Epigenetic Regulation of the Hippocampus, with Special Reference to Radiation Exposure
Abstract
:1. Introduction
1.1. Epigenetics
1.2. DNA Methylation
1.3. Histone Modifications
1.4. Non-Coding RNA
2. Epigenetic Regulation of Hippocampal Neurogenesis
3. Epigenetic Regulation of Hippocampal Principal Neurons
4. Epigenetic Regulation of Hippocampal Interneurons
5. Epigenetic Regulation of Hippocampal Glial Cells (Astrocytes, Microglia and Oligodendrocytes)
5.1. Astrocytes
5.2. Microglia
5.3. Oligodendrocytes
6. Radiation-Induced Epigenetic Changes in the Hippocampus
7. Concluding Remarks
Funding
Acknowledgments
Conflicts of Interest
References
- Knierim, J.J. The hippocampus. Curr. Biol. 2015, 25, R1116–R1121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tatu, L.; Vuillier, F. Structure and vascularization of the human hippocampus. In The Hippocampus in Clinical Neuroscience; Karger Publishers: Basel, Switzerland, 2014; pp. 18–25. [Google Scholar]
- Fink, G. Stress: Physiology, Biochemistry, and Pathology: Handbook of Stress Series; Academic Press: New York, NY, USA, 2019; Volume 3. [Google Scholar]
- Squire, L.R.; Genzel, L.; Wixted, J.T.; Morris, R.G. Memory consolidation. Cold Spring Harb. Perspect. Biol. 2015, 7, a021766. [Google Scholar] [CrossRef] [PubMed]
- Bledsoe, J.C. Effects of cranial radiation on structural and functional brain development in pediatric brain tumors. J. Pediatr. Neuropsychol. 2016, 2, 3–13. [Google Scholar] [CrossRef] [Green Version]
- Yang, B.; Ren, B.X.; Tang, F.R. Prenatal irradiation–induced brain neuropathology and cognitive impairment. Brain Dev. 2017, 39, 10–22. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.R.; Loke, W.K.; Khoo, B.C. Low-dose or low-dose-rate ionizing radiation–induced bioeffects in animal models. J. Radiat. Res. 2017, 58, 165–182. [Google Scholar] [CrossRef]
- Amador-Arjona, A.; Cimadamore, F.; Huang, C.-T.; Wright, R.; Lewis, S.; Gage, F.H.; Terskikh, A.V. SOX2 primes the epigenetic landscape in neural precursors enabling proper gene activation during hippocampal neurogenesis. Proc. Natl. Acad. Sci. USA 2015, 112, E1936–E1945. [Google Scholar] [CrossRef] [Green Version]
- Mews, P.; Donahue, G.; Drake, A.M.; Luczak, V.; Abel, T.; Berger, S.L. Acetyl-CoA synthetase regulates histone acetylation and hippocampal memory. Nature 2017, 546, 381–386. [Google Scholar] [CrossRef] [Green Version]
- Zhong, H.; Rong, J.; Zhu, C.; Liang, M.; Li, Y.; Zhou, R. Epigenetic modifications of GABAergic interneurons contributes to the deficits in adult hippocampus neurogenesis and depression-like behavior in prenatally stressed mice. Int. J. Neuropsychopharmacol. 2020, 23, 274–285. [Google Scholar] [CrossRef]
- Covic, M.; Karaca, E.; Lie, D. Epigenetic regulation of neurogenesis in the adult hippocampus. Heredity 2010, 105, 122–134. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, Y.; Abe, H.; Nakajima, K.; Ideta-Otsuka, M.; Igarashi, K.; Woo, G.-H.; Yoshida, T.; Shibutani, M. Aberrant epigenetic gene regulation in GABAergic interneuron subpopulations in the hippocampal dentate gyrus of mouse offspring following developmental exposure to hexachlorophene. Toxicol. Sci. 2018, 163, 13–25. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, T.; Nakajima, K.; Masubuchi, Y.; Ito, Y.; Kikuchi, S.; Ideta-Ohtsuka, M.; Woo, G.-H.; Yoshida, T.; Igarashi, K.; Shibutani, M. Aberrant epigenetic gene regulation in hippocampal neurogenesis of mouse offspring following maternal exposure to 3, 3’-iminodipropionitrile. J. Toxicol. Sci. 2019, 44, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.J.; Sun, A.B.; Liu, L. Epigenetic modulation during hippocampal development. Biomed. Rep. 2018, 9, 463–473. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Wang, B.; Wang, H.; Meng, L.; Zhao, Q.; Li, X.; Xin, Y.; Jiang, X. Radiation-Induced Normal Tissue Damage: Oxidative Stress and Epigenetic Mechanisms. Oxid. Med. Cell. Longev. 2019, 2019, 3010342. [Google Scholar] [CrossRef] [PubMed]
- Meiliana, A.; Dewi, N.M.; Wijaya, A. Nutritional Influences on Epigenetics, Aging and Disease. Indones. Biomed. J. 2019, 11, 16–29. [Google Scholar] [CrossRef]
- Cui, D.; Xu, X. DNA methyltransferases, DNA methylation, and age-associated cognitive function. Int. J. Mol. Sci. 2018, 19, 1315. [Google Scholar] [CrossRef] [Green Version]
- John, R.M.; Rougeulle, C. Developmental epigenetics: Phenotype and the flexible epigenome. Front. Cell Dev. Biol. 2018, 6, 130. [Google Scholar] [CrossRef]
- Jones, P.A.; Issa, J.-P.J.; Baylin, S. Targeting the cancer epigenome for therapy. Nat. Rev. Genet. 2016, 17, 630. [Google Scholar] [CrossRef]
- Schachtschneider, K.M.; Welge, M.E.; Auvil, L.S.; Chaki, S.; Rund, L.A.; Madsen, O.; Elmore, M.R.; Johnson, R.W.; Groenen, M.A.; Schook, L.B. Altered hippocampal epigenetic regulation underlying reduced cognitive development in response to early life environmental insults. Genes 2020, 11, 162. [Google Scholar] [CrossRef] [Green Version]
- Nebbioso, A.; Tambaro, F.P.; Dell’Aversana, C.; Altucci, L. Cancer epigenetics: Moving forward. PLoS Genet. 2018, 14, e1007362. [Google Scholar] [CrossRef] [Green Version]
- Berson, A.; Nativio, R.; Berger, S.L.; Bonini, N.M. Epigenetic regulation in neurodegenerative diseases. Trends Neurosci. 2018, 41, 587–598. [Google Scholar] [CrossRef]
- Stoccoro, A.; Coppedè, F. Role of epigenetics in Alzheimer’s disease pathogenesis. Neurodegener. Dis. Manag. 2018, 8, 181–193. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.F.; Levine, R.L. Genetic and epigenetic determinants of AML pathogenesis. In Seminars in Hematology; Elsevier: Amsterdam, The Netherlands, 2019. [Google Scholar]
- Moore-Morris, T.; van Vliet, P.P.; Andelfinger, G.; Puceat, M. Role of epigenetics in cardiac development and congenital diseases. Physiol. Rev. 2018, 98, 2453–2475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, J.; Sun, J.; Ming, G.l.; Song, H. Epigenetic regulation of neurogenesis in the adult mammalian brain. Eur. J. Neurosci. 2011, 33, 1087–1093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kotandeniya, D.; Seiler, C.; Fernandez, J.; Pujari, S.; Curwick, L.; Murphy, K.; Wickramaratne, S.; Yan, S.; Murphy, D.; Sham, Y.Y. Can 5-methylcytosine analogues with extended alkyl side chains guide DNA methylation? Chem. Commun. 2018, 54, 1061–1064. [Google Scholar] [CrossRef] [PubMed]
