Maize microRNA166 Inactivation Confers Plant Development and Abiotic Stress Resistance
Abstract
:1. Introduction
2. Results
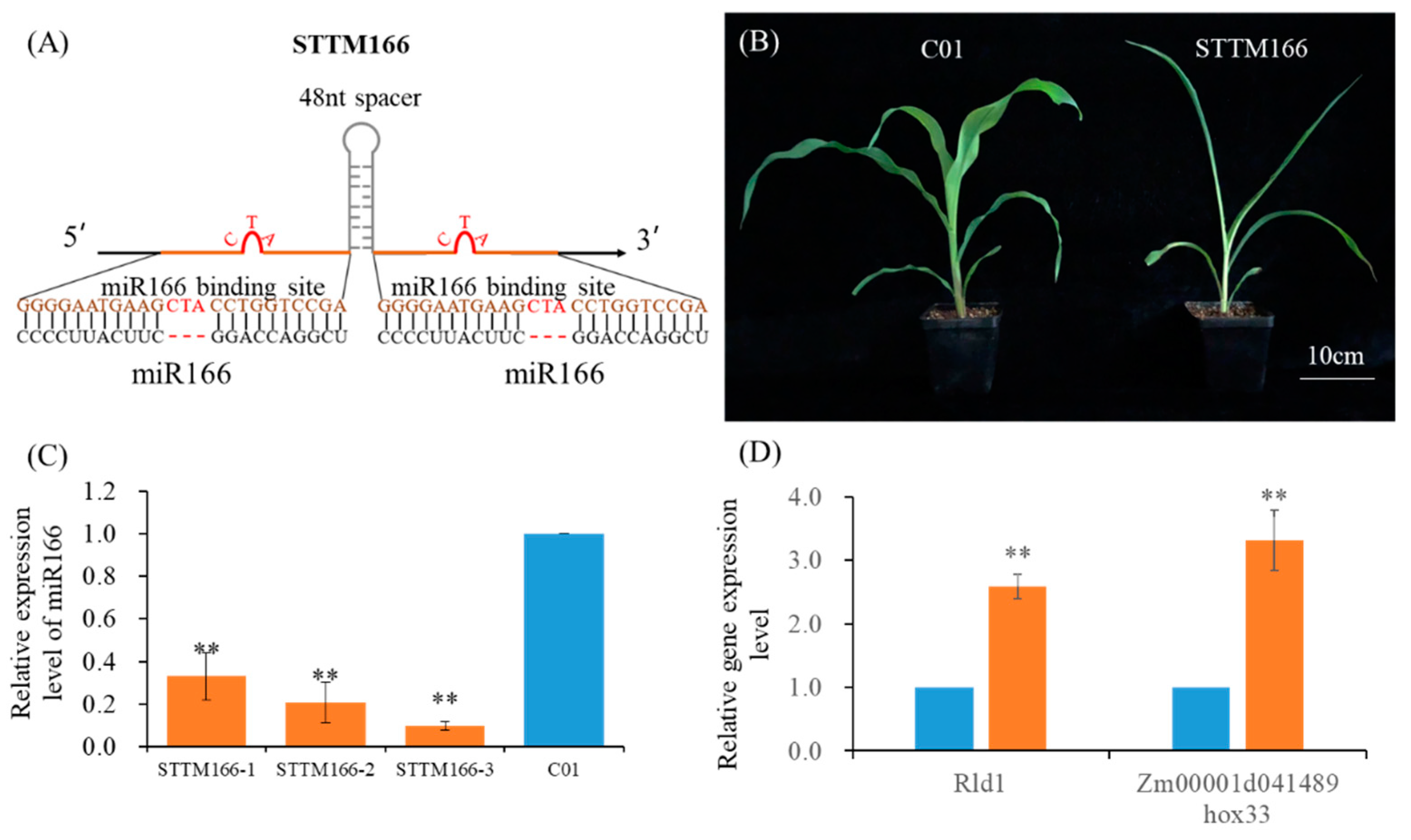
2.1. Functional Blockage of Maize miR166 Family Using STTM Technology
2.2. miR166 Knockdown Mediates Maize Agronomic Traits Phenotypic Alterations
2.3. Maize STTM166 Displays Enhanced Abiotic Stresses Resistance
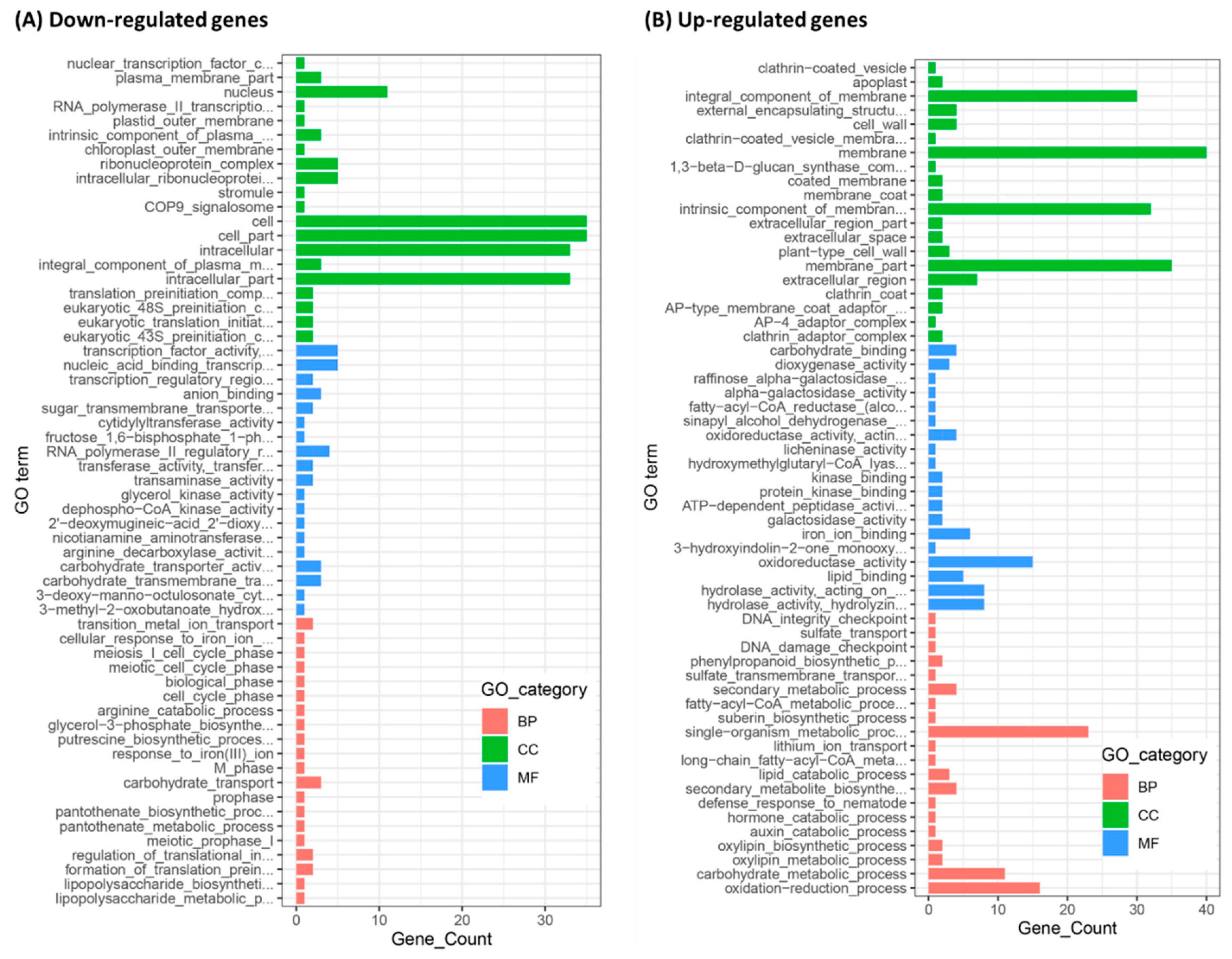
2.4. Differentially Expressed Genes (DEGs) in C01 and STTM166 Plants Revealed by RNA-Seq
2.5. Expressions of Key Regulatory Genes in ABA and Auxin Biogenesis and Signaling Pathways
3. Discussion
4. Materials and Methods
4.1. Maize STTM166 Construction
- STTM common real-PF: 5′-CATTTGGAGAGGACAGCCCAAG-3′
- STTM common real-PR: 5′-CTGGTGATTTCAGCGTACCGAA-3′
4.2. Plant Transformation, Transgenic Plants Screening, Genotyping and Phenotyping
4.3. Histological Analysis
4.4. Total RNA Extraction, Transcriptome Sequencing Analysis, qRT-PCR
4.5. Plant Hormone Content Measurement
4.6. Drought, Salt, Heat Stress Treatment
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| STTM | Short Tandem Target Mimic |
| miR166 | microRNA166 |
| HD-ZIP III | Class III homeodomain/Leu zipper |
| DEGs | Differentially expressed genes |
References
- Chen, X. Small RNAs in development-insights from plants. Curr. Opin. Genet. Dev. 2012, 22, 361–367. [Google Scholar] [CrossRef] [Green Version]
- Voinnet, O. Origin, biogenesis, and activity of plant microRNAs. Cell 2009, 136, 669–687. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Tang, C.; Yu, T.; Zhang, R.; Zheng, H.; Yan, W. MicroRNAs control mRNA fate by compartmentalization based on 3′ UTR length in male germ cells. Genome Biol. 2017, 18, 1–18. [Google Scholar] [CrossRef]
- Chen, X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 2004, 303, 2022–2025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, J.; Gu, Y.; Jia, X.; Kang, W.; Pan, S.; Tang, X.; Chen, X.; Tang, G. Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. Plant Cell 2012, 24, 415–427. [Google Scholar] [CrossRef] [Green Version]
- Song, X.; Li, Y.; Cao, X.; Qi, Y. MicroRNAs and their regulatory roles in plant-environment interactions. Annu. Rev. Plant Biol. 2019, 70, 489–525. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Axtell, M.J. Long-range genomic enrichment, sequencing, and assembly to determine unknown sequences flanking a known microRNA. PLoS ONE 2013, 8, e83721. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Chu, C. MicroRNAs in crop improvement: Fine-tuners for complex traits. Nat. Plants 2017, 3, 17077. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Teotia, S.; Tang, J.; Tang, G. Perspectives on microRNAs and phased small interfering RNAs in maize (Zea mays L.): Functions and big Impact on agronomic traits enhancement. Plants 2019, 8, 170. [Google Scholar] [CrossRef] [Green Version]
