cardiac substrate uptake and metabolism in obesity and type-2 diabetes: Role of sarcolemmal substrate transporters. Mol. Cell. Biochem. 2007, 299, 5–18. [Google Scholar] [CrossRef] [PubMed]
- Holloway, G.P.; Benton, C.R.; Mullen, K.L.; Yoshida, Y.; Snook, L.A.; Han, X.X.; Glatz, J.F.C.; Luiken, J.J.F.P.; Lally, J.; Dyck, D.J.; et al. In obese rat muscle transport of palmitate is increased and is channeled to triacylglycerol storage despite an increase in mitochondrial palmitate oxidation. Am. J. Physiol. Endocrinol. Metab. 2009, 296, 738–747. [Google Scholar] [CrossRef] [PubMed]
- Steinbusch, L.K.M.; Schwenk, R.W.; Ouwens, D.M.; Diamant, M.; Glatz, J.F.C.; Luiken, J.J.F.P. Subcellular trafficking of the substrate transporters GLUT4 and CD36 in cardiomyocytes. Cell. Mol. Life Sci. 2011, 68, 2525–2538. [Google Scholar] [CrossRef]
- Martin, B.R.; Wang, C.; Adibekian, A.; Tully, S.E.; Cravatt, B.F. Global profiling of dynamic protein palmitoylation. Nat. Methods 2012, 9, 84–89. [Google Scholar] [CrossRef]
- Cordeddu, V.; Di Schiavi, E.; Pennacchio, L.A.; Ma’ayan, A.; Sarkozy, A.; Fodale, V.; Cecchetti, S.; Cardinale, A.; Martin, J.; Schackwitz, W.; et al. Mutation of SHOC2 promotes aberrant protein N-myristoylation and causes Noonan-like syndrome with loose anagen hair. Nat. Genet. 2009, 41, 1022–1026. [Google Scholar] [CrossRef]
- Palsuledesai, C.C.; Distefano, M.D. Protein Prenylation: Enzymes, Therapeutics, and Biotechnology Applications. ACS Chem. Biol. 2015, 10, 51–62. [Google Scholar] [CrossRef]
- Udenwobele, D.I.; Su, R.C.; Good, S.V.; Ball, T.B.; Shrivastav, S.V.; Shrivastav, A. Myristoylation: An important protein modification in the immune response. Front. Immunol. 2017, 8, 1–16. [Google Scholar] [CrossRef]
- Montersino, A.; Thomas, G.M. Slippery signaling: Palmitoylation-dependent control of neuronal kinase localization and activity. Mol. Membr. Biol. 2015, 32, 179–188. [Google Scholar] [CrossRef][Green Version]
- Cho, E.; Park, M. Palmitoylation in Alzheimer’s disease and other neurodegenerative diseases. Pharmacol. Res. 2016, 111, 133–151. [Google Scholar] [CrossRef]
- Spinelli, M.; Fusco, S.; Grassi, C. Nutrient-dependent changes of protein palmitoylation: Impact on nuclear enzymes and regulation of gene expression. Int. J. Mol. Sci. 2018, 19, 3820. [Google Scholar] [CrossRef] [PubMed]
- Chamberlain, L.H.; Shipston, M.J. The physiology of protein s-acylation. Physiol. Rev. 2015, 95, 341–376. [Google Scholar] [CrossRef] [PubMed]
- Smotrys, J.E.; Linder, M.E. Palmitoylation of Intracellular Signaling Proteins: Regulation and function. Annu. Rev. Biochem. 2004, 73, 559–587. [Google Scholar] [CrossRef] [PubMed]
- Zaballa, M.E.; van der Goot, F.G. The molecular era of protein S-acylation: Spotlight on structure, mechanisms, and dynamics. Crit. Rev. Biochem. Mol. Biol. 2018, 53, 420–451. [Google Scholar] [CrossRef]
- Linder, M.E.; Deschenes, R.J. Palmitoylation: Policing protein stability and traffic. Nat. Rev. Mol. Cell Biol. 2007, 8, 74–84. [Google Scholar] [CrossRef]
- Bertrand, L.; Horman, S.; Beauloye, C.; Vanoverschelde, J.L. Insulin signalling in the heart. Cardiovasc. Res. 2008, 79, 238–248. [Google Scholar] [CrossRef]
- Yamamoto, M.; Toya, Y.; Schwencke, C.; Lisanti, M.P.; Myers, M.G.; Ishikawa, Y. Caveolin is an activator of insulin receptor signaling. J. Biol. Chem. 1998, 273, 26962–26968. [Google Scholar] [CrossRef]
- Krishna, A.; Sengupta, D. Interplay between Membrane Curvature and Cholesterol: Role of Palmitoylated Caveolin-1. Biophys. J. 2019, 116, 69–78. [Google Scholar] [CrossRef]
- Way, M.; Parton, R.G. M-caveolin, a muscle-specific caveolin-related protein. FEBS Lett. 1995, 376, 108–112. [Google Scholar] [CrossRef]
- Talukder, M.A.H.; Preda, M.; Ryzhova, L.; Prudovsky, I.; Pinz, I.M. Heterozygous caveolin-3 mice show increased susceptibility to palmitate-induced insulin resistance. Physiol. Rep. 2016, 4, 1–14. [Google Scholar] [CrossRef]
- Riehle, C.; Abel, E.D. Insulin Signaling and Heart Failure. Circ. Res. 2016, 118, 1151–1169. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.; Sun, H.Q.; Wang, H.; Barylko, B.; Fukata, Y.; Fukata, M.; Albanesi, J.P.; Yin, H.L. Phosphatidylinositol 4-kinase IIα is palmitoylated by golgi-localized palmitoyltransferases in cholesterol-dependent manner. J. Biol. Chem. 2012, 287, 21856–21865. [Google Scholar] [CrossRef] [PubMed]
- Jung, G.; Wang, J.; Wlodarski, P.; Barylko, B.; Binns, D.D.; Shu, H.; Yin, H.L.; Albanesi, J.P. Molecular determinants of activation and membrane targeting of phosphoinositol 4-kinase IIβ. Biochem. J. 2008, 409, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Du, K. ClipR-59 Interacts with Akt and Regulates Akt Cellular Compartmentalization. Mol. Cell. Biol. 2009, 29, 1459–1471. [Google Scholar] [CrossRef] [PubMed]