- Lim, W.-J.; Kim, K.H.; Kim, J.-Y.; Jeong, S.; Kim, N. Identification of DNA-methylated CpG islands associated with gene silencing in the adult body tissues of the Ogye chicken using RNA-Seq and reduced representation bisulfite sequencing. Front. Genet. 2019, 10, 346. [Google Scholar] [CrossRef]
- Podobinska, M.; Szablowska-Gadomska, I.; Augustyniak, J.; Sandvig, I.; Sandvig, A.; Buzanska, L. Epigenetic modulation of stem cells in neurodevelopment: The role of methylation and acetylation. Front. Cell. Neurosci. 2017, 11, 23. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Zhang, Y. TET-mediated active DNA demethylation: Mechanism, function and beyond. Nat. Rev. Genet. 2017, 18, 517. [Google Scholar] [CrossRef]
- Niehrs, C.; Schäfer, A. Active DNA demethylation by Gadd45 and DNA repair. Trends Cell Biol. 2012, 22, 220–227. [Google Scholar] [CrossRef]
- Zhang, R.-p.; Shao, J.-z.; Xiang, L.-x. GADD45A protein plays an essential role in active DNA demethylation during terminal osteogenic differentiation of adipose-derived mesenchymal stem cells. J. Biol. Chem. 2011, 286, 41083–41094. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Gu, T.-P.; Weber, A.R.; Shen, J.-Z.; Li, B.-Z.; Xie, Z.-G.; Yin, R.; Guo, F.; Liu, X.; Tang, F. Gadd45a promotes DNA demethylation through TDG. Nucleic Acids Res. 2015, 43, 3986–3997. [Google Scholar] [CrossRef]
- Zhou, L.; Wang, W.; Yang, C.; Zeng, T.; Hu, M.; Wang, X.; Li, N.; Sun, K.; Wang, C.; Zhou, J. GADD45a promotes active DNA demethylation of the MMP-9 promoter via base excision repair pathway in AGEs-treated keratinocytes and in diabetic male rat skin. Endocrinology 2018, 159, 1172–1186. [Google Scholar] [CrossRef] [PubMed]
- Sultan, F.A.; Wang, J.; Tront, J.; Liebermann, D.A.; Sweatt, J.D. Genetic deletion of Gadd45b, a regulator of active DNA demethylation, enhances long-term memory and synaptic plasticity. J. Neurosci. 2012, 32, 17059–17066. [Google Scholar] [CrossRef] [PubMed]
- Labonté, B.; Jeong, Y.H.; Parise, E.; Issler, O.; Fatma, M.; Engmann, O.; Cho, K.-A.; Neve, R.; Nestler, E.J.; Koo, J.W. Gadd45b mediates depressive-like role through DNA demethylation. Sci. Rep. 2019, 9, 4615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cooper, G.M. Chromosomes and Chromatin. The Cell: A Molecular Approach, 2nd ed.; Sinauer Associates: Sunderland, MA, USA, 2000. [Google Scholar]
- Strahl, B.D.; Allis, C.D. The language of covalent histone modifications. Nature 2000, 403, 41–45. [Google Scholar] [CrossRef]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- Jambhekar, A.; Dhall, A.; Shi, Y. Roles and regulation of histone methylation in animal development. Nat. Rev. Mol. Cell Biol. 2019, 20, 625–641. [Google Scholar] [CrossRef]
- Hyun, K.; Jeon, J.; Park, K.; Kim, J. Writing, erasing and reading histone lysine methylations. Exp. Mol. Med. 2017, 49, e324. [Google Scholar] [CrossRef] [Green Version]
- Collins, B.E.; Sweatt, J.D.; Greer, C.B. Broad domains of histone 3 lysine 4 trimethylation are associated with transcriptional activation in CA1 neurons of the hippocampus during memory formation. Neurobiol. Learn. Mem. 2019, 161, 149–157. [Google Scholar] [CrossRef]
- Rossetto, D.; Avvakumov, N.; Côté, J. Histone phosphorylation: A chromatin modification involved in diverse nuclear events. Epigenetics 2012, 7, 1098–1108. [Google Scholar] [CrossRef] [Green Version]
- Cao, J.; Yan, Q. Histone ubiquitination and deubiquitination in transcription, DNA damage response, and cancer. Front. Oncol. 2012, 2, 26. [Google Scholar] [CrossRef] [Green Version]
- Shiio, Y.; Eisenman, R.N. Histone sumoylation is associated with transcriptional repression. Proc. Natl. Acad. Sci. USA 2003, 100, 13225–13230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palazzo, L.; Mikolčević, P.; Mikoč, A.; Ahel, I. ADP-ribosylation signalling and human disease. Open Biol. 2019, 9, 190041. [Google Scholar] [CrossRef] [Green Version]
- Srijyothi, L.; Ponne, S.; Prathama, T.; Ashok, C.; Baluchamy, S. Roles of Non-Coding RNAs in Transcriptional Regulation. Transcriptional and Post-Transcriptional Regulation; IntechOpen: Rijeka, Croatia, 2018; p. 55. [Google Scholar]
- MacFarlane, L.-A.; Murphy, P.R. MicroRNA: Biogenesis, function and role in cancer. Curr. Genom. 2010, 11, 537–561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Felekkis, K.; Touvana, E.; Stefanou, C.; Deltas, C. microRNAs: A newly described class of encoded molecules that play a role in health and disease. Hippokratia 2010, 14, 236. [Google Scholar] [PubMed]
- Cai, Y.; Yu, X.; Hu, S.; Yu, J. A brief review on the mechanisms of miRNA regulation. Genom. Proteom. Bioinf. 2009, 7, 147–154. [Google Scholar] [CrossRef] [Green Version]
- Ayub, A.L.P.; Papaiz, D.D.A.; da Silva Soares, R.; Jasiulionis, M.G. The Function of lncRNAs as Epigenetic Regulators. In Non-Coding RNAs; IntechOpen: Rijeka, Croatia, 2019. [Google Scholar]
- Wei, J.-W.; Huang, K.; Yang, C.; Kang, C.-S. Non-coding RNAs as regulators in epigenetics. Oncol. Rep. 2017, 37, 3–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuramochi-Miyagawa, S.; Watanabe, T.; Gotoh, K.; Totoki, Y.; Toyoda, A.; Ikawa, M.; Asada, N.; Kojima, K.; Yamaguchi, Y.; Ijiri, T.W. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008, 22, 908–917. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.; Deng, W.; Gage, F.H. Mechanisms and functional implications of adult neurogenesis. Cell 2008, 132, 645–660. [Google Scholar] [CrossRef] [Green Version]
- Altman, J. Are new neurons formed in the brains of adult mammals? Science 1962, 135, 1127–1128. [Google Scholar] [CrossRef] [Green Version]
- Eriksson, P.S.; Perfilieva, E.; Björk-Eriksson, T.; Alborn, A.-M.; Nordborg, C.; Peterson, D.A.; Gage, F.H. Neurogenesis in the adult human hippocampus. Nat. Med. 1998, 4, 1313–1317. [Google Scholar] [CrossRef]