- Sakaguchi, J.; Watanabe, Y. miR165/166 and the development of land plants. Dev. Growth Differ. 2012, 54, 93–99. [Google Scholar] [CrossRef]
- Ding, Y.; Gong, S.; Wang, Y.; Wang, F.; Bao, H.; Sun, J.; Cai, C.; Yi, K.; Chen, Z.; Zhu, C. MicroRNA166 modulates Cadmium tolerance and accumulation in rice. Plant Physiol. 2018, 177, 1691–1703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maher, C.G.; Stein, L.; Ware, D. Evolution of Arabidopsis microRNA families through duplication events. Genome Res. 2006, 16, 510–519. [Google Scholar] [CrossRef] [Green Version]
- Taylor, R.S.; Tarver, J.E.; Hiscock, S.J.; Donoghue, P.C.J. Evolutionary history of plant microRNAs. Trends Plant Sci. 2014, 19, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, K.; Miyashima, S.; Sato-Nara, K.; Yamada, T.; Nakajima, K. Functionally diversified members of the MIR165/6 gene family regulate ovule morphogenesis in Arabidopsis thaliana. Plant Cell Physiol. 2018, 59, 1017–1026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McConnell, J.R.; Emery, J.; Eshed, Y.; Bao, N.; Bowman, J.; Barton, M.K. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 2001, 411, 709–713. [Google Scholar] [CrossRef] [PubMed]
- Ursache, R.; Miyashima, S.; Chen, Q.; Vaten, A.; Nakajima, K.; Carlsbecker, A.; Zhao, Y.; Helariutta, Y.; Dettmer, J. Tryptophan-dependent auxin biosynthesis is required for HD-ZIP III-mediated xylem patterning. Development 2014, 141, 1250–1260. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.; Roy, S.; Singh, S.; Das, S.S.; Gautam, V.; Yadav, S.; Kumar, A.; Singh, A.; Samantha, S.; Sarkar, A.K. Phytohormonal crosstalk modulates the expression of miR166/165s, target Class. III HD-ZIPs, and KANADI genes during root growth in Arabidopsis thaliana. Sci. Rep. 2017, 7, 3408. [Google Scholar] [CrossRef] [Green Version]
- Ramachandran, P.; Wang, G.; Augstein, F.; de Vries, J.; Carlsbecker, A. Continuous root xylem formation and vascular acclimation to water deficit involves endodermal ABA signalling via miR165. Development 2018, 145. [Google Scholar] [CrossRef] [Green Version]
- Todesco, M.; Rubiosomoza, I.; Pazares, J.; Weigel, D. A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet. 2010, 6. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Honda, M.; Zhu, H.; Zhang, Z.; Guo, X.; Li, T.; Li, Z.; Peng, X.; Nakajima, K.; Duan, L. Spatiotemporal sequestration of miR165/166 by Arabidopsis Argonaute10 promotes shoot apical meristem maintenance. Cell Rep. 2015, 10, 1819–1827. [Google Scholar] [CrossRef] [Green Version]