- Glatz, J.F.C.; Nabben, M.; Heather, L.C.; Bonen, A.; Luiken, J.J.F.P. Regulation of the subcellular trafficking of CD36, a major determinant of cardiac fatty acid utilization. Biochim. Biophys. Acta-Mol. Cell Biol. Lipids 2016, 1861, 1461–1471. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Jhala, U.S.; Du, K. Proteomic analysis of protein palmitoylation in adipocytes. Adipocyte 2013, 2, 17–27. [Google Scholar] [CrossRef]
- Lianos, E.; Hager, S.; April, R. Insulin stimulates fatty acid acylation of adipocyte proteins. Biochem. Biophys. Res. Commun. 1991, 177, 797. [Google Scholar]
- Zhao, L.; Zhang, C.; Luo, X.; Wang, P.; Zhou, W.; Zhong, S.; Xie, Y.; Jiang, Y.; Yang, P.; Tang, R.; et al. CD36 palmitoylation disrupts free fatty acid metabolism and promotes tissue inflammation in non-alcoholic steatohepatitis. J. Hepatol. 2018, 69, 705–717. [Google Scholar] [CrossRef]
- Schwenk, R.W.; Dirkx, E.; Coumans, W.A.; Bonen, A.; Klip, A.; Glatz, J.F.C.; Luiken, J.J.F.P. Requirement for distinct vesicle-associated membrane proteins in insulin- and AMP-activated protein kinase (AMPK)-induced translocation of GLUT4 and CD36 in cultured cardiomyocytes. Diabetologia 2010, 53, 2209–2219. [Google Scholar] [CrossRef]
- Veit, M.; Becher, A.; Ahnert-Hilger, G. Synaptobrevin 2 is palmitoylated in synaptic vesicles prepared from adult, but not from embryonic brain. Mol. Cell. Neurosci. 2000, 15, 408–416. [Google Scholar] [CrossRef]
- Luiken, J.J.F.P.; Nabben, M.; Neumann, D.; Glatz, J.F.C. Understanding the distinct subcellular trafficking of CD36 and GLUT4 during the development of myocardial insulin resistance. Biochim. Biophys. Acta-Mol. Basis Dis. 2020, 1866, 165775. [Google Scholar] [CrossRef]
- Zeigerer, A.; Lampson, M.A.; Karylovsky, O.; Sabatini, D.D.; Adesnik, M.; Ren, M.; McGraW, T.E. GLUT4 Retention in Adipocytes Requires Two Intracellular Insulin-regulated Transport Steps. Mol. Biol. Cell 2002, 13, 2421–2435. [Google Scholar] [CrossRef] [PubMed]
- Steinbusch, L.K.M.; Wijnen, W.; Schwenk, R.W.; Coumans, W.A.; Hoebers, N.T.H.; Ouwens, D.M.; Coumans, W.A.; Hoebers, N.T.H.; Diamant, M.; Bonen, A.; et al. Differential regulation of cardiac glucose and fatty acid uptake by endosomal pH and actin filaments. Am. J. Physiol. Cell Physiol. 2010, 298, 1549–1559. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Steinbusch, L.K.M.; Nabben, M.; Kapsokalyvas, D.; Van Zandvoort, M.; Schönleitner, P.; Antoons, G.; Simons, P.J.; Coumans, W.A.; Geomini, A.; et al. Palmitate-induced vacuolar-type H+-ATPase inhibition feeds forward into insulin resistance and contractile dysfunction. Diabetes 2017, 66, 1521–1534. [Google Scholar] [CrossRef] [PubMed]
- Schenk, S.; Saberi, M.; Olefsky, J.M. Personal perspective Insulin sensitivity: Modulation by nutrients and inflammation. J. Clin. Investig. 2008, 118, 2992–3002. [Google Scholar] [CrossRef]
- Dasgupta, S.; Bhattacharya, S.; Maitra, S.; Pal, D.; Majumdar, S.S.; Datta, A.; Bhattacharya, S. Mechanism of lipid induced insulin resistance: Activated PKCε is a key regulator. Biochim. Biophys. Acta-Mol. Basis Dis. 2011, 1812, 495–506. [Google Scholar] [CrossRef]
- Stratford, S.; DeWald, D.B.; Summers, S.A. Ceramide dissociates 3′-phosphoinositide production from pleckstrin homology domain translocation. Biochem. J. 2001, 354, 359–368. [Google Scholar] [CrossRef]
- Itani, S.I.; Ruderman, N.B.; Schmieder, F.; Boden, G. Lipid-induced insulin resistance in human muscle is associated with changes in diacylglycerol, protein kinase C, and IκB-α. Diabetes 2002, 51, 2005–2011. [Google Scholar] [CrossRef]
- Straczkowski, M.; Kowalska, I.; Baranowski, M.; Nikolajuk, A.; Otziomek, E.; Zabielski, P.; Adamska, A.; Blachnio, A.; Gorski, J.; Gorska, M. Increased skeletal muscle ceramide level in men at risk of developing type 2 diabetes. Diabetologia 2007, 50, 2366–2373. [Google Scholar] [CrossRef]
- Coen, P.M.; Hames, K.C.; Leachman, E.M.; Delany, J.P.; Ritov, V.B.; Menshikova, E.V.; Dubé, J.J.; Stefanovic-Racic, M.; Toledo, F.G.S.; Goodpaster, B.H. Reduced skeletal muscle oxidative capacity and elevated ceramide but not diacylglycerol content in severe obesity. Obesity 2013, 21, 2362–2371. [Google Scholar] [CrossRef]
- Ussher, J.R.; Koves, T.R.; Cadete, V.J.J.; Zhang, L.; Jaswal, J.S.; Swyrd, S.J.; Lopaschuk, D.G.; Proctor, S.D.; Keung, W.; Muoio, D.M.; et al. Inhibition of de novo ceramide synthesis reverses diet-induced insulin resistance and enhances whole-body oxygen consumption. Diabetes 2010, 59, 2453–2464. [Google Scholar] [CrossRef] [PubMed]
- Levin, M.C.; Monetti, M.; Watt, M.J.; Sajan, M.P.; Stevens, R.D.; Bain, J.R.; Newgard, C.B.; Farese, R.V.; Farese, R.V. Increased lipid accumulation and insulin resistance in transgenic mice expressing DGAT2 in glycolytic (type II) muscle. Am. J. Physiol. Endocrinol. Metab. 2007, 293, 1772–1781. [Google Scholar] [CrossRef] [PubMed]
- Anastasiou, C.A.; Kavouras, S.A.; Lentzas, Y.; Gova, A.; Sidossis, L.S.; Melidonis, A. Diabetes mellitus is associated with increased intramyocellular triglyceride, but not diglyceride, content in obese humans. Metabolism 2009, 58, 1636–1642. [Google Scholar] [CrossRef] [PubMed]
