Genetic Factors in Antiphospholipid Syndrome: Preliminary Experience with Whole Exome Sequencing
Abstract
1. Introduction
2. Genetics of APS: What We Know
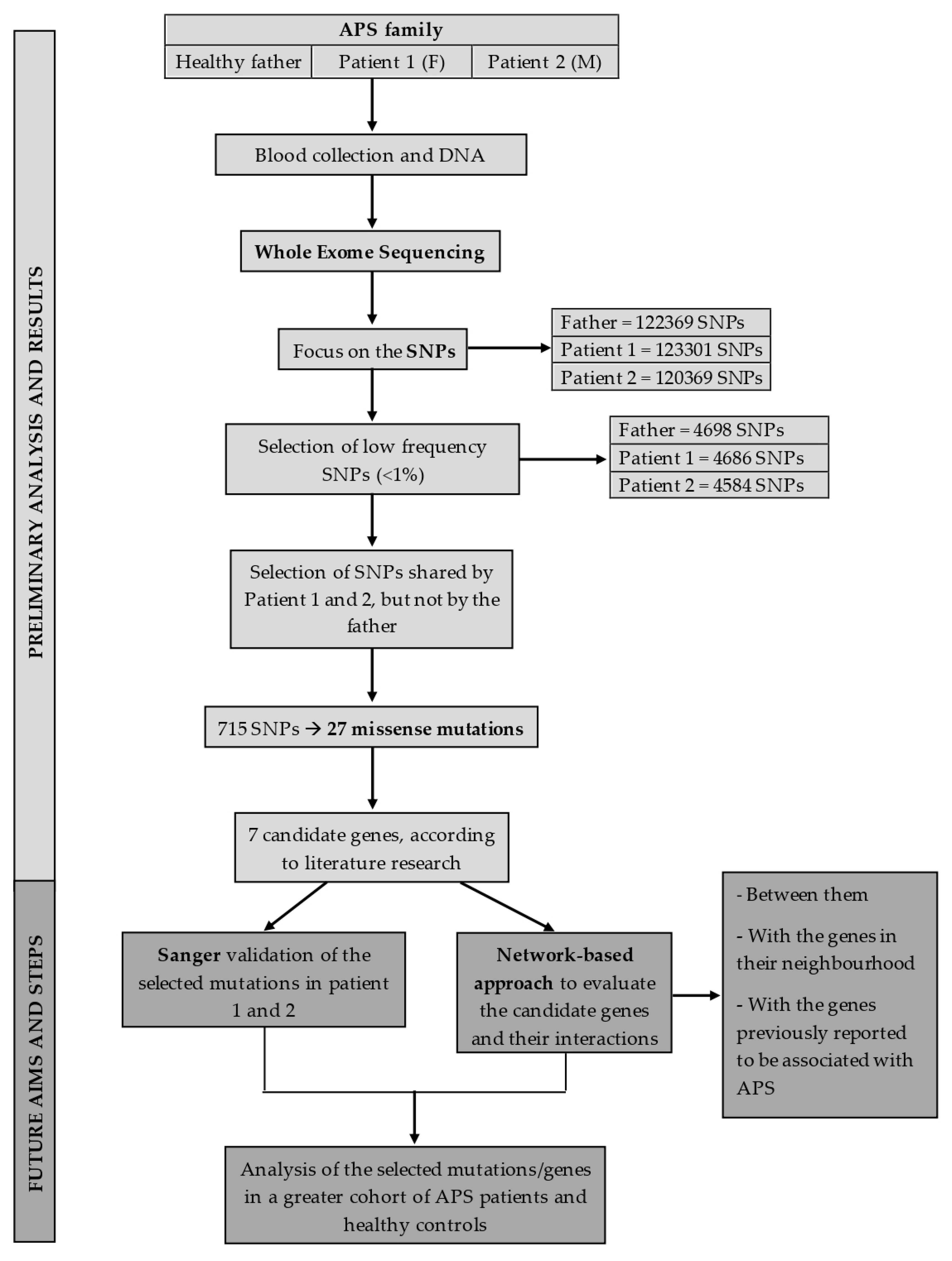
3. APS Family Study
3.1. Methods
3.1.1. Patients Selection
3.1.2. Exome Sequencing
3.1.3. Variants Filtering, Annotation, and Prioritization
3.2. Results
3.2.1. SNPs of Interest Found in Patient 1 and Patient 2
PLA2G6
HSPG2
BCL3
ZFAT
ATP2B2
CRTC3
ADCY3
3.2.2. Analysis of Genes Previously Associated with Thrombosis
3.2.3. Analysis of B2GPI Gene
4. Discussion and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| aCL | Anti-cardiolipin antibodies |
| ADCY3 | Adenylate cyclase 3 |
| aPL | Antiphospholipid Antibodies |
| APOH | Apolipoprotein H |
| APS | Antiphospholipid Syndrome |
| aPS/PT | Anti-phosphatidylserine/prothrombin antibodies |
| ATP2B2 | ATPase plasma membrane Ca2+ transporting 2 |
| BCL3 | BCL3 transcription coactivator |
| BLK | BLK proto-oncogene, Src family tyrosine kinase |
| BWA | Burrows–Wheeler Alignment |
| cAMP | Cyclic adenosine monophosphate |
| CAPS | Catastrophic Antiphospholipid Syndrome |
| CREB | cAMP response element-binding protein |
| CRTC3 | CREB regulated transcription coactivator 3 |
| eNOS | Endothelial nitric oxide synthase |
| FBG | Fibrinogen Beta chain |
| FLT1 | FMS-related tyrosine kinase 1 |
| F2 | Coagulation factor II |
| F2R | Coagulation factor II receptor |
| F2RL1 | Coagulation factor II receptor-like 1 |
| F3 | Coagulation factor III |
| F5 | Coagulation factor V |
| F13A1 | Coagulation factor XIII A chain |
| GATK | Genome Analysis Toolkit |
| GPIa/ITGA2 | Integrin subunit alpha 2 |
| GPIIIa/ITGB3 | Integrin subunit beta 3 |
| GP1BA | Glycoprotein Ib platelet subunit alpha |
| HLA | Human leukocytes antigen |
| HSPG2 | Heparan sulfate proteoglycan 2 |
| HUVEC | Human umbilical vein endothelial cells |
| IFNα | Interferon α |
| IGV | Integrative Genomics Viewer |
| iKB | Inhibitors of kB proteins |
| IRF5 | Interferon regulatory factor 5 |
| LA | Lupus Anticoagulant |
| LPS | Lipopolysaccharide |
| MTHFR | Methylenetetrahydrofolate reductase |
| NGS | Next-generation sequencing |
| PAPS | Primary Antiphospholipid Syndrome |
| PF4V1 | Platelet factor 4, variant 1 |
| F13A1 | Coagulation factor XIII A chain |
| MTHFR | Methylenetetrahydrofolate reductase |
| PLA2G6 | Phospholipase A2 group VI |
| PROCR | Protein C receptor |
| PROS1 | Protein S |
| PTPN22 | Protein tyrosine phosphatase non-receptor type 22 |
| SAPS | Secondary Antiphospholipid Syndrome |
| SELP | Selectin P |