- Merelo, P.; Ram, H.; Caggiano, M.P.; Ohno, C.; Ott, F.; Straub, D.; Graeff, M.; Cho, S.K.; Yang, S.W.; Wenkel, S. Regulation of MIR165/166 by class II and class III homeodomain leucine zipper proteins establishes leaf polarity. Proc. Natl. Acad. Sci. USA 2016, 113, 11973–11978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, Q.; Wang, H. The role of HD-ZIP III transcription factors and miR165/166 in vascular development and secondary cell wall formation. Plant Signal. Behav. 2015, 10, e1078955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, X.; Ding, N.; Fan, W.; Yan, J.; Gu, Y.; Tang, X.; Li, R.; Tang, G. Functional plasticity of miR165/166 in plant development revealed by small tandem target mimic. Plant Sci. 2015, 233, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Carlsbecker, A.; Lee, J.Y.; Roberts, C.J.; Dettmer, J.; Lehesranta, S.; Zhou, J.; Lindgren, O.; Moreno-Risueno, M.A.; Vaten, A.; Thitamadee, S.; et al. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 2010, 465, 316–321. [Google Scholar] [CrossRef] [Green Version]
- Yan, J.; Zhao, C.; Zhou, J.; Yang, Y.; Wang, P.; Zhu, X.; Tang, G.; Bressan, R.A.; Zhu, J.K. The miR165/166 mediated regulatory module plays critical roles in ABA homeostasis and response in Arabidopsis thaliana. PLoS Genet. 2016, 12, e1006416. [Google Scholar] [CrossRef]
- Kidner, C.A.; Martienssen, R.A. Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 2004, 428, 81–84. [Google Scholar] [CrossRef]
- Zhu, H.; Hu, F.; Wang, R.; Zhou, X.; Sze, S.H.; Liou, L.W.; Barefoot, A.; Dickman, M.; Zhang, X. Arabidopsis Argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell 2011, 145, 242–256. [Google Scholar] [CrossRef] [Green Version]
- Tang, G.; Yan, J.; Gu, Y.; Qiao, M.; Fan, R.; Mao, Y.; Tang, X. Construction of short tandem target mimic (STTM) to block the functions of plant and animal microRNAs. Methods 2012, 58, 118–125. [Google Scholar] [CrossRef] [Green Version]
- Xue, T.; Dai, X.; Wang, R.; Wang, J.; Liu, Z.; Xiang, F. ARGONAUTE10 inhibits in vitro shoot regeneration via repression of miR165/166 in Arabidopsis thaliana. Plant Cell Physiol. 2017, 58, 1789–1800. [Google Scholar] [CrossRef] [Green Version]
- D’Ario, M.; Griffiths-Jones, S.; Kim, M. Small RNAs: Big impact on plant development. Trends Plant Sci. 2017, 22, 1056–1068. [Google Scholar] [CrossRef] [Green Version]