- Perreault, L.; Bergman, B.C.; Hunerdosse, D.M.; Eckel, R.H. Altered intramuscular lipid metabolism relates to diminished insulin action in men, but not women, in progression to diabetes. Obesity 2010, 18, 2093–2100. [Google Scholar] [CrossRef]
- Jocken, J.W.E.; Moro, C.; Goossens, G.H.; Hansen, D.; Mairal, A.; Hesselink, M.K.C.; Langin, D.; Van Loon, L.J.C.; Blaak, E.E. Skeletal muscle lipase content and activity in obesity and type 2 diabetes. J. Clin. Endocrinol. Metab. 2010, 95, 5449–5453. [Google Scholar] [CrossRef]
- Holland, W.L.; Brozinick, J.T.; Wang, L.P.; Hawkins, E.D.; Sargent, K.M.; Liu, Y.; Narra, K.; Hoehn, K.L.; Knotts, T.A.; Siesky, A.; et al. Inhibition of Ceramide Synthesis Ameliorates Glucocorticoid-, Saturated-Fat-, and Obesity-Induced Insulin Resistance. Cell Metab. 2007, 5, 167–179. [Google Scholar] [CrossRef]
- Skovbro, M.; Baranowski, M.; Skov-Jensen, C.; Flint, A.; Dela, F.; Gorski, J.; Helge, J.W. Human skeletal muscle ceramide content is not a major factor in muscle insulin sensitivity. Diabetologia 2008, 51, 1253–1260. [Google Scholar] [CrossRef]
- Adams, J.M.; Pratipanawatr, T.; Berria, R.; Wang, E.; DeFronzo, R.A.; Sullards, M.C.; Mandarino, L.J. Ceramide Content Is Increased in Skeletal Muscle from Obese Insulin-Resistant Humans. Diabetes 2004, 53, 25–31. [Google Scholar] [CrossRef]
- Bosma, M.; Kersten, S.; Hesselink, M.K.C.; Schrauwen, P. Progress in Lipid Research Re-evaluating lipotoxic triggers in skeletal muscle: Relating intramyocellular lipid metabolism to insulin sensitivity. Prog. Lipid Res. 2012, 51, 36–49. [Google Scholar] [CrossRef]
- Greaves, J.; Chamberlain, L.H. Palmitoylation-dependent protein sorting. J. Cell Biol. 2007, 176, 249–254. [Google Scholar] [CrossRef]
- Fernández-Hernando, C.; Fukata, M.; Bernatchez, P.N.; Fukata, Y.; Lin, M.I.; Bredt, D.S.; Sessa, W.C. Identification of Golgi-localized acyl transferases that palmitoylate and regulate endothelial nitric oxide synthase. J. Cell Biol. 2006, 174, 369–377. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.M.; Tsou, L.K.; Charron, G.; Raghavan, A.S.; Hang, H.C. Tandem fluorescence imaging of dynamic S-acylation and protein turnover. Proc. Natl. Acad. Sci. USA 2010, 107, 8627–8632. [Google Scholar] [CrossRef] [PubMed]
- Resh, M.D. Fatty acylation of proteins: New insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim. Biophys. Acta-Mol. Cell Res. 1999, 1451, 1–16. [Google Scholar] [CrossRef]
- Putilina, T.; Wong, P.; Gentleman, S. The DHHC domain: A new highly conserved cysteine-rich motif. Mol. Cell. Biochem. 1999, 195, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Rocks, O.; Gerauer, M.; Vartak, N.; Koch, S.; Huang, Z.P.; Pechlivanis, M.; Kuhlmann, J.; Brunsveld, L.; Chandra, A.; Ellinger, B.; et al. The palmitoylation machinery is a spatially organizing system for peripheral membrane proteins. Cell 2010, 141, 458–471. [Google Scholar] [CrossRef] [PubMed]
- Hirano, T.; Kishi, M.; Sugimoto, H.; Taguchi, R.; Obinata, H.; Ohshima, N.; Tatei, K.; Izumi, T. Thioesterase activity and subcellular localization of acylprotein thioesterase 1/lysophospholipase 1. Biochim. Biophys. Acta-Mol. Cell Biol. Lipids 2009, 1791, 797–805. [Google Scholar] [CrossRef] [PubMed]
- Toyoda, T.; Sugimoto, H.; Yamashita, S. Sequence, expression in Escherichia coli, and characterization of lysophospholipase II. Biochim. Biophys. Acta-Mol. Cell Biol. Lipids 1999, 1437, 182–193. [Google Scholar] [CrossRef]
- Vartak, N.; Papke, B.; Grecco, H.E.; Rossmannek, L.; Waldmann, H.; Hedberg, C.; Bastiaens, P.I.H. The autodepalmitoylating activity of APT maintains the spatial organization of palmitoylated membrane proteins. Biophys. J. 2014, 106, 93–105. [Google Scholar] [CrossRef]
- Verkruyse, L.A.; Hofmann, S.L. Lysosomal targeting of palmitoyl-protein thioesterase. J. Biol. Chem. 1996, 271, 15831–15836. [Google Scholar] [CrossRef]
- Dawson, G.; Schroeder, C.; Dawson, P.E. Palmitoyl: Protein thioesterase (PPT1) inhibitors can act as pharmacological chaperones in infantile Batten Disease. Biochem. Biophys. Res. Commun. 2010, 23, 66–69. [Google Scholar] [CrossRef]
- Howie, J.; Reilly, L.; Fraser, N.J.; Walker, J.M.V.; Wypijewski, K.J.; Ashford, M.L.J.; Calaghan, S.C.; McClafferty, H.; Tian, L.; Shipston, M.J.; et al. Substrate recognition by the cell surface palmitoyl transferase DHHC5. Proc. Natl. Acad. Sci. USA 2014, 111, 17534–17539. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.J.; Fine, M.; Lu, J.Y.; Hofmann, S.L.; Frazier, G.; Hilgemann, D.W. Massive palmitoylation-dependent endocytosis during reoxygenation of anoxic cardiac muscle. Elife 2013, 2013, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.J.; Marsden, A.N.; Scott, C.A.; Akimzhanov, A.M.; Boehning, D. DHHC5 Mediates β-Adrenergic Signaling in Cardiomyocytes by Targeting Gα Proteins. Biophys. J. 2020, 118, 826–835. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Li, J.; Zhao, P.; Liu, H.; Jia, D.; Jia, H.; He, L.; Cang, Y.; Boast, S.; Chen, Y.H.; et al. Palmitoyl acyltransferase Aph2 in cardiac function and the development of cardiomyopathy. Proc. Natl. Acad. Sci. USA 2015, 112, 15666–15671. [Google Scholar] [CrossRef] [PubMed]