- Knoth, R.; Singec, I.; Ditter, M.; Pantazis, G.; Capetian, P.; Meyer, R.P.; Horvat, V.; Volk, B.; Kempermann, G. Murine features of neurogenesis in the human hippocampus across the lifespan from 0 to 100 years. PLoS ONE 2010, 5, e8809. [Google Scholar] [CrossRef] [PubMed]
- Roy, N.S.; Wang, S.; Jiang, L.; Kang, J.; Benraiss, A.; Harrison-Restelli, C.; Fraser, R.A.; Couldwell, W.T.; Kawaguchi, A.; Okano, H. In vitro neurogenesis by progenitor cells isolated from the adult human hippocampus. Nat. Med. 2000, 6, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Balu, D.T.; Lucki, I. Adult hippocampal neurogenesis: Regulation, functional implications, and contribution to disease pathology. Neurosci. Biobehav. Rev. 2009, 33, 232–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, D.K.; Jang, M.-H.; Guo, J.U.; Kitabatake, Y.; Chang, M.-l.; Pow-Anpongkul, N.; Flavell, R.A.; Lu, B.; Ming, G.-l.; Song, H. Neuronal activity–induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science 2009, 323, 1074–1077. [Google Scholar] [CrossRef] [Green Version]
- Gao, Z.; Ure, K.; Ding, P.; Nashaat, M.; Yuan, L.; Ma, J.; Hammer, R.E.; Hsieh, J. The master negative regulator REST/NRSF controls adult neurogenesis by restraining the neurogenic program in quiescent stem cells. J. Neurosci. 2011, 31, 9772–9786. [Google Scholar] [CrossRef] [Green Version]
- MacDonald, J.L.; Roskams, A.J. Histone deacetylases 1 and 2 are expressed at distinct stages of neuro-glial development. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2008, 237, 2256–2267. [Google Scholar] [CrossRef]
- Galluzzi, L.; Vitale, I.; Aaronson, S.A.; Abrams, J.M.; Adam, D.; Agostinis, P.; Alnemri, E.S.; Altucci, L.; Amelio, I.; Andrews, D.W. Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018, 25, 486–541. [Google Scholar] [CrossRef]
- Montgomery, R.L.; Hsieh, J.; Barbosa, A.C.; Richardson, J.A.; Olson, E.N. Histone deacetylases 1 and 2 control the progression of neural precursors to neurons during brain development. Proc. Natl. Acad. Sci. USA 2009, 106, 7876–7881. [Google Scholar] [CrossRef] [Green Version]
- Niklison-Chirou, M.V.; Agostini, M.; Amelio, I.; Melino, G. Regulation of Adult Neurogenesis in Mammalian Brain. Int. J. Mol. Sci. 2020, 21, 4869. [Google Scholar] [CrossRef]
- Lim, D.A.; Huang, Y.-C.; Swigut, T.; Mirick, A.L.; Garcia-Verdugo, J.M.; Wysocka, J.; Ernst, P.; Alvarez-Buylla, A. Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature 2009, 458, 529–533. [Google Scholar] [CrossRef] [Green Version]
- Boyer, L.A.; Plath, K.; Zeitlinger, J.; Brambrink, T.; Medeiros, L.A.; Lee, T.I.; Levine, S.S.; Wernig, M.; Tajonar, A.; Ray, M.K. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 2006, 441, 349–353. [Google Scholar] [CrossRef] [PubMed]
- Lim, D.A.; Alvarez-Buylla, A. The adult ventricular–subventricular zone (V-SVZ) and olfactory bulb (OB) neurogenesis. Cold Spring Harb. Perspect. Biol. 2016, 8, a018820. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, J.; Eisch, A.J. Epigenetics, hippocampal neurogenesis, and neuropsychiatric disorders: Unraveling the genome to understand the mind. Neurobiol. Dis. 2010, 39, 73–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jobe, E.M.; Zhao, X. DNA methylation and adult neurogenesis. Brain Plast. 2017, 3, 5–26. [Google Scholar] [CrossRef] [Green Version]
- Noguchi, H.; Kimura, A.; Murao, N.; Matsuda, T.; Namihira, M.; Nakashima, K. Expression of DNMT1 in neural stem/precursor cells is critical for survival of newly generated neurons in the adult hippocampus. Neurosci. Res. 2015, 95, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Noguchi, H.; Murao, N.; Kimura, A.; Matsuda, T.; Namihira, M.; Nakashima, K. DNA methyltransferase 1 is indispensable for development of the hippocampal dentate gyrus. J. Neurosci. 2016, 36, 6050–6068. [Google Scholar] [CrossRef]
- Wu, H.; Coskun, V.; Tao, J.; Xie, W.; Ge, W.; Yoshikawa, K.; Li, E.; Zhang, Y.; Sun, Y.E. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science 2010, 329, 444–448. [Google Scholar] [CrossRef] [Green Version]
- Mohan, K.N. Stem cell models to investigate the role of DNA methylation machinery in development of neuropsychiatric disorders. Stem Cells Int. 2016, 2016, 4379425. [Google Scholar] [CrossRef] [Green Version]
- Schouten, M.; Bielefeld, P.; Garcia-Corzo, L.; Passchier, E.; Gradari, S.; Jungenitz, T.; Pons-Espinal, M.; Gebara, E.; Martín-Suárez, S.; Lucassen, P.J. Circadian glucocorticoid oscillations preserve a population of adult hippocampal neural stem cells in the aging brain. Mol. Psychiatry 2020, 25, 1382–1405. [Google Scholar] [CrossRef] [Green Version]
- Namihira, M.; Nakashima, K.; Taga, T. Developmental stage dependent regulation of DNA methylation and chromatin modification in a immature astrocyte specific gene promoter. FEBS Lett. 2004, 572, 184–188. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.; Hong, E.J.; Cohen, S.; Zhao, W.-n.; Ho, H.-y.H.; Schmidt, L.; Chen, W.G.; Lin, Y.; Savner, E.; Griffith, E.C. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron 2006, 52, 255–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinowich, K.; Hattori, D.; Wu, H.; Fouse, S.; He, F.; Hu, Y.; Fan, G.; Sun, Y.E. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science 2003, 302, 890–893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smrt, R.D.; Eaves-Egenes, J.; Barkho, B.Z.; Santistevan, N.J.; Zhao, C.; Aimone, J.B.; Gage, F.H.; Zhao, X. Mecp2 deficiency leads to delayed maturation and altered gene expression in hippocampal neurons. Neurobiol. Dis. 2007, 27, 77–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, X.; Ueba, T.; Christie, B.R.; Barkho, B.; McConnell, M.J.; Nakashima, K.; Lein, E.S.; Eadie, B.D.; Willhoite, A.R.; Muotri, A.R. Mice lacking methyl-CpG binding protein 1 have deficits in adult neurogenesis and hippocampal function. Proc. Natl. Acad. Sci. USA 2003, 100, 6777–6782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allan, A.M.; Liang, X.; Luo, Y.; Pak, C.; Li, X.; Szulwach, K.E.; Chen, D.; Jin, P.; Zhao, X. The loss of methyl-CpG binding protein 1 leads to autism-like behavioral deficits. Hum. Mol. Genet. 2008, 17, 2047–2057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szulwach, K.E.; Li, X.; Smrt, R.D.; Li, Y.; Luo, Y.; Lin, L.; Santistevan, N.J.; Li, W.; Zhao, X.; Jin, P. Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J. Cell Biol. 2010, 189, 127–141. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Teng, Z.-Q.; Santistevan, N.J.; Szulwach, K.E.; Guo, W.; Jin, P.; Zhao, X. Epigenetic regulation of miR-184 by MBD1 governs neural stem cell proliferation and differentiation. Cell Stem Cell 2010, 6, 433–444. [Google Scholar] [CrossRef] [Green Version]