- Yang, T.; Wang, Y.; Teotia, S.; Wang, Z.; Shi, C.; Sun, H.; Gu, Y.; Zhang, Z.; Tang, G. The interaction between miR160 and miR165/166 in the control of leaf development and drought tolerance in Arabidopsis. Sci. Rep. 2019, 9, 2832. [Google Scholar] [CrossRef] [PubMed]
- Peng, T.; Qiao, M.; Liu, H.; Teotia, S.; Zhang, Z.; Zhao, Y.; Wang, B.; Zhao, D.; Shi, L.; Zhang, C.; et al. A resource for inactivation of microRNAs using short tandem target mimic technology in model and crop plants. Mol. Plant 2018, 11, 1400–1417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Zhang, H.; Srivastava, A.K.; Pan, Y.; Bai, J.; Fang, J.; Shi, H.; Zhu, J.K. Knockdown of rice microRNA166 confers drought resistance by causing leaf rolling and altering stem xylem development. Plant Physiol. 2018, 176, 2082–2094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gore, M.A.; Chia, J.M.; Elshire, R.J.; Sun, Q.; Ersoz, E.S.; Hurwitz, B.L.; Peiffer, J.A.; Mcmullen, M.D.; Grills, G.; Rossibarra, J. A first-generation haplotype map of maize. Science 2009, 326, 1115–1117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juarez, M.T.; Kui, J.S.; Thomas, J.; Heller, B.A.; Timmermans, M.C. microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 2004, 428, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Juarez, M.T.; Twigg, R.W.; Timmermans, M.C.P. Specification of adaxial cell fate during maize leaf development. Development 2004, 131, 4533–4544. [Google Scholar] [CrossRef] [Green Version]
- Douglas, R.N.; Wiley, D.; Sarkar, A.K.; Springer, N.M.; Timmermans, M.C.P.; Scanlon, M.J. ragged seedling2 encodes an ARGONAUTE7-Like protein required for mediolateral expansion, but not dorsiventrality, of maize leaves. Plant Cell 2010, 22, 1441–1451. [Google Scholar] [CrossRef] [Green Version]
- Johnston, R.; Candela, H.; Hake, S.; Foster, T. The maize milkweed pod1 mutant reveals a mechanism to modify organ morphology. Genesis 2010, 48, 416–423. [Google Scholar] [CrossRef]
- Nogueira, F.T.S.; Chitwood, D.H.; Madi, S.; Ohtsu, K.; Schnable, P.S.; Scanlon, M.J.; Timmermans, M.C.P. Regulation of small RNA accumulation in the maize shoot apex. PLoS Genet. 2009, 5. [Google Scholar] [CrossRef] [Green Version]
- Itoh, J.-I.; Hibara, K.-I.; Sato, Y.; Nagato, Y. Developmental role and auxin responsiveness of Class III homeodomain leucine zipper gene family members in rice. Plant Physiol. 2008, 147, 1960–1975. [Google Scholar] [CrossRef] [Green Version]