- Essandoh, K.; Philippe, J.M.; Jenkins, P.M.; Brody, M.J. Palmitoylation: A Fatty Regulator of Myocardial Electrophysiology. Front. Physiol. 2020, 11, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Jennings, B.C.; Linder, M.E. DHHC protein S-acyltransferases use similar ping-pong kinetic mechanisms but display different Acyl-CoA specificities. J. Biol. Chem. 2012, 287, 7236–7245. [Google Scholar] [CrossRef]
- Lobo, S.; Greentree, W.K.; Linder, M.E.; Deschenes, R.J. Identification of a Ras palmitoyltransferase in Saccharomyces cerevisiae. J. Biol. Chem. 2002, 277, 41268–41273. [Google Scholar] [CrossRef]
- Bartels, D.J.; Mitchell, D.A.; Dong, X.; Deschenes, R.J. Erf2, a Novel Gene Product That Affects the Localization and Palmitoylation of Ras2 in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999, 19, 6775–6787. [Google Scholar] [CrossRef]
- Roth, A.F.; Feng, Y.; Chen, L.; Davis, N.G. The yeast DHHC cysteine-rich domain protein Akr1p is a palmitoyl transferase. J. Cell Biol. 2002, 159, 23–28. [Google Scholar] [CrossRef]
- Mitchell, D.A.; Mitchell, G.; Ling, Y.; Budde, C.; Deschenes, R.J. Mutational analysis of Saccharomyces cerevisiae Erf2 reveals a two-step reaction mechanism for protein palmitoylation by DHHC enzymes. J. Biol. Chem. 2010, 285, 38104–38114. [Google Scholar] [CrossRef]
- Verardi, R.; Kim, J.S.; Ghirlando, R.; Banerjee, A. Structural Basis for Substrate Recognition by the Ankyrin Repeat Domain of Human DHHC17 Palmitoyltransferase. Physiol. Behav. 2017, 176, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Lemonidis, K.; Gorleku, O.A.; Sanchez-Perez, M.C.; Grefen, C.; Chamberlain, L.H. The Golgi S-acylation machinery comprises zDHHC enzymes with major differences in substrate affinity and S-acylation activity. Mol. Biol. Cell 2014, 25, 3870–3883. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Di Vizio, D.; Kirchner, M.; Steen, H.; Freeman, M.R. Proteome scale characterization of human S-acylated proteins in lipid raft-enriched and non-raft membranes. Mol. Cell. Proteom. 2010, 9, 54–70. [Google Scholar] [CrossRef] [PubMed]
- Spinelli, M.; Fusco, S.; Mainardi, M.; Scala, F.; Natale, F.; Lapenta, R.; Mattera, A.; Rinaudo, M.; Li Puma, D.D.; Ripoli, C.; et al. Brain insulin resistance impairs hippocampal synaptic plasticity and memory by increasing GluA1 palmitoylation through. Nat. Commun. 2017, 8, 1–14. [Google Scholar] [CrossRef]
- Gottlieb, C.D.; Zhang, S.; Linder, M.E. The cysteine-rich domain of the DHHC3 palmitoyltransferase is palmitoylated and contains tightly bound zinc. J. Biol. Chem. 2015, 290, 29259–29269. [Google Scholar] [CrossRef]
- Segal-Salto, M.; Sapir, T.; Reiner, O. Reversible cysteine acylation regulates the activity of human palmitoyl-protein thioesterase 1 (PPT1). PLoS ONE 2016, 11, e0146466. [Google Scholar] [CrossRef]
- Abrami, L.; Dallavilla, T.; Sandoz, P.A.; Demir, M.; Kunz, B.; Savoglidis, G.; Hatzimanikatis, V.; van Der Goot, F.G. Identification and dynamics of the human ZDHHC16-ZDHHC6 palmitoylation cascade. Elife 2017, 6, 1–24. [Google Scholar] [CrossRef]
- Galil, D.A.; Cherkas, V.; Ronkina, N.; Lafera, J.; Gaestel, M.; Ponimaskin, E. ZDHHC3 Tyrosine Phosphorylation Regulates Neural Cell Adhesion Molecule Palmitoylation. Mol. Cell. Biol. 2016, 36, 2208–2225. [Google Scholar] [CrossRef]
- Wei, X.; Adak, S.; Zayed, M.; Yin, L.; Feng, C.; Speck, S.L.; Rahul, S.; Zhang, Q.; Dickinson, B.C.; Semenkovich, C.F. Endothelial Palmitoylation Cycling Coordinates Vessel Remodeling in Peripheral Artery Disease. Circ. Res. 2020. [Google Scholar] [CrossRef]
- Parat, M.O. Chapter 4 The Biology of Caveolae. Achievements and Perspectives. Int. Rev. Cell Mol. Biol. 2009, 273, 117–162. [Google Scholar] [CrossRef]
- Parton, R.G.; Del Pozo, M.A. Caveolae as plasma membrane sensors, protectors and organizers. Nat. Rev. Mol. Cell Biol. 2013, 14, 98–112. [Google Scholar] [CrossRef] [PubMed]
- Xiao, B.W.; Lee, H.; Capozza, F.; Marmon, S.; Sotgia, F.; Brooks, J.W.; Campos-Gonzalez, R.; Lisanti, M.P. Tyrosine phosphorylation of caveolin-2 at residue 27: Differences in the spatial and temporal behavior of phospho-Cav-2 (pY19 and pY27). Biochemistry 2004, 43, 13694–13706. [Google Scholar] [CrossRef]
- Mastick, C.C.; Brady, M.J.; Saltiel, A.R. Insulin stimulates the tyrosine phosphorylation of caveolin. J. Cell Biol. 1995, 129, 1523–1531. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Scherer, P.E.; Okamoto, T.; Song, K.; Chu, C.; Kohtz, D.S.; Nishimoto, I.; Lodish, H.F.; Lisanti, M.P. Molecular cloning of caveolin-3, a novel member of the caveolin gene family expressed predominantly in muscle. J. Biol. Chem. 1996, 271, 2255–2261. [Google Scholar] [CrossRef]