- Khandelwal, N.; Dey, S.K.; Chakravarty, S.; Kumar, A. miR-30 family miRNAs mediate the effect of chronic social defeat stress on hippocampal neurogenesis in mouse depression model. Front. Mol. Neurosci. 2019, 12, 188. [Google Scholar] [CrossRef] [Green Version]
- Antonini, D.; Russo, M.T.; De Rosa, L.; Gorrese, M.; Del Vecchio, L.; Missero, C. Transcriptional repression of miR-34 family contributes to p63-mediated cell cycle progression in epidermal cells. J. Investig. Dermatol. 2010, 130, 1249–1257. [Google Scholar] [CrossRef] [Green Version]
- Smrt, R.D.; Szulwach, K.E.; Pfeiffer, R.L.; Li, X.; Guo, W.; Pathania, M.; Teng, Z.Q.; Luo, Y.; Peng, J.; Bordey, A. MicroRNA miR-137 regulates neuronal maturation by targeting ubiquitin ligase mind bomb-1. Stem Cells 2010, 28, 1060–1070. [Google Scholar] [CrossRef] [Green Version]
- Han, J.; Kim, H.J.; Schafer, S.T.; Paquola, A.; Clemenson, G.D.; Toda, T.; Oh, J.; Pankonin, A.R.; Lee, B.S.; Johnston, S.T. Functional implications of miR-19 in the migration of newborn neurons in the adult brain. J. Exp. Med. 2016, 213, 667–675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deo, M.; Yu, J.Y.; Chung, K.H.; Tippens, M.; Turner, D.L. Detection of mammalian microRNA expression by in situ hybridization with RNA oligonucleotides. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2006, 235, 2538–2548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, C.; Kim, T.; Chang, K.T.; Min, K.T. DSCR1-mediated TET1 splicing regulates miR-124 expression to control adult hippocampal neurogenesis. EMBO J. 2019, 38, e101293. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Ambigapathy, G.; Keifer, J. MeCP2 regulates Tet1-catalyzed demethylation, CTCF binding, and learning-dependent alternative splicing of the BDNF gene in Turtle. eLife 2017, 6, e25384. [Google Scholar] [CrossRef]
- Pons-Espinal, M.; de Luca, E.; Marzi, M.J.; Beckervordersandforth, R.; Armirotti, A.; Nicassio, F.; Fabel, K.; Kempermann, G.; Tonelli, D.D.P. Synergic functions of miRNAs determine neuronal fate of adult neural stem cells. Stem Cell Rep. 2017, 8, 1046–1061. [Google Scholar] [CrossRef] [Green Version]
- Bielefeld, P.; Mooney, C.; Henshall, D.C.; Fitzsimons, C.P. miRNA-mediated regulation of adult hippocampal neurogenesis; implications for epilepsy. Brain Plast. 2017, 3, 43–59. [Google Scholar] [CrossRef] [Green Version]
- Stappert, L.; Klaus, F.; Brüstle, O. MicroRNAs engage in complex circuits regulating adult neurogenesis. Front. Neurosci. 2018, 12, 707. [Google Scholar] [CrossRef]
- Nicoll, R.A. A brief history of long-term potentiation. Neuron 2017, 93, 281–290. [Google Scholar] [CrossRef] [Green Version]
- Sun, C.; Yang, W.; Martin, J.; Tonegawa, S. Hippocampal neurons represent events as transferable units of experience. Nat. Neurosci. 2020, 23, 651–663. [Google Scholar] [CrossRef]
- Chen, Y.; Ozturk, N.C.; Zhou, F.C. DNA methylation program in developing hippocampus and its alteration by alcohol. PLoS ONE 2013, 8, e60503. [Google Scholar] [CrossRef]
- Karnay, A.; Karisetty, B.C.; Beaver, M.; Elefant, F. Hippocampal stimulation promotes intracellular Tip60 dynamics with concomitant genome reorganization and synaptic gene activation. Mol. Cell. Neurosci. 2019, 101, 103412. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.A.; Sweatt, J.D. Covalent modification of DNA regulates memory formation. Neuron 2007, 53, 857–869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marrocco, J.; Gray, J.D.; Kogan, J.F.; Einhorn, N.R.; O’Cinneide, E.M.; Rubin, T.G.; Carroll, T.S.; Schmidt, E.F.; McEwen, B.S. Early life stress restricts translational reactivity in CA3 neurons associated with altered stress responses in adulthood. Front. Behav. Neurosci. 2019, 13, 157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kerimoglu, C.; Agis-Balboa, R.C.; Kranz, A.; Stilling, R.; Bahari-Javan, S.; Benito-Garagorri, E.; Halder, R.; Burkhardt, S.; Stewart, A.F.; Fischer, A. Histone-methyltransferase MLL2 (KMT2B) is required for memory formation in mice. J. Neurosci. 2013, 33, 3452–3464. [Google Scholar] [CrossRef] [Green Version]
- Gupta, S.; Kim, S.Y.; Artis, S.; Molfese, D.L.; Schumacher, A.; Sweatt, J.D.; Paylor, R.E.; Lubin, F.D. Histone methylation regulates memory formation. J. Neurosci. 2010, 30, 3589–3599. [Google Scholar] [CrossRef]
- Jie, J.; Xu, X.; Xia, J.; Tu, Z.; Guo, Y.; Li, C.; Zhang, X.; Wang, H.; Song, W.; Xie, P. Memory impairment induced by borna disease virus 1 infection is associated with reduced H3K9 acetylation. Cell. Physiol. Biochem. 2018, 49, 381–394. [Google Scholar] [CrossRef]
- Jury, N.; Abarzua, S.; Diaz, I.; Guerra, M.V.; Ampuero, E.; Cubillos, P.; Martinez, P.; Herrera-Soto, A.; Arredondo, C.; Rojas, F. Widespread loss of the silencing epigenetic mark H3K9me3 in astrocytes and neurons along with hippocampal-dependent cognitive impairment in C9orf72 BAC transgenic mice. Clin. Epigenet. 2020, 12, 32. [Google Scholar] [CrossRef] [Green Version]
- Hu, E.; Du, H.; Zhu, X.; Wang, L.; Shang, S.; Wu, X.; Lu, H.; Lu, X. Beta-hydroxybutyrate promotes the expression of BDNF in hippocampal neurons under adequate glucose supply. Neuroscience 2018, 386, 315–325. [Google Scholar] [CrossRef]
- Gonzalez, M.C.; Radiske, A.; Cammarota, M. On the involvement of BDNF signaling in memory reconsolidation. Front. Cell. Neurosci. 2019, 13, 383. [Google Scholar] [CrossRef] [Green Version]
- Gu, X.; Xu, Y.; Xue, W.-Z.; Wu, Y.; Ye, Z.; Xiao, G.; Wang, H.-L. Interplay of miR-137 and EZH2 contributes to the genome-wide redistribution of H3K27me3 underlying the Pb-induced memory impairment. Cell Death Dis. 2019, 10, 671. [Google Scholar] [CrossRef]