- Nogueira, F.T.; Madi, S.; Chitwood, D.H.; Juarez, M.T.; Timmermans, M.C. Two small regulatory RNAs establish opposing fates of a developmental axis. Genes Dev. 2007, 21, 750–755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Zhang, J.; Yan, J.; Gou, F.; Mao, Y.; Tang, G.; Botella, J.R.; Zhu, J. Short tandem target mimic rice lines uncover functions of miRNAs in regulating important agronomic traits. Proc. Natl. Acad. Sci. USA 2017, 114, 5277–5282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujita, Y.; Fujita, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 2011, 124, 509–525. [Google Scholar] [CrossRef] [PubMed]
- Peleg, Z.; Blumwald, E. Hormone balance and abiotic stress tolerance in crop plants. Curr. Opin. Plant Biol. 2011, 14, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.H.; Park, C.M. MIR166/165 genes exhibit dynamic expression patterns in regulating shoot apical meristem and floral development in Arabidopsis. Planta 2007, 225, 1327–1338. [Google Scholar] [CrossRef] [PubMed]
- Boualem, A.; Laporte, P.; Jovanovic, M.; Laffont, C.; Plet, J.; Combier, J.P.; Niebel, A.; Crespi, M.; Frugier, F. MicroRNA166 controls root and nodule development in Medicago truncatula. Plant J. Cell Mol. Biol. 2008, 54, 876–887. [Google Scholar] [CrossRef]
- Wang, T.; Li, C.; Wu, Z.; Jia, Y.; Wang, H.; Sun, S.; Mao, C.; Wang, X. Abscisic acid regulates auxin homeostasis in rice root tips to promote root hair elongation. Front. Plant Sci. 2017, 8, 1121. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [Green Version]
- Vishwakarma, K.; Upadhyay, N.; Kumar, N.; Yadav, G.; Singh, J.; Mishra, R.K.; Kumar, V.; Verma, R.; Upadhyay, R.G.; Pandey, M.; et al. Abscisic acid signaling and abiotic stress tolerance in plants: A review on current knowledge and future prospects. Front. Plant Sci. 2017, 8, 161. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.H.; Wang, H. Molecular mechanisms forvascular development and secondary cell wall formation. Front. Plant Sci. 2016, 7, 356. [Google Scholar] [CrossRef] [Green Version]
- Kubo, M.; Udagawa, M.; Nishikubo, N.; Horiguchi, G.; Yamaguchi, M.; Ito, J.; Mimura, T.; Fukuda, H.; Demura, T. Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 2005, 19, 1855–1860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruzicka, K.; Ursache, R.; Hejatko, J.; Helariutta, Y. Xylem development—From the cradle to the grave. New Phytol. 2015, 207, 519–535. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.K.; Kubo, M.; Zhong, R.; Demura, T.; Ye, Z.H. Overexpression of miR165 affects apical meristem formation, organ polarity establishment and vascular development in Arabidopsis. Plant Cell Physiol. 2007, 48, 391–404. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Jung, J.-H.; Reyes, J.L.; Kim, Y.-S.; Kim, S.-Y.; Chung, K.-S.; Kim, J.A.; Lee, M.; Lee, Y.; Narry Kim, V.; et al. microRNA-directed cleavage of ATHB15 mRNA regulates vascular development in Arabidopsis inflorescence stems. Plant J. Cell Mol. Biol. 2005, 42, 84–94. [Google Scholar] [CrossRef] [Green Version]