| SERPINE1 | Serpin family E member 1 |
| SLE | Systemic Lupus Erythematosus |
| SLOs | Secondary lymphoid organs |
| SNP | Single nucleotide polymorphism |
| STAT4 | Signal transducer and activator of transcription 4 |
| TFPI | Tissue factor pathway inhibitor |
| TLR2 | Toll-like receptor 2 |
| TLR4 | Toll-like receptor 4 |
| TNF | Tumor necrosis factor |
| VEGFA | Vascular endothelial growth factor A |
| VSMCs | Vascular smooth muscle cells |
| WES | Whole exome sequencing |
| ZFAT | Zinc finger and AT-hook domain containing |
| ZPI | Protein Z-dependent inhibitor |
References
- Clark, K.E.N.; Giles, I. Antiphospholipid syndrome. Medicine 2018, 46, 118–125. [Google Scholar] [CrossRef]
- Sciascia, S.; Amigo, M.; Roccatello, D.; Khamashta, M. Diagnosing antiphospholipid. Nat. Rev. Rheumatol. 2017, 13, 548–560. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-García, V.; Ioannou, Y.; Fernández-Nebro, A.; Isenberg, D.A.; Giles, I.P. Examining the prevalence of non-criteria anti-phospholipid antibodies in patients with anti-phospholipid syndrome: A systematic review. Rheumatology 2015, 54, 2042–2050. [Google Scholar] [CrossRef] [PubMed]
- Sciascia, S.; Sanna, G.; Murru, V.; Roccatello, D.; Khamashta, M.A.; Bertolaccini, M.L. Anti-prothrombin (aPT) and anti-phosphatidylserine/prothrombin (aPS/PT) antibodies and the risk of thrombosis in the antiphospholipid syndrome a systematic review. Thromb. Haemost. 2013, 111, 354–364. [Google Scholar] [CrossRef]
- Asherson, R.A.; Cervera, R.; De Groot, P.G.; Erkan, D.; Boffa, M.C.; Piette, J.C.; Khamashta, M.A.; Shoenfeld, Y. Catastrophic Antiphospholipid Syndrome Registry Project Group. Catastrophic antiphospholipid syndrome: International consensus statement on classification criteria and treatment guidelines. Lupus 2003, 12, 530–534. [Google Scholar] [CrossRef]
- Teruel, M.; Alarcón-Riquelme, M.E. The genetic basis of systemic lupus erythematosus: What are the risk factors and what have we learned. J. Autoimmun. 2016, 74, 161–175. [Google Scholar] [CrossRef]
- Karami, J.; Aslani, S.; Jamshidi, A.; Garshasbi, M.; Mahmoudi, M. Genetic implications in the pathogenesis of rheumatoid arthritis; an updated review. Gene 2019, 702, 8–16. [Google Scholar] [CrossRef]
- Yamamoto, K.; Okada, Y. Shared genetic factors and their causality in autoimmune diseases. Ann. Rheum. Dis. 2019, 78, 1449–1451. [Google Scholar] [CrossRef]
- Ramos, P.S.; Shedlock, A.M.; Langefeld, C.D. Genetics of autoimmune diseases: Insights from population genetics. J. Hum. Genet. 2015, 60, 657–664. [Google Scholar] [CrossRef]