- Jarome, T.J.; Perez, G.A.; Hauser, R.M.; Hatch, K.M.; Lubin, F.D. EZH2 methyltransferase activity controls Pten expression and mTOR signaling during fear memory reconsolidation. J. Neurosci. 2018, 38, 7635–7648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gafford, G.M.; Parsons, R.G.; Helmstetter, F.J. Consolidation and reconsolidation of contextual fear memory requires mammalian target of rapamycin-dependent translation in the dorsal hippocampus. Neuroscience 2011, 182, 98–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.R.; Ho, M.-F.; Vega, M.C.S.; Burne, T.H.; Chong, S. Prenatal ethanol exposure alters adult hippocampal VGLUT2 expression with concomitant changes in promoter DNA methylation, H3K4 trimethylation and miR-467b-5p levels. Epigenet. Chromatin 2015, 8, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pelkey, K.A.; Chittajallu, R.; Craig, M.T.; Tricoire, L.; Wester, J.C.; McBain, C.J. Hippocampal GABAergic inhibitory interneurons. Physiol. Rev. 2017, 97, 1619–1747. [Google Scholar] [CrossRef] [PubMed]
- Booker, S.A.; Vida, I. Morphological diversity and connectivity of hippocampal interneurons. Cell Tissue Res. 2018, 373, 619–641. [Google Scholar] [CrossRef] [Green Version]
- Ruzicka, W.B.; Subburaju, S.; Coyle, J.T.; Benes, F.M. Location matters: Distinct DNA methylation patterns in GABAergic interneuronal populations from separate microcircuits within the human hippocampus. Hum. Mol. Genet. 2018, 27, 254–265. [Google Scholar] [CrossRef]
- Bond, A.M.; VanGompel, M.J.; Sametsky, E.A.; Clark, M.F.; Savage, J.C.; Disterhoft, J.F.; Kohtz, J.D. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat. Neurosci. 2009, 12, 1020. [Google Scholar] [CrossRef] [Green Version]
- Magloire, V.; Mercier, M.S.; Kullmann, D.M.; Pavlov, I. GABAergic interneurons in seizures: Investigating causality with optogenetics. Neuroscientist 2019, 25, 344–358. [Google Scholar] [CrossRef] [Green Version]
- Matrisciano, F.; Tueting, P.; Dalal, I.; Kadriu, B.; Grayson, D.R.; Davis, J.M.; Nicoletti, F.; Guidotti, A. Epigenetic modifications of GABAergic interneurons are associated with the schizophrenia-like phenotype induced by prenatal stress in mice. Neuropharmacology 2013, 68, 184–194. [Google Scholar] [CrossRef] [Green Version]
- Satta, R.; Maloku, E.; Zhubi, A.; Pibiri, F.; Hajos, M.; Costa, E.; Guidotti, A. Nicotine decreases DNA methyltransferase 1 expression and glutamic acid decarboxylase 67 promoter methylation in GABAergic interneurons. Proc. Natl. Acad. Sci. USA 2008, 105, 16356–16361. [Google Scholar] [CrossRef] [Green Version]
- Von Bartheld, C.S.; Bahney, J.; Herculano-Houzel, S. The search for true numbers of neurons and glial cells in the human brain: A review of 150 years of cell counting. J. Comp. Neurol. 2016, 524, 3865–3895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jäkel, S.; Dimou, L. Glial cells and their function in the adult brain: A journey through the history of their ablation. Front. Cell. Neurosci. 2017, 11, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hertz, L.; Chen, Y. all 3 types of glial cells are important for memory formation. Front. Integr. Neurosci. 2016, 10, 31. [Google Scholar] [CrossRef] [Green Version]
- Saw, G.; Krishna, K.; Gupta, N.; Soong, T.W.; Mallilankaraman, K.; Sajikumar, S.; Dheen, S.T. Epigenetic regulation of microglial phosphatidylinositol 3-kinase pathway involved in long-term potentiation and synaptic plasticity in rats. Glia 2020, 68, 656–669. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, T.; Avignone, E.; Nägerl, U.V. Induction of hippocampal long-term potentiation increases the morphological dynamics of microglial processes and prolongs their contacts with dendritic spines. Sci. Rep. 2016, 6, 32422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keyser, D.O.; Pellmar, T.C. Synaptic transmission in the hippocampus: Critical role for glial cells. Glia 1994, 10, 237–243. [Google Scholar] [CrossRef]
- Ota, Y.; Zanetti, A.T.; Hallock, R.M. The role of astrocytes in the regulation of synaptic plasticity and memory formation. Neural Plast. 2013, 2013, 185463. [Google Scholar] [CrossRef] [Green Version]
- Koeppen, J.; Nguyen, A.Q.; Nikolakopoulou, A.M.; Garcia, M.; Hanna, S.; Woodruff, S.; Figueroa, Z.; Obenaus, A.; Ethell, I.M. Functional consequences of synapse remodeling following astrocyte-specific regulation of Ephrin-B1 in the adult hippocampus. J. Neurosci. 2018, 38, 5710–5726. [Google Scholar] [CrossRef] [Green Version]
- Zuchero, J.B.; Barres, B.A. Glia in mammalian development and disease. Development 2015, 142, 3805–3809. [Google Scholar] [CrossRef] [Green Version]
- Bellot-Saez, A.; Kekesi, O.; Morley, J.W.; Buskila, Y. Astrocytic modulation of neuronal excitability through K+ spatial buffering. Neurosci. Biobehav. Rev. 2017, 77, 87–97. [Google Scholar] [CrossRef]
- Matias, I.; Morgado, J.; Gomes, F.C.A. Astrocyte heterogeneity: Impact to brain aging and disease. Front. Aging Neurosci. 2019, 11, 59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moroni, R.F.; Inverardi, F.; Regondi, M.C.; Pennacchio, P.; Frassoni, C. Developmental expression of Kir4.1 in astrocytes and oligodendrocytes of rat somatosensory cortex and hippocampus. Int. J. Dev. Neurosci. 2015, 47, 198–205. [Google Scholar] [CrossRef] [PubMed]
- Nwaobi, S.E.; Olsen, M.L. Correlating gene-specific DNA methylation changes with expression and transcriptional activity of astrocytic KCNJ10 (Kir4. 1). JoVE J. Vis. Exp. 2015, e52406. [Google Scholar] [CrossRef]
- Myung, N.-H.; Zhu, X.; Kruman, I.I.; Castellani, R.J.; Petersen, R.B.; Siedlak, S.L.; Perry, G.; Smith, M.A.; Lee, H.-g. Evidence of DNA damage in Alzheimer disease: Phosphorylation of histone H2AX in astrocytes. Age 2008, 30, 209–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, C.; Zhu, L.; Ma, R.; Ren, J.; Wang, J.; Gao, S.; Yang, D.; Ning, K.; Ling, B.; Lu, B. Astrocytic miR-324-5p is essential for synaptic formation by suppressing the secretion of CCL5 from astrocytes. Cell Death Dis. 2019, 10, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, P.; Bin, H.; Chen, W. Inhibition of microRNA-103a inhibits the activation of astrocytes in hippocampus tissues and improves the pathological injury of neurons of epilepsy rats by regulating BDNF. Cancer Cell Int. 2019, 19, 109. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Liu, L.; Shi, J.; Tan, M.; Xiong, J.; Li, X.; Hu, Q.; Yi, Z. MicroRNA-34b mediates hippocampal astrocyte apoptosis in a rat model of recurrent seizures. BMC Neurosci. 2016, 17, 56. [Google Scholar] [CrossRef] [Green Version]