- Dietzen, D.J.; Hastings, W.R.; Lublin, D.M. Caveolin is palmitoylated on multiple cysteine residues. Palmitoylation is not necessary for localization of caveolin to caveolae. J. Biol. Chem. 1995, 270, 6838–6842. [Google Scholar] [CrossRef]
- Tonn Eisinger, K.R.; Woolfrey, K.M.; Swanson, S.P.; Schnell, S.A.; Meitzen, J.; Dell’Acqua, M.; Mermelstein, P.G. Palmitoylation of caveolin-1 is regulated by the same DHHC acyltransferases that modify steroid hormone receptors. J. Biol. Chem. 2018, 293, 15901–15911. [Google Scholar] [CrossRef]
- Rodriguez-Walker, M.; Daniotti, J.L. Human Sialidase Neu3 is S-Acylated and Behaves Like an Integral Membrane Protein. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef]
- Sergeeva, O.A.; Gisou van der Goot, F. Anthrax toxin requires ZDHHC5-mediated palmitoylation of its surface-processing host enzymes. Proc. Natl. Acad. Sci. USA 2019, 116, 1279–1288. [Google Scholar] [CrossRef]
- Sada, R.; Kimura, H.; Fukata, Y.; Fukata, M.; Yamamoto, H.; Kikuchi, A. Dynamic palmitoylation controls the microdomain localization of the DKK1 receptors CKAP4 and LRP6. Sci. Signal. 2019, 12, 1–15. [Google Scholar] [CrossRef]
- Lee, H.; Woodman, S.E.; Engelman, J.A.; Volonte, D.; Galbiati, F.; Kaufman, H.L.; Lublin, D.M.; Lisanti, M.P. Palmitoylation of Caveolin-1 at a Single Site (Cys-156) Controls its Coupling to the c-Src Tyrosine Kinase: Targeting of dually acylated molecules (Gpi-linked, transmembrane, or cytoplasmic) to caveolae effectively uncouples c-Src and caveolin-1 (Tyr-14). J. Biol. Chem. 2001, 276, 35150–35158. [Google Scholar] [CrossRef]
- Li, S.; Seitz, R.; Lisanti, M.P. Phosphorylation of caveolin by Src tyrosine kinases: The α-isoform of caveolin is selectively phosphorylated by v-Src in vivo. J. Biol. Chem. 1996, 271, 3863–3868. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Volonte, D.; Galbiati, F.; Iyengar, P.; Lublin, D.M.; Bregman, D.B.; Wilson, M.T.; Campos-Gonzalez, R.; Bouzahzah, B.; Pestell, R.G.; et al. Constitutive and growth factor-regulated phosphorylation of caveolin-1 occurs at the same site (Tyr-14) in vivo: Identification of a c-Src/Cav-1/Grb7 signaling cassette. Mol. Endocrinol. 2000, 14, 1750–1775. [Google Scholar] [CrossRef] [PubMed]
- Rosenzweig, T.; Aga-Mizrachi, S.; Bak, A.; Sampson, S.R. Src tyrosine kinase regulates insulin-induced activation of protein kinase C (PKC) δ in skeletal muscle. Cell. Signal. 2004, 16, 1299–1308. [Google Scholar] [CrossRef] [PubMed]
- Braiman, L.; Alt, A.; Kuroki, T.; Ohba, M.; Bak, A.; Tennenbaum, T.; Sampson, S.R. Protein kinase Cδ mediates insulin-induced glucose transport in primary cultures of rat skeletal muscle. Mol. Endocrinol. 1999, 13, 2002–2012. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Courchesne, W.E.; Mastick, C.C. A phosphotyrosine-dependent protein interaction screen reveals a role for phosphorylation of caveolin-1 on tyrosine 14. Recruitment of C-terminal Src kinase. J. Biol. Chem. 2002, 277, 8771–8774. [Google Scholar] [CrossRef]
- Kwon, H.; Lee, J.; Jeong, K.; Jang, D.; Pak, Y. Fatty acylated caveolin-2 is a substrate of insulin receptor tyrosine kinase for insulin receptor substrate-1-directed signaling activation. Biochim. Biophys. Acta-Mol. Cell Res. 2015, 1853, 1022–1034. [Google Scholar] [CrossRef]
- Scherer, P.E.; Lewis, Y.; Volonte, D.; Engelman, J.A.; Galbiati, F.; Couet, J.; Kohtz, D.S.; Van Donselaar, E.; Peters, P.; Lisanti, M.P. Cell-type and Tissue-specific Expression of Caveolin-2. J. Biol. Chem. 1997, 272, 29337–29346. [Google Scholar] [CrossRef]
- Ahn, M.; Kim, H.; Matsumoto, Y.; Shin, T. Increased expression of caveolin-1 and -2 in the hearts of Lewis rats with experimental autoimmune myocarditis. Autoimmunity 2006, 39, 489–495. [Google Scholar] [CrossRef]
- Rybin, V.O.; Grabham, P.W.; Elouardighi, H.; Steinberg, S.F. Caveolae-associated proteins in cardiomyocytes: Caveolin-2 expression and interactions with caveolin-3. Am. J. Physiol. Heart Circ. Physiol. 2003, 285, 325–332. [Google Scholar] [CrossRef]
- Tulloch, L.B.; Howie, J.; Wypijewski, K.J.; Wilson, C.R.; Bernard, W.G.; Shattock, M.J.; Fuller, W. The inhibitory effect of phospholemman on the sodium pump requires its palmitoylation. J. Biol. Chem. 2011, 286, 36020–36031. [Google Scholar] [CrossRef]
- Virkamäki, A.; Ueki, K.; Kahn, C.R. Protein-protein interaction in insulin signaling and the molecular mechanisms of insulin resistance. J. Clin. Investig. 1999, 103, 931–943. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Li, J.; Yu, H.; Zhai, Y.; Gao, Z.; Liu, Y.; Pang, X.; Zhang, L.; Schulten, K.; Sun, F.; et al. Molecular insights into the membrane-associated phosphatidylinositol 4-kinase IIα. Nat. Commun. 2014, 5, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Barylko, B.; Mao, Y.S.; Wlodarski, P.; Jung, G.; Binns, D.D.; Sun, H.Q.; Yin, H.L.; Albanesi, J.P. Palmitoylation controls the catalytic activity and subcellular distribution of phosphatidylinositol 4-kinase IIα. J. Biol. Chem. 2009, 284, 9994–10003. [Google Scholar] [CrossRef] [PubMed]