- Alarcón-Riquelme, M.E. Recent advances in the genetics of autoimmune diseases. Ann. N. Y. Acad. Sci. 2007, 1110, 1–9. [Google Scholar] [CrossRef]
- Iuliano, A.; Galeazzi, M.; Sebastiani, G.D. Antiphospholipid syndrome’s genetic and epigenetic aspects. Autoimmun. Rev. 2019, 18, 102352. [Google Scholar] [CrossRef] [PubMed]
- Domenico Sebastiani, G.; Minisola, G.; Galeazzi, M. HLA class II alleles and genetic predisposition to the antiphospholipid syndrome. Autoimmun. Rev. 2003, 2, 387–394. [Google Scholar] [CrossRef]
- Tanimura, K.; Jin, H.; Suenaga, T.; Morikami, S.; Arase, N.; Kishida, K.; Hirayasu, K.; Kohyama, M.; Ebina, Y.; Yasuda, S.; et al. β2-Glycoprotein I/HLA class II complexes are novel autoantigens in antiphospholipid syndrome. Blood 2015, 125, 2835–2844. [Google Scholar] [CrossRef] [PubMed]
- Asherson, R.A.; Doherty, D.G.; Vergani, D.; Khamashta, M.A.; Hughes, G.R.V. Major histocompatibility complex associations with primary antiphospholipid syndrome. Arthritis Rheum 1992, 35, 124–125. [Google Scholar] [CrossRef]
- Camps, M.T.; Cuadrado, M.J.; Ocón, P.; Alonso, A.; Gutierrez, A.; Guil, M.; Grana, M.I.; de Ramón, E. ssociation between HLA class II antigens and primary antiphospholipid syndrome from the South of Spain. Lupus 1995, 4, 51–55. [Google Scholar] [CrossRef]
- Arnett, F.C.; Thiagarajan, P.; Ahn, C.; Reveille, J.D. Associations of anti-beta2-glycoprotein I Autoantibodies with HLA Class II Alleles in Three Ethnic Groups. Arthritis Rheum 1999, 42, 268–274. [Google Scholar] [CrossRef]
- Bertolaccini, M.L.; Atsumi, T.; Caliz, A.R.; Amengual, O.; Khamashta, M.A.; Hughes, G.R.; Koike, T. Association of antiphosphatidylserine/prothrombin Autoantibodies with HLA Class II Genes. Arthritis Rheum 2000, 43, 683–688. [Google Scholar] [CrossRef]
- Sanchez, M.L.; Katsumata, K.; Atsumi, T.; Romero, F.I.; Bertolaccini, M.L.; Funke, A.; Amengual, O.; Kondeatis, E.; Vaughan, R.W.; Cox, A.; et al. Association of HLA-DM polymorphism with the production of antiphospholipid antibodies. Ann. Rheum. Dis. 2004, 63, 1645–1648. [Google Scholar] [CrossRef]
- Sebastiani, G.D.; Iuliano, A.; Cantarini, L.; Galeazzi, M. Genetic aspects of the antiphospholipid syndrome: An update. Autoimmun. Rev. 2016, 15, 433–439. [Google Scholar] [CrossRef]
- Ortiz-Fernández, L.; Sawalha, A.H. Genetics of Antiphospholipid Syndrome. Curr. Rheumatol. Rep. 2019, 21, 65. [Google Scholar] [CrossRef]
- Sacharidou, A.; Shaul, P.W.; Mineo, C. New Insights in the Pathophysiology of Antiphospholipid Syndrome. Semin. Thromb. Hemost. 2018, 44, 475–482. [Google Scholar] [CrossRef] [PubMed]