- Vollbrecht, E.; Schmidt, R.J. Development of the Inflorescences. In Handbook of Maize: Its Biology; Bennetzen, J.L., Hake, S.C., Eds.; Springer: New York, NY, USA, 2009. [Google Scholar]
- McSteen, P.; Hake, S. barren inflorescence2 regulates axillary meristem development in the maize inflorescence. Development 2001, 128, 2881–2891. [Google Scholar]
- Taguchi-Shiobara, F.; Yuan, Z.; Hake, S.; Jackson, D. The fasciated ear2 gene encodes a leucine-rich repeat receptor-like protein that regulates shoot meristem proliferation in maize. Genes Dev. 2001, 15, 2755–2766. [Google Scholar] [CrossRef] [Green Version]
- Bommert, P.; Nagasawa, N.S.; Jackson, D. Quantitative variation in maize kernel row number is controlled by the FASCIATED EAR2 locus. Nat. Genet. 2013, 45, 334–337. [Google Scholar] [CrossRef]
- Chuck, G.; Whipple, C.; Jackson, D.; Hake, S. The maize SBP-box transcription factor encoded by tasselsheath4 regulates bract development and the establishment of meristem boundaries. Development 2010, 137, 1243–1250. [Google Scholar] [CrossRef] [Green Version]
- Chuck, G.S.; Brown, P.J.; Meeley, R.; Hake, S. Maize SBP-box transcription factors unbranched2 and unbranched3 affect yield traits by regulating the rate of lateral primordia initiation. Proc. Natl. Acad. Sci. USA 2014, 111, 18775–18780. [Google Scholar] [CrossRef] [Green Version]
- Chuck, G.; Meeley, R.; Hake, S. Floral meristem initiation and meristem cell fate are regulated by the maize AP2 genes ids1 and sid1. Development 2008, 135, 3013–3019. [Google Scholar] [CrossRef] [Green Version]
- Nagasaki, H.; Itoh, J.; Hayashi, K.; Hibara, K.; Satoh-Nagasawa, N.; Nosaka, M.; Mukouhata, M.; Ashikari, M.; Kitano, H.; Matsuoka, M.; et al. The small interfering RNA production pathway is required for shoot meristem initiation in rice. Proc. Natl. Acad. Sci. USA 2007, 104, 14867–14871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, W.; Xiang, X.; Zhai, L.; Zhang, D.; Cao, Z.; Liu, L.; Zhang, Z. AGO18b negatively regulates determinacy of spikelet meristems on the tassel central spike in maize. J. Integr. Plant Biol. 2018, 60, 65–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Morgan, M.; Carlson, M.; Tenenbaum, D.; Arora, S. AnnotationHub: Client to access AnnotationHub Resources. R Package Version. 2017. Available online: https://bioconductor.org/packages/release/bioc/html/AnnotationHub.html (accessed on 11 December 2020).