- Lallemand-Breitenbach, V.; Quesnoit, M.; Braun, V.; El Marjou, A.; Poüs, C.; Goud, B.; Perez, F. CLIPR-59 is a lipid raft-associated protein containing a cytoskeleton-associated protein glycine-rich domain (CAP-Gly) that perturbs microtubule dynamics. J. Biol. Chem. 2004, 279, 41168–41178. [Google Scholar] [CrossRef]
- Ren, W.; Sun, Y.; Du, K. DHHC17 Palmitoylates ClipR-59 and Modulates ClipR-59 Association with the Plasma Membrane. Mol. Cell. Biol. 2013, 33, 4255–4265. [Google Scholar] [CrossRef]
- Chen, B.; Zheng, B.; DeRan, M.; Jarugumilli, G.K.; Fu, J.; Brooks, Y.S.; Wu, X. ZDHHC7-Mediated S-Palmitoylation of Scribble Regulates Cell Polarity. Nat. Chem. Biol. 2016, 12, 686–693. [Google Scholar] [CrossRef]
- Liao, P.; Wang, W.; Li, Y.; Wang, R.; Jin, J.; Pang, W.; Chen, Y.; Shen, M.; Wang, X.; Jiang, D.; et al. Palmitoylated SCP1 is targeted to the plasma membrane and negatively regulates angiogenesis. Elife 2017, 6, 1–24. [Google Scholar] [CrossRef]
- Du, K.; Murakami, S.; Sun, Y.; Kilpatrick, C.L.; Luscher, B. DHHC7 palmitoylates glucose transporter 4 (Glut4) and regulates Glut4 membrane translocation. J. Biol. Chem. 2017, 292, 2979–2991. [Google Scholar] [CrossRef]
- Vogel, K.; Roche, P.A. SNAP-23 and SNAP-25 are palmitoylated in vivo. Biochem. Biophys. Res. Commun. 1999, 258, 407–410. [Google Scholar] [CrossRef]
- Greaves, J.; Gorleku, O.A.; Salaun, C.; Chamberlain, L.H. Palmitoylation of the SNAP25 protein family: Specificity and regulation by DHHC palmitoyl transferases. J. Biol. Chem. 2010, 285, 24629–24638. [Google Scholar] [CrossRef]
- Salaün, C.; Gould, G.W.; Chamberlain, L.H. The SNARE proteins SNAP-25 and SNAP-23 display different affinities for lipid rafts in PC12 cells: Regulation by distinct cysteine-rich domains. J. Biol. Chem. 2005, 280, 1236–1240. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Zhang, Z.; Sarkar, C.; Tsai, P.C.; Lee, Y.C.; Dye, L.; Mukherjee, A.B. Palmitoyl protein thioesterase-1 deficiency impairs synaptic vesicle recycling at nerve terminals, contributing to neuropathology in humans and mice. J. Clin. Investig. 2008, 118, 3075–3086. [Google Scholar] [CrossRef] [PubMed]
- Foster, L.J.; Yaworsky, K.; Trimble, W.S.; Klip, A. SNAP23 promotes insulin-dependent glucose uptake in 3T3-L1 adipocytes: Possible interaction with cytoskeleton. Am. J. Physiol. Cell Physiol. 1999, 276. [Google Scholar] [CrossRef] [PubMed]
- Peters, C.G.; Miller, D.F.; Giovannucci, D.R. Identification, localization and interaction of SNARE proteins in atrial cardiac myocytes. J. Mol. Cell. Cardiol. 2006, 40, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Bowman, P.R.T.; Smith, G.L.; Gould, G.W. Cardiac SNARE Expression in Health and Disease. Front. Endocrinol. (Lausanne) 2019, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Abel, E.D.; Peroni, O.; Kim, J.K.K.Y.; Boss, O.; Hadro, E.; Minnemann, T.; Shulman, G.I.; Kahn, B.B. Adipose-selective targeting of the GLUT4 gene impairs insulin action in muscle and liver. Nature 2001, 409, 729–733. [Google Scholar] [CrossRef]
- Alkhateeb, H.; Chabowski, A.; Glatz, J.F.C.; Luiken, J.F.P.; Bonen, A. Two phases of palmitate-induced insulin resistance in skeletal muscle: Impaired GLUT4 translocation is followed by a reduced GLUT4 intrinsic activity. Am. J. Physiol. Endocrinol. Metab. 2007, 293, 783–793. [Google Scholar] [CrossRef]
- Kern, M.; Wells, J.A.; Stephens, J.M.; Elton, C.W.; Friedman, J.E.; Tapscott, E.B.; Pekala, P.H.; Dohm, G.L. Insulin responsiveness in skeletal muscle is determined by glucose transporter (Glut4) protein level. Biochem. J. 1990, 270, 397–400. [Google Scholar] [CrossRef]
- Heather, L.C.; Pates, K.M.; Atherton, H.J.; Cole, M.A.; Ball, D.R.; Evans, R.D.; Glatz, J.F.; Luiken, J.J.; Griffin, J.L.; Clarke, K. Differential translocation of the fatty acid transporter, FAT/CD36, and the glucose transporter, GLUT4, coordinates changes in cardiac substrate metabolism during ischemia and reperfusion. Circ. Heart Fail. 2013, 6, 1058–1066. [Google Scholar] [CrossRef]
- Ren, W.; Sun, Y.; Du, K. Glut4 palmitoylation at Cys223 plays a critical role in Glut4 membrane trafficking. Biochem. Biophys. Res. Commun. 2015, 460, 709–714. [Google Scholar] [CrossRef]
- Perrini, S.; Natalicchio, A.; Laviola, L.; Belsanti, G.; Montrone, C.; Cignarelli, A.; Minielli, V.; Grano, M.; De Pergola, G.; Giorgino, R.; et al. Dehydroepiandrosterone Stimulates Glucose Uptake in Human and Murine Adipocytes by Inducing GLUT1 and GLUT4 Translocation to the Plasma Membrane. Diabetes 2004, 53, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Geraets, I.M.E.; Glatz, J.F.C.; Luiken, J.J.F.P.; Nabben, M. Pivotal role of membrane substrate transporters on the metabolic alterations in the pressure-overloaded heart. Cardiovasc. Res. 2019, 115, 1000–1012. [Google Scholar] [CrossRef] [PubMed]
- Stapel, B.; Gorinski, N.; Gmahl, N.; Rhein, M.; Preuss, V.; Hilfiker-Kleiner, D.; Frieling, H.; Bleich, S.; Ponimaskin, E.; Kahl, K.G. Fluoxetine induces glucose uptake and modifies glucose transporter palmitoylation in human peripheral blood mononuclear cells. Expert Opin. Ther. Targets 2019, 23, 883–891. [Google Scholar] [CrossRef] [PubMed]