- Korotkov, A.; Broekaart, D.W.; Banchaewa, L.; Pustjens, B.; van Scheppingen, J.; Anink, J.J.; Baayen, J.C.; Idema, S.; Gorter, J.A.; van Vliet, E.A. microRNA-132 is overexpressed in glia in temporal lobe epilepsy and reduces the expression of pro-epileptogenic factors in human cultured astrocytes. Glia 2020, 68, 60–75. [Google Scholar] [CrossRef] [Green Version]
- Griffiths, B.B.; Ouyang, Y.-B.; Xu, L.; Sun, X.; Giffard, R.G.; Stary, C.M. Postinjury Inhibition of miR-181a Promotes Restoration of Hippocampal CA1 Neurons after Transient Forebrain Ischemia in Rats. eNeuro 2019, 6. [Google Scholar] [CrossRef]
- Norden, D.M.; Godbout, J.P. Microglia of the aged brain: Primed to be activated and resistant to regulation. Neuropathol. Appl. Neurobiol. 2013, 39, 19–34. [Google Scholar] [CrossRef]
- Nimmerjahn, A.; Kirchhoff, F.; Helmchen, F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science 2005, 308, 1314–1318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wake, H.; Moorhouse, A.J.; Jinno, S.; Kohsaka, S.; Nabekura, J. Resting microglia directly monitor the functional state of synapses in vivo and determine the fate of ischemic terminals. J. Neurosci. 2009, 29, 3974–3980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diaz-Aparicio, I.; Paris, I.; Sierra-Torre, V.; Plaza-Zabala, A.; Rodríguez-Iglesias, N.; Márquez-Ropero, M.; Beccari, S.; Huguet, P.; Abiega, O.; Alberdi, E. Microglia actively remodel adult hippocampal neurogenesis through the phagocytosis secretome. J. Neurosci. 2020, 40, 1453–1482. [Google Scholar] [CrossRef] [PubMed]
- Gemma, C.; Bachstetter, A.D. The role of microglia in adult hippocampal neurogenesis. Front. Cell. Neurosci. 2013, 7, 229. [Google Scholar] [CrossRef] [Green Version]
- De Lucia, C.; Rinchon, A.; Olmos-Alonso, A.; Riecken, K.; Fehse, B.; Boche, D.; Perry, V.H.; Gomez-Nicola, D. Microglia regulate hippocampal neurogenesis during chronic neurodegeneration. Brain Behav. Immun. 2016, 55, 179–190. [Google Scholar] [CrossRef] [Green Version]
- Sierra, A.; Encinas, J.M.; Deudero, J.J.; Chancey, J.H.; Enikolopov, G.; Overstreet-Wadiche, L.S.; Tsirka, S.E.; Maletic-Savatic, M. Microglia shape adult hippocampal neurogenesis through apoptosis-coupled phagocytosis. Cell Stem Cell 2010, 7, 483–495. [Google Scholar] [CrossRef] [Green Version]
- Butovsky, O.; Weiner, H.L. Microglial signatures and their role in health and disease. Nat. Rev. Neurosci. 2018, 19, 622–635. [Google Scholar] [CrossRef]
- Man, H.-Y.; Wang, Q.; Lu, W.-Y.; Ju, W.; Ahmadian, G.; Liu, L.; D’Souza, S.; Wong, T.; Taghibiglou, C.; Lu, J. Activation of PI3-kinase is required for AMPA receptor insertion during LTP of mEPSCs in cultured hippocampal neurons. Neuron 2003, 38, 611–624. [Google Scholar] [CrossRef] [Green Version]
- Parkhurst, C.N.; Yang, G.; Ninan, I.; Savas, J.N.; Yates III, J.R.; Lafaille, J.J.; Hempstead, B.L.; Littman, D.R.; Gan, W.-B. Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell 2013, 155, 1596–1609. [Google Scholar] [CrossRef] [Green Version]
- Zhou, L.-J.; Peng, J.; Xu, Y.-N.; Zeng, W.-J.; Zhang, J.; Wei, X.; Mai, C.-L.; Lin, Z.-J.; Liu, Y.; Murugan, M. Microglia are indispensable for synaptic plasticity in the spinal dorsal horn and chronic pain. Cell Rep. 2019, 27, 3844–3859.e6. [Google Scholar] [CrossRef] [Green Version]
- Hu, Q.-P.; Mao, D.-A. Histone deacetylase inhibitor SAHA attenuates post-seizure hippocampal microglia TLR4/MYD88 signaling and inhibits TLR4 gene expression via histone acetylation. BMC Neurosci. 2016, 17, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Datta, M.; Staszewski, O.; Raschi, E.; Frosch, M.; Hagemeyer, N.; Tay, T.L.; Blank, T.; Kreutzfeldt, M.; Merkler, D.; Ziegler-Waldkirch, S. Histone deacetylases 1 and 2 regulate microglia function during development, homeostasis, and neurodegeneration in a context-dependent manner. Immunity 2018, 48, 514–529.e6. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.-Z.; Zhang, D.-F.; Yang, J.-T.; Liu, Q.-H.; Wang, Y.-R.; Wang, W.-S. Overexpression of microRNA-301b accelerates hippocampal microglia activation and cognitive impairment in mice with depressive-like behavior through the NF-κB signaling pathway. Cell Death Dis. 2019, 10, 316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woodbury, M.E.; Freilich, R.W.; Cheng, C.J.; Asai, H.; Ikezu, S.; Boucher, J.D.; Slack, F.; Ikezu, T. miR-155 is essential for inflammation-induced hippocampal neurogenic dysfunction. J. Neurosci. 2015, 35, 9764–9781. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Dong, R.; Lu, Y.; Zhou, Y.; Li, K.; Zhang, Z.; Peng, M. MicroRNA-146a protects against cognitive decline induced by surgical trauma by suppressing hippocampal neuroinflammation in mice. Brain Behav. Immun. 2019, 78, 188–201. [Google Scholar] [CrossRef]
- Varol, D.; Mildner, A.; Blank, T.; Shemer, A.; Barashi, N.; Yona, S.; David, E.; Boura-Halfon, S.; Segal-Hayoun, Y.; Chappell-Maor, L. Dicer deficiency differentially impacts microglia of the developing and adult brain. Immunity 2017, 46, 1030–1044.e8. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Hong, W.; Wang, X.; Zhang, P.; Körner, H.; Tu, J.; Wei, W. MicroRNAs in Microglia: How do MicroRNAs Affect Activation, Inflammation, Polarization of Microglia and Mediate the Interaction Between Microglia and Glioma? Front. Mol. Neurosci. 2019, 12, 125. [Google Scholar] [CrossRef]
- Han, C.-L.; Ge, M.; Liu, Y.-P.; Zhao, X.-M.; Wang, K.-L.; Chen, N.; Meng, W.-J.; Hu, W.; Zhang, J.-G.; Li, L. LncRNA H19 contributes to hippocampal glial cell activation via JAK/STAT signaling in a rat model of temporal lobe epilepsy. J. Neuroinflamm. 2018, 15, 103. [Google Scholar] [CrossRef] [Green Version]