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Forcat, S.; Bennett, M.H.; Mansfield, J.W.; Grant, M.R. A rapid and robust method for simultaneously measuring changes in the phytohormones ABA, JA and SA in plants following biotic and abiotic stress. Plant Methods 2008, 4, 16. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Wang, H.; Liu, S.; Ferjani, A.; Li, J.; Yan, J.; Yang, X.; Qin, F. Genetic variation in ZmVPP1 contributes to drought tolerance in maize seedlings. Nat. Genet. 2016, 48, 1233–1241. [Google Scholar] [CrossRef]
- Ding, D.; Xiao, Z.; Xiao, H.; Xia, T.; Zheng, Y.; Qiu, F. Revelation of the early responses of salt tolerance in maize via SSH libraries. Genes Genom 2012, 34, 265–273. [Google Scholar] [CrossRef]








| Gene | logFC | p-Value | C01-FPKM | STTM166-FPKM | Description |
|---|---|---|---|---|---|
| Up-regulated genes | |||||
| Zm00001d040952 | 7.96 | 4.86 × 10−10 | 0.03 | 8.26 | Homeobox-leucine zipper protein TF1 |
| Zm00001d013699 | 5.52 | 3.07 × 10−21 | 1.91 | 103.01 | Homeobox-leucine zipper protein HOX32 |
| Zm00001d036638 | 4.9 | 1.32 × 10−4 | 0.1 | 2.79 | MYB family transcription factor |
| Zm00001d021777 | 4.55 | 1.01 × 10−5 | 0.19 | 5.5 | Vacuolar protein 8-like |
| Zm00001d026510 | 4.38 | 1.77 × 10−3 | 0.1 | 1.92 | Transcription factor IBH1 |
| Zm00001d041489 | 3.74 | 8.30 × 10−7 | 0.43 | 5.61 | Homeobox-leucine zipper protein HOX33 |
| Zm00001d036242 | 3.71 | 5.39 × 10−3 | 0.25 | 3.11 | Flowering locus T protein |
| Zm00001d031061 | 3.46 | 2.66 × 10−4 | 0.19 | 2.03 | Homeobox-leucine zipper protein HOX33 |
| Zm00001d033518 | 3.22 | 5.02 × 10−3 | 0.06 | 0.54 | Protein Argonaute 12-like |
| GRMZM2G083717 | 3.19 | 6.80 × 10−3 | 0.26 | 2.51 | Probable WRKY transcription factor 14 |
| Zm00001d037343 | 3.15 | 1.98 × 10−4 | 1.35 | 10.35 | ABA-induced protein |
| Zm00001d027957 | 2.95 | 5.41 × 10−3 | 3.83 | 33.24 | MADS-box transcription factor 47-like isoform x1 |
| Zm00001d033246 | 2.87 | 2.13 × 10−9 | 5.16 | 36.31 | Homeobox-leucine zipper protein HOX32 |
| AC212859.3_FG008, NAC103 | 2.85 | 1.30 × 10−3 | 0.73 | 5.21 | NAC domain-containing protein 7 |
| Zm00001d035211 | 2.4 | 7.24 × 10−4 | 3.58 | 15.93 | E3 ubiquitin-protein ligase RGLG2 |
| Zm00001d027317, rld2 | 2.15 | 7.00 × 10−6 | 3.78 | 16.54 | Homeobox-leucine zipper protein HOX10 |
| Zm00001d036900 | 1.81 | 7.83 × 10−3 | 1.88 | 6. 41 | Cellulose synthase a catalytic subunit 11 |
| Zm00001d048527, rld1 | 1.59 | 2.34 × 10−3 | 2.96 | 8.84 | Homeobox-leucine zipper protein HOX10 |