- Pouliot, J.F.; Béliveau, R. Palmitoylation of the glucose transporter in blood-brain barrier capillaries. Biochim. Biophys. Acta (BBA)-Biomembr. 1995, 1234, 191–196. [Google Scholar] [CrossRef]
- Tao, N.; Wagner, S.J.; Lublin, D.M. CD36 is palmitoylated on both N- and C-terminal cytoplasmic tails. J. Biol. Chem. 1996, 271, 22315–22320. [Google Scholar] [CrossRef]
- Thorne, R.F.; Ralston, K.J.; de Bock, C.E.; Mhaidat, N.M.; Zhang, X.D.; Boyd, A.W.; Burns, G.F. Palmitoylation of CD36/FAT regulates the rate of its post-transcriptional processing in the endoplasmic reticulum. Biochim. Biophys. Acta-Mol. Cell Res. 2010, 1803, 1298–1307. [Google Scholar] [CrossRef]
- Wang, J.; Hao, J.W.; Wang, X.; Guo, H.; Sun, H.H.; Lai, X.Y.; Liu, L.Y.; Zhu, M.; Wang, H.Y.; Li, Y.F.; et al. DHHC4 and DHHC5 Facilitate Fatty Acid Uptake by Palmitoylating and Targeting CD36 to the Plasma Membrane. Cell Rep. 2019, 26, 209–221.e5. [Google Scholar] [CrossRef]
- Glatz, J.F.C.; Luiken, J.J.F.P. Time for a détente in the war on the mechanism of cellular fatty acid uptake. J. Lipid Res. 2020, 61, 1300–1303. [Google Scholar] [CrossRef]
- Hao, J.W.; Wang, J.; Guo, H.; Zhao, Y.Y.; Sun, H.H.; Li, Y.F.; Lai, X.Y.; Zhao, N.; Wang, X.; Xie, C.; et al. CD36 facilitates fatty acid uptake by dynamic palmitoylation-regulated endocytosis. Nat. Commun. 2020, 11, 1–16. [Google Scholar] [CrossRef]
- Van Oort, M.M.; Drost, R.; Janßen, L.; Van Doorn, J.M.; Kerver, J.; Van Der Horst, D.J.; Luiken, J.J.F.P.; Rodenburg, K.C.W. Each of the four intracellular cysteines of CD36 is essential for insulin- or AMP-activated protein kinase-induced CD36 translocation. Arch. Physiol. Biochem. 2014, 120, 40–49. [Google Scholar] [CrossRef]
- D’Andrea-Merrins, M.; Chang, L.; Lam, A.D.; Ernst, S.A.; Stuenkel, E.L. Munc18c interaction with syntaxin 4 monomers and SNARE complex intermediates in GLUT4 vesicle trafficking. J. Biol. Chem. 2007, 282, 16553–16566. [Google Scholar] [CrossRef] [PubMed]
- Jochen, A.; Hays, J. Purification of the major substrate for palmitoylation in rat adipocytes: N-terminal homology with CD36 and evidence for cell surface acylation. J. Lipid Res. 1993, 34, 1783–1792. [Google Scholar] [PubMed]
- Yang, J.; Gibson, B.; Snider, J.; Jenkins, C.M.; Han, X.; Gross, R.W. Submicromolar concentrations of palmitoyl-CoA specifically thioesterify cysteine 244 in glyceraldehyde-3-phosphate dehydrogenase inhibiting enzyme activity: A novel mechanism potentially underlying fatty acid induced insulin resistance. Biochemistry 2005, 44, 11903–11912. [Google Scholar] [CrossRef] [PubMed]
- Schaap, D.; Parker, P.J. Expression, purification, and characterization of protein kinase C-ε. J. Biol. Chem. 1990, 265, 7301–7307. [Google Scholar] [PubMed]
- Ping, P.; Zhang, J.; Pierce, W.M.; Bolli, R. Functional proteomic analysis of protein kinase C ε signaling complexes in the normal heart and during cardioprotection. Circ. Res. 2001, 88, 59–62. [Google Scholar] [CrossRef] [PubMed]
- Noland, T.A.; Guo, X.; Raynor, R.L.; Jideama, N.M.; Averyhart-fullard, V.; Solaro, R.J.; Kuo, J.F. Cardiac Troponin I Mutants mutants Phosphorylation by protein kinases C and A and regulation of Ca2+-stimulated MgATPase of reconstituted actomyosin S-1. J. Biol. Chem. 1995, 270, 25445–25454. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, Y.; Olsen, G.S.; Ziv, E.; Hansen, L.L.; Busch, A.K.; Hansen, B.F.; Shafrir, E.; Mosthaf-Seedorf, L. Cellular mechanism of nutritionally induced insulin resistance in Psammomys obesus: Overexpression of protein kinase Cε in skeletal muscle precedes the onset of hyperinsulinemia and hyperglycemia. Diabetes 2001, 50, 584–592. [Google Scholar] [CrossRef]
- Dey, D.; Basu, D.; Roy, S.S.; Bandyopadhyay, A.; Bhattacharya, S. Involvement of novel PKC isoforms in FFA induced defects in insulin signaling. Mol. Cell. Endocrinol. 2006, 246, 60–64. [Google Scholar] [CrossRef]
- Dey, D.; Mukherjee, M.; Basu, D.; Datta, M.; Roy, S.S.; Bandyopadhyay, A.; Bhattacharya, S. Inhibition of insulin receptor gene expression and insulin signaling by fatty acids: Interplay of PKC isoforms therein. Cell. Physiol. Biochem. 2005, 16, 217–228. [Google Scholar] [CrossRef]
- Ramzan, R.; Weber, P.; Linne, U.; Vogt, S. GAPDH: The missing link between glycolysis and mitochondrial oxidative phosphorylation? Biochem. Soc. Trans. 2013, 41, 1294–1297. [Google Scholar] [CrossRef]
- Terasaki, J.; Anai, M.; Funaki, M.; Shibata, T.; Inukai, K.; Ogihara, T.; Ishihara, H.; Katagiri, H.; Onishi, Y.; Sakoda, H.; et al. Role of JTT-501, a new insulin sensitiser, in restoring impaired GLUT4 translocation in adipocytes of rats fed a high fat diet. Diabetologia 1998, 41, 400–409. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Carvalho, E.; Rondinone, C.; Smith, U. Insulin resistance in fat cells from obese Zucker rats—Evidence for an impaired activation and translocation of protein kinase B and glucose transporter 4. Mol. Cell. Biochem. 2000, 206, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wong, L.Y.; Neumann, D.; Liu, Y.; Sun, A.; Antoons, G.; Strzelecka, A.; Glatz, J.F.C.; Nabben, M.; Luiken, J.J.F.P. Augmenting vacuolar H+-ATPase function prevents cardiomyocytes from lipid-overload induced dysfunction. Int. J. Mol. Sci. 2020, 21, 1520. [Google Scholar] [CrossRef] [PubMed]