- Phipps, A.J.; Vickers, J.C.; Taberlay, P.C.; Woodhouse, A. Neurofilament-labeled pyramidal neurons and astrocytes are deficient in DNA methylation marks in Alzheimer’s disease. Neurobiol. Aging 2016, 45, 30–42. [Google Scholar] [CrossRef]
- Cheray, M.; Joseph, B. Epigenetics control microglia plasticity. Front. Cell. Neurosci. 2018, 12, 243. [Google Scholar] [CrossRef] [Green Version]
- Bercury, K.K.; Macklin, W.B. Dynamics and mechanisms of CNS myelination. Dev. Cell 2015, 32, 447–458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bergles, D.E.; Richardson, W.D. Oligodendrocyte development and plasticity. Cold Spring Harb. Perspect. Biol. 2016, 8, a020453. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Choi, Y.-S.; Obrietan, K.; Dudek, S.M. Synaptic plasticity (and the lack thereof) in hippocampal CA2 neurons. J. Neurosci. 2007, 27, 12025–12032. [Google Scholar] [CrossRef] [PubMed]
- Tzakis, N.; Holahan, M.R. Social memory and the role of the hippocampal CA2 region. Front. Behav. Neurosci. 2019, 13, 233. [Google Scholar] [CrossRef]
- Yamazaki, Y. Oligodendrocyte Physiology Modulating Axonal Excitability and Nerve Conduction. In Myelin; Springer: Singapore, 2019; pp. 123–144. [Google Scholar]
- Li, Q.; Mathena, R.P.; Xu, J.; O’Rukevwe, N.E.; Wen, J.; Mintz, C.D. Early postnatal exposure to isoflurane disrupts oligodendrocyte development and myelin formation in the mouse hippocampus. Anesthesiol. J. Am. Soc. Anesthesiol. 2019, 131, 1077–1091. [Google Scholar] [CrossRef]
- Chomyk, A.M.; Volsko, C.; Tripathi, A.; Deckard, S.A.; Trapp, B.D.; Fox, R.J.; Dutta, R. DNA methylation in demyelinated multiple sclerosis hippocampus. Sci. Rep. 2017, 7, 8696. [Google Scholar] [CrossRef] [Green Version]
- Pedre, X.; Mastronardi, F.; Bruck, W.; López-Rodas, G.; Kuhlmann, T.; Casaccia, P. Changed histone acetylation patterns in normal-appearing white matter and early multiple sclerosis lesions. J. Neurosci. 2011, 31, 3435–3445. [Google Scholar] [CrossRef] [Green Version]
- Koreman, E.; Sun, X.; Lu, Q.R. Chromatin remodeling and epigenetic regulation of oligodendrocyte myelination and myelin repair. Mol. Cell. Neurosci. 2018, 87, 18–26. [Google Scholar] [CrossRef]
- Dai, J.; Bercury, K.K.; Jin, W.; Macklin, W.B. Olig1 acetylation and nuclear export mediate oligodendrocyte development. J. Neurosci. 2015, 35, 15875–15893. [Google Scholar] [CrossRef]
- Lin, S.-T.; Huang, Y.; Zhang, L.; Heng, M.Y.; Ptáček, L.J.; Fu, Y.-H. MicroRNA-23a promotes myelination in the central nervous system. Proc. Natl. Acad. Sci. USA 2013, 110, 17468–17473. [Google Scholar] [CrossRef] [Green Version]
- Dutta, R.; Chomyk, A.M.; Chang, A.; Ribaudo, M.V.; Deckard, S.A.; Doud, M.K.; Edberg, D.D.; Bai, B.; Li, M.; Baranzini, S.E. Hippocampal demyelination and memory dysfunction are associated with increased levels of the neuronal microRNA miR-124 and reduced AMPA receptors. Ann. Neurol. 2013, 73, 637–645. [Google Scholar] [CrossRef] [PubMed]
- Dawson, M.R.; Polito, A.; Levine, J.M.; Reynolds, R. NG2-expressing glial progenitor cells: An abundant and widespread population of cycling cells in the adult rat CNS. Mol. Cell. Neurosci. 2003, 24, 476–488. [Google Scholar] [CrossRef]
- Ong, W.; Levine, J. A light and electron microscopic study of NG2 chondroitin sulfate proteoglycan-positive oligodendrocyte precursor cells in the normal and kainate-lesioned rat hippocampus. Neuroscience 1999, 92, 83–95. [Google Scholar] [CrossRef]
- Du, X.; Zhang, Z.; Zhou, H.; Zhou, J. Differential Modulators of NG2-Glia Differentiation into Neurons and Glia and Their Crosstalk. Neuroscience 2020. [Google Scholar] [CrossRef] [PubMed]
- Merrifield, M.; Kovalchuk, O. Epigenetics in radiation biology: A new research frontier. Front. Genet. 2013, 4, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kovalchuk, A.; Kolb, B. Low dose radiation effects on the brain–from mechanisms and behavioral outcomes to mitigation strategies. Cell Cycle 2017, 16, 1266–1270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Impey, S.; Pelz, C.; Tafessu, A.; Marzulla, T.; Turker, M.S.; Raber, J. Proton irradiation induces persistent and tissue-specific DNA methylation changes in the left ventricle and hippocampus. BMC Genom. 2016, 17, 273. [Google Scholar] [CrossRef] [Green Version]
- Torres, E.R.S.; Hall, R.; CHOI, J.; Impey, S.; Pelz, C.; Lindner, J.R.; Stevens, J.F.; Raber, J. Integrated metabolomics-DNA methylation analysis reveals significant long-term tissue-dependent directional alterations in aminoacyl-tRNA biosynthesis in the left ventricle of the heart and hippocampus following proton irradiation. Front. Mol. Biosci. 2019, 6, 77. [Google Scholar] [CrossRef]
- Impey, S.; Jopson, T.; Pelz, C.; Tafessu, A.; Fareh, F.; Zuloaga, D.; Marzulla, T.; Riparip, L.-K.; Stewart, B.; Rosi, S. Short-and long-term effects of 56 Fe irradiation on cognition and hippocampal DNA methylation and gene expression. BMC Genom. 2016, 17, 825. [Google Scholar] [CrossRef] [Green Version]
- Impey, S.; Jopson, T.; Pelz, C.; Tafessu, A.; Fareh, F.; Zuloaga, D.; Marzulla, T.; Riparip, L.-K.; Stewart, B.; Rosi, S. Bi-directional and shared epigenomic signatures following proton and 56 Fe irradiation. Sci. Rep. 2017, 7, 10227. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, F.; Takashima, N.; Murayama, A.; Inokuchi, K. Decreased postnatal neurogenesis in the hippocampus combined with stress experience during adolescence is accompanied by an enhanced incidence of behavioral pathologies in adult mice. Mol. Brain 2008, 1, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Acharya, M.M.; Al Anoud, D.B.; Kawashita, T.; Allen, B.D.; Syage, A.R.; Nguyen, T.H.; Yoon, N.; Giedzinski, E.; Yu, L.; Parihar, V.K. Epigenetic determinants of space radiation-induced cognitive dysfunction. Sci. Rep. 2017, 7, 42885. [Google Scholar] [CrossRef] [PubMed]
- Alaghband, Y.; Bredy, T.W.; Wood, M.A. The role of active DNA demethylation and Tet enzyme function in memory formation and cocaine action. Neurosci. Lett. 2016, 625, 40–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marshall, P.; Bredy, T.W. Cognitive neuroepigenetics: The next evolution in our understanding of the molecular mechanisms underlying learning and memory? NPJ Sci. Learn. 2016, 1, 16014. [Google Scholar] [CrossRef] [Green Version]