| GRMZM2G079632 | 1.54 | 4.79 × 10−3 | 7.83 | 22.4 | NAC1 |
| GRMZM2G088964 | 1.52 | 7.89 × 10−3 | 3.84 | 11.48 | Probable potassium transporter 17 |
| Zm00001d042809 | 1.15 | 9.42 × 10−3 | 12.47 | 27.61 | Auxin transporter-like protein 1 |
| Down-regulated genes | |||||
| Zm00001d036613 | −6.73 | 1.98 × 10−21 | 48.8 | 0.45 | Receptor-like protein kinase AT3G47110 |
| Zm00001d050350, KAN3 | −5.16 | 2.11 × 10−4 | 3.71 | 0.09 | Probable transcription factor Rl9 |
| Zm00001d023311 | −5.1 | 4.28 × 10−20 | 17.08 | 0.5 | Disease resistance protein RGA3 |
| Zm00001d042062 | −4.72 | 1.49 × 10−7 | 20.16 | 0.8 | Transcription factor bHLH100-like |
| Zm00001d018667 | −3.42 | 5.64 × 10−4 | 7.95 | 0.71 | Mads-box transcription factor 15 |
| Zm00001d035343 | −3.37 | 1.73 × 10−3 | 6.17 | 0.5 | Wall-associated receptor kinase 2-like |
| Zm00001d032249, KAN1 | −3.30 | 1.13 × 10−4 | 5.76 | 0.56 | Probable transcription factor Rl9 |
| Zm00001d020495 | −2.87 | 6.20 × 10−4 | 14.87 | 2.03 | Probable WRKY transcription factor 40 |
| Zm00001d011847 | −2.04 | 3.02 × 10−3 | 267.89 | 68.32 | Transcription factor bHLH100-like |
| Zm00001d010752, ZCN8 | −1.79 | 4.14 × 10−4 | 91.07 | 26.51 | Flowering locus T like protein |
| Zm00001d03099, bZIP111 | −1.58 | 5.45 × 10−3 | 746.06 | 237.9 | bZIP transcription factor superfamily protein |
| GRMZM2G014558 | −1.42 | 9.32 × 10−3 | 123.94 | 47.05 | Cellulose synthase-like protein E6 |
| Zm00001d037354 | −1.38 | 3.00 × 10−3 | 31.73 | 12.35 | Calmodulin-binding heat-shock protein |
| Zm00001d004248 | −1.29 | 9.04 × 10−3 | 99.98 | 41.04 | Cytokinin-o-glucosyltransferase 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, N.; Yang, T.; Guo, Z.; Wang, Q.; Chai, M.; Wu, M.; Li, X.; Li, W.; Li, G.; Tang, J.; et al. Maize microRNA166 Inactivation Confers Plant Development and Abiotic Stress Resistance. Int. J. Mol. Sci. 2020, 21, 9506. https://doi.org/10.3390/ijms21249506
Li N, Yang T, Guo Z, Wang Q, Chai M, Wu M, Li X, Li W, Li G, Tang J, et al. Maize microRNA166 Inactivation Confers Plant Development and Abiotic Stress Resistance. International Journal of Molecular Sciences. 2020; 21(24):9506. https://doi.org/10.3390/ijms21249506
Chicago/Turabian StyleLi, Na, Tianxiao Yang, Zhanyong Guo, Qiusheng Wang, Mao Chai, Mingbo Wu, Xiaoqi Li, Weiya Li, Guangxian Li, Jihua Tang, and et al. 2020. "Maize microRNA166 Inactivation Confers Plant Development and Abiotic Stress Resistance" International Journal of Molecular Sciences 21, no. 24: 9506. https://doi.org/10.3390/ijms21249506