- Sharma, C.; Rabinovitz, I.; Hemler, M.E. Palmitoylation by DHHC3 is critical for the function, expression, and stability of integrin α6β4. Cell. Mol. Life Sci. 2012, 69, 2233–2244. [Google Scholar] [CrossRef] [PubMed]
- Ducker, C.E.; Stettler, E.M.; French, K.J.; Upson, J.J.; Smith, C.D. Huntingtin interacting protein 14 is an oncogenic human protein: Palmitoyl acyltransferase. Oncogene 2004, 23, 9230–9237. [Google Scholar] [CrossRef] [PubMed]
- De, I.; Sadhukhan, S. Emerging Roles of DHHC-mediated Protein S-palmitoylation in Physiological and Pathophysiological Context. Eur. J. Cell Biol. 2018, 97, 319–338. [Google Scholar] [CrossRef]

| Palmitoylated Protein | Localization of the Palmitoylated Protein | Palmitoyl Acyl Transferase | Effect of the Basally Palmitoylated Protein on Insulin Signaling and Substrates Uptake | Cell Type |
|---|---|---|---|---|
| Caveolin-1 | Plasma membrane [18] | DHHC7, DHHC21 [86] | (Possible) Negative regulation Mitigate insulin-stimulated glucose uptake | Adipocytes, HEK293 |
| Caveolin-2 | Plasma membrane [96] | Unknown | Positive regulation Facilitates phosphorylation of IRS-1 by IR [96] | Adipocytes |
| Caveolin-3 | Plasma membrane [100] | Unknown | Unknown | Cardiomyocytes |
| PI4KIIα | Golgi [22,102,103] | DHHC3, DHHC7 [22] | (Possible) Positive regulation Increases PI4P (PIP2 precursor) content | COS-7, HeLa |
| Clip59-R | Plasma membrane [104] | DHHC17 [105] | Positive regulation Facilitates phosphorylation of Akt kinase [105] | Adipocytes |
| RabGAP AS160 [26] | Unknown | Unknown | Unknown | Adipocytes |
| SCRIB | Plasma membrane [106] | DHHC7 [106] | (Possible) Negative regulation Inhibits PI3K/Akt signaling in response to EGF [106] | HEK293, MCF10A |
| SCP1 | Plasma membrane [107] | Unknown | Negative regulation Inhibits Akt phosphorylation in response to insulin [107] | HEK293, MEF |
| VAMP2 (v-SNARE) | GLUT4 vesicles [108] | DHHC7 [108] | Positive regulation Mediate GLUT4-mediated glucose uptake [108] | Adipocytes |
| SNAP23 (t-SNARE) | Plasma membrane [109,111] | DHHC 2,3,7 and 17 [110] | (Possible) Positive regulation Might increase insulin-stimulated glucose uptake | PC12, COS-7 |
| IRAP | GLUT4 vesicles [108] | DHHC7 [108] | (Possible) Positive regulation Mediate insulin-stimulated glucose uptake [108] | Adipocytes |
| MUNC18 | Unknown | Unknown | Unknown | Adipocytes |
| GLUT4 | Plasma membrane [108,120] | DHHC7 [108] | Positive regulation Necessary for insulin-stimulated stimulated glucose uptake [108] | Adipocytes |
| CD36 | Plasma membrane [126,127,132] | DHHC4, DHHC5 [127,129] | Positive regulation Necessary for fatty acids uptake [127,129] | Adipocytes, COS7 |
| Palmitoylated Protein | Localization of the Aberrantly Palmitoylated Protein | Effect of the Aberrant Palmitoylation | Cell Type |
|---|---|---|---|
| GAPDH | Cell membranes [133] | Decreases GAPDH enzymatic activity and glucose utilization [133] | Rabbit muscle |
| PKCε | Actin filament [36] | Increases phosphorylation of S-Acylated PKCε, leading to downregulation of IR transcription [36] | Skeletal muscle Adipocytes |
| GLUT4 | Unknown | Might impair GLUT4 trafficking to the plasma membrane [26] | Adipocytes |
| CD36 | Increased presence at the plasma membrane [28] | Increases both CD36 translocation to the plasma membrane and fatty acids uptake, leading to NASH [28] | Hepatocytes |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schianchi, F.; Glatz, J.F.C.; Navarro Gascon, A.; Nabben, M.; Neumann, D.; Luiken, J.J.F.P. Putative Role of Protein Palmitoylation in Cardiac Lipid-Induced Insulin Resistance. Int. J. Mol. Sci. 2020, 21, 9438. https://doi.org/10.3390/ijms21249438
Schianchi F, Glatz JFC, Navarro Gascon A, Nabben M, Neumann D, Luiken JJFP. Putative Role of Protein Palmitoylation in Cardiac Lipid-Induced Insulin Resistance. International Journal of Molecular Sciences. 2020; 21(24):9438. https://doi.org/10.3390/ijms21249438
Chicago/Turabian StyleSchianchi, Francesco, Jan F. C. Glatz, Artur Navarro Gascon, Miranda Nabben, Dietbert Neumann, and Joost J. F. P. Luiken. 2020. "Putative Role of Protein Palmitoylation in Cardiac Lipid-Induced Insulin Resistance" International Journal of Molecular Sciences 21, no. 24: 9438. https://doi.org/10.3390/ijms21249438
APA StyleSchianchi, F., Glatz, J. F. C., Navarro Gascon, A., Nabben, M., Neumann, D., & Luiken, J. J. F. P. (2020). Putative Role of Protein Palmitoylation in Cardiac Lipid-Induced Insulin Resistance. International Journal of Molecular Sciences, 21(24), 9438. https://doi.org/10.3390/ijms21249438