- Rudenko, A.; Tsai, L.-H. Epigenetic modifications in the nervous system and their impact upon cognitive impairments. Neuropharmacology 2014, 80, 70–82. [Google Scholar] [CrossRef]
- Kaas, G.A.; Zhong, C.; Eason, D.E.; Ross, D.L.; Vachhani, R.V.; Ming, G.-l.; King, J.R.; Song, H.; Sweatt, J.D. TET1 controls CNS 5-methylcytosine hydroxylation, active DNA demethylation, gene transcription, and memory formation. Neuron 2013, 79, 1086–1093. [Google Scholar] [CrossRef] [Green Version]
- Ji, S.; Sun, R.; Zhang, L.; Tian, Y. Whole Brain Irradiation-induced Decrease of Histone H3 Acetylation in Hippocampus of Rats. Int. J. Radiat. Oncol. Biol. Phys. 2012, 84, S665. [Google Scholar] [CrossRef]
- Kang, S.; Son, Y.; Lee, S.; Kim, J.; Kim, J.-C.; Kim, J.-S.; Jung, U.; Kim, S.-H.; Yang, M.; Moon, C. Changes in epigenetic markers, DNMT1 and HDAC1/2, in the adult mouse hippocampus after cranial irradiation. Neurosci. Lett. 2017, 657, 113–119. [Google Scholar] [CrossRef]
- Nelson, E.D.; Kavalali, E.T.; Monteggia, L.M. Activity-dependent suppression of miniature neurotransmission through the regulation of DNA methylation. J. Neurosci. 2008, 28, 395–406. [Google Scholar] [CrossRef] [Green Version]
- Shi, L.; Adams, M.M.; Long, A.; Carter, C.C.; Bennett, C.; Sonntag, W.E.; Nicolle, M.M.; Robbins, M.; D’Agostino, R., Jr.; Brunso-Bechtold, J.K. Spatial learning and memory deficits after whole-brain irradiation are associated with changes in NMDA receptor subunits in the hippocampus. Radiat. Res. 2006, 166, 892–899. [Google Scholar] [CrossRef] [Green Version]
- Wu, P.H.; Coultrap, S.; Pinnix, C.; Davies, K.D.; Tailor, R.; Ang, K.K.; Browning, M.D.; Grosshans, D.R. Radiation induces acute alterations in neuronal function. PLoS ONE 2012, 7, e37677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parihar, V.K.; Pasha, J.; Tran, K.K.; Craver, B.M.; Acharya, M.M.; Limoli, C.L. Persistent changes in neuronal structure and synaptic plasticity caused by proton irradiation. Brain Struct. Funct. 2015, 220, 1161–1171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sokolova, I.V.; Schneider, C.J.; Bezaire, M.; Soltesz, I.; Vlkolinsky, R.; Nelson, G.A. Proton radiation alters intrinsic and synaptic properties of CA1 pyramidal neurons of the mouse hippocampus. Radiat. Res. 2015, 183, 208–218. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-H.; Dudok, B.; Parihar, V.K.; Jung, K.-M.; Zöldi, M.; Kang, Y.-J.; Maroso, M.; Alexander, A.L.; Nelson, G.A.; Piomelli, D. Neurophysiology of space travel: Energetic solar particles cause cell type-specific plasticity of neurotransmission. Brain Struct. Funct. 2017, 222, 2345–2357. [Google Scholar] [CrossRef] [Green Version]
- Koturbash, I.; Zemp, F.; Kolb, B.; Kovalchuk, O. Sex-specific radiation-induced microRNAome responses in the hippocampus, cerebellum and frontal cortex in a mouse model. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2011, 722, 114–118. [Google Scholar] [CrossRef]
- Kempf, S.J.; Casciati, A.; Buratovic, S.; Janik, D.; von Toerne, C.; Ueffing, M.; Neff, F.; Moertl, S.; Stenerlöw, B.; Saran, A. The cognitive defects of neonatally irradiated mice are accompanied by changed synaptic plasticity, adult neurogenesis and neuroinflammation. Mol. Neurodegener. 2014, 9, 57. [Google Scholar] [CrossRef] [Green Version]
- Cui, M.; Xiao, H.; Li, Y.; Dong, J.; Luo, D.; Li, H.; Feng, G.; Wang, H.; Fan, S. Total abdominal irradiation exposure impairs cognitive function involving miR-34a-5p/BDNF axis. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2017, 1863, 2333–2341. [Google Scholar] [CrossRef]
- Zhao, L.; Sun, C.; Xiong, L.; Yang, Y.; Gao, Y.; Wang, L.; Zuo, H.; Xu, X.; Dong, J.; Zhou, H. MicroRNAs: Novel mechanism involved in the pathogenesis of microwave exposure on rats’ hippocampus. J. Mol. Neurosci. 2014, 53, 222–230. [Google Scholar] [CrossRef]
- Kumar, R.; Deshmukh, P.S.; Sharma, S.; Banerjee, B.D. Effect of mobile phone signal radiation on epigenetic modulation in the hippocampus of Wistar rat. Environ. Res. 2020, 192, 110297. [Google Scholar] [CrossRef]
- Zhang, J.; Li, J.; Zhu, Y.; Miao, Z.; Tian, Y. Forced running exercise mitigates radiation-induced cognitive deficits via regulated DNA hydroxymethylation. Epigenomics 2020, 12, 385–396. [Google Scholar] [CrossRef]
- Ji, S.; Tian, Y.; Lu, Y.; Sun, R.; Ji, J.; Zhang, L.; Duan, S. Irradiation-induced hippocampal neurogenesis impairment is associated with epigenetic regulation of bdnf gene transcription. Brain Res. 2014, 1577, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Harris, C.J.; Davis, B.A.; Zweig, J.A.; Nevonen, K.A.; Quinn, J.F.; Carbone, L.; Gray, N.E. Age-associated DNA methylation patterns are shared between the hippocampus and peripheral blood cells. Front. Genet. 2020, 11, 111. [Google Scholar] [CrossRef] [PubMed] [Green Version]

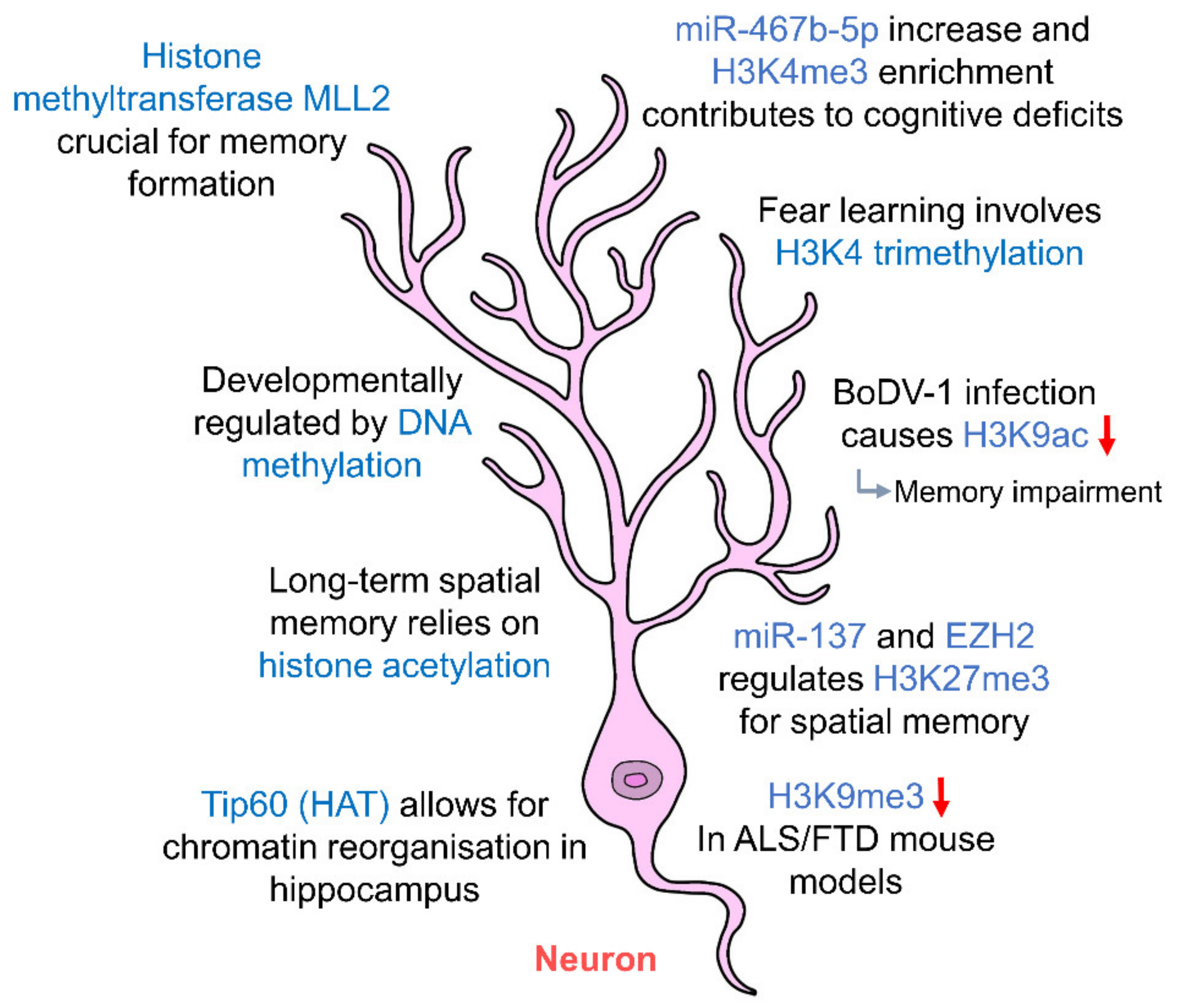

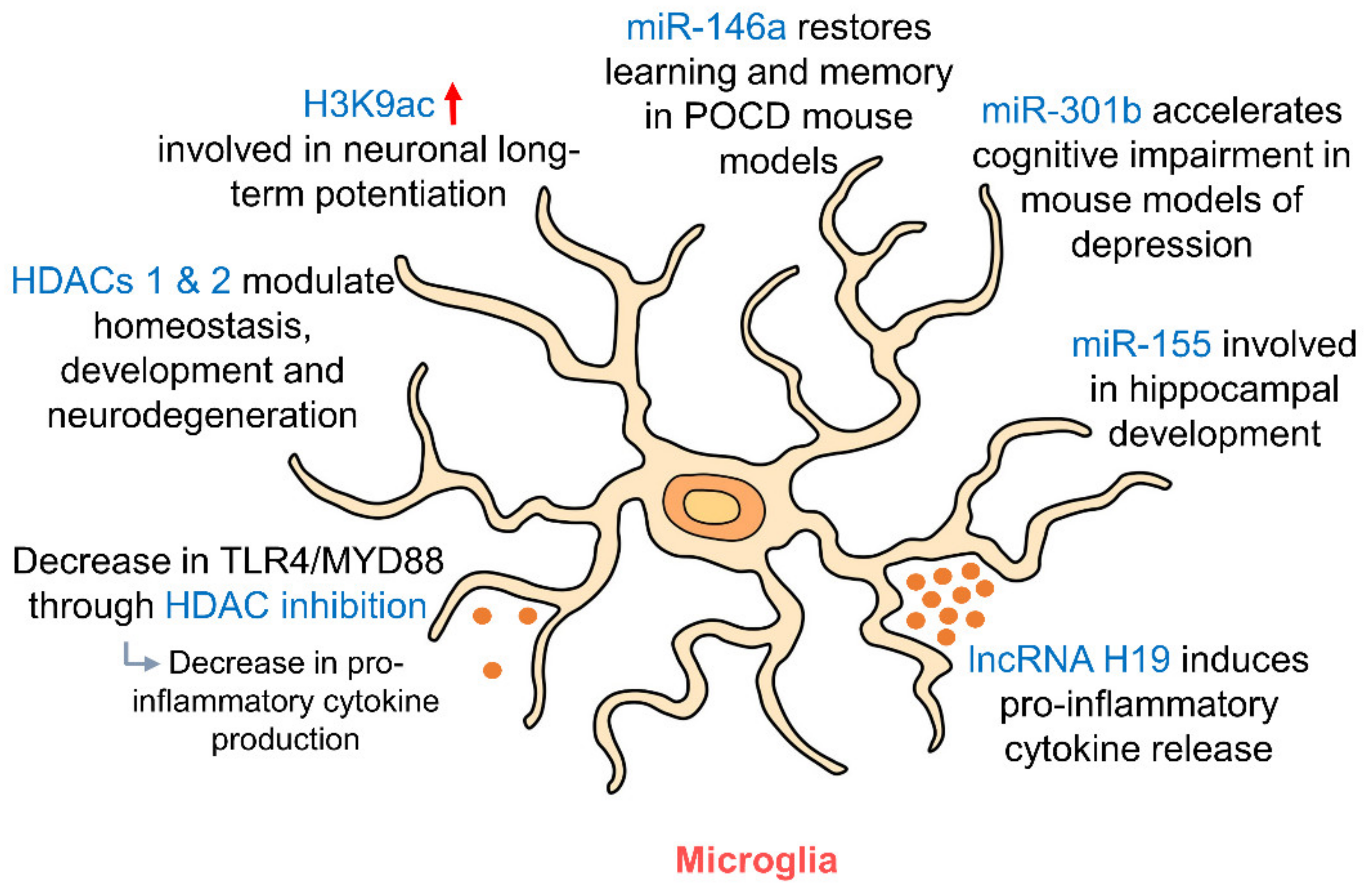

| Cell Type | DNA Methylation | Histone Modifications | Non-Coding RNAs (ncRNAs) |
|---|---|---|---|
| Principal neurons |
|
|
|
| Interneurons |
|
| |
| Astrocytes |
|
|
|
| Microglia |
|
|
|
| Oligodendrocytes |
|
|
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saw, G.; Tang, F.R. Epigenetic Regulation of the Hippocampus, with Special Reference to Radiation Exposure. Int. J. Mol. Sci. 2020, 21, 9514. https://doi.org/10.3390/ijms21249514
Saw G, Tang FR. Epigenetic Regulation of the Hippocampus, with Special Reference to Radiation Exposure. International Journal of Molecular Sciences. 2020; 21(24):9514. https://doi.org/10.3390/ijms21249514
Chicago/Turabian StyleSaw, Genevieve, and Feng Ru Tang. 2020. "Epigenetic Regulation of the Hippocampus, with Special Reference to Radiation Exposure" International Journal of Molecular Sciences 21, no. 24: 9514. https://doi.org/10.3390/ijms21249514
APA StyleSaw, G., & Tang, F. R. (2020). Epigenetic Regulation of the Hippocampus, with Special Reference to Radiation Exposure. International Journal of Molecular Sciences, 21(24), 9514. https://doi.org/10.3390/ijms21249514