- Castro-Marrero, J.; Balada, E.; Vilardell-Tarrés, M.; Ordi-Ros, J. Genetic risk factors of thrombosis in the antiphospholipid syndrome. Br. J. Haematol. 2009, 147, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Exner, T.; Barber, S.; Kronenberg, H.; Rickard, K.A. Familial Association of the Lupus Anticoagulant. Br. J. Haematol. 1980, 45, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Matthey, F.; Walshe, K.; Mackie, I.J.; Machin, S.J. Familial occurrence of the antiphospholipid syndrome. J. Clin. Pathol. 1989, 42, 495–497. [Google Scholar] [CrossRef] [PubMed]
- Dagenais, P.; Urowitz, M.B.; Gladman, D.D.; Norman, C.S. A family study of the antiphospholipid syndrome associated with other autoimmune diseases. J. Rheumatol. 1992, 19, 1393–1396. [Google Scholar]
- Papalardo, E.; Romay-Penabad, Z.; Willis, R.; Christadoss, P.; Carrera-Marin, A.L.; Reyes-Maldonado, E.; Rudrangi, R.; Alfieri-Papalardo, S.; Garcia-Latorre, E.; Blank, M.; et al. Major Histocompatibility Complex Class II Alleles Influence Induction of Pathogenic Antiphospholipid Antibodies in a Mouse Model of Thrombosis. Arthritis Rheumatol. 2017, 69, 2052–2061. [Google Scholar] [CrossRef]
- Lee, Y.H.; Choi, S.J.; Ji, J.D.; Song, G.G. Association between the valine/leucine247 polymorphism of β2-glycoprotein i and susceptibility to anti-phospholipid syndrome: A meta-analysis. Lupus 2012, 21, 865–871. [Google Scholar] [CrossRef]
- Yasuda, S.; Atsumi, T.; Matsuura, E.; Kaihara, K.; Yamamoto, D.; Ichikawa, K.; Koike, T. Significance of valine/leucine247 polymorphism of β2-glycoprotein I in antiphospholipid syndrome: Increased reactivity of anti-β2-glycoprotein I autoantibodies to the valine247 β2-glycoprotein I variant. Arthritis Rheum. 2005, 52, 212–218. [Google Scholar] [CrossRef]
- Gorji, A.E.; Roudbari, Z.; Alizadeh, A.; Sadeghi, B. Investigation of systemic lupus erythematosus (SLE) with integrating transcriptomics and genome wide association information. Gene 2019, 706, 181–187. [Google Scholar] [CrossRef]
- Beltrán Ramírez, O.; Mendoza Rincón, J.F.; Barbosa Cobos, R.E.; Alemán Ávila, I.; Ramírez Bello, J. STAT4 confers risk for rheumatoid arthritis and systemic lupus erythematosus in Mexican patients. Immunol. Lett. 2016, 175, 40–43. [Google Scholar] [CrossRef]
- Goropevšek, A.; Holcar, M.; Avčin, T. The Role of STAT Signaling Pathways in the Pathogenesis of Systemic Lupus Erythematosus. Clin. Rev. Allergy Immunol. 2017, 52, 164–181. [Google Scholar] [CrossRef] [PubMed]
- Kariuki, S.N.; Kirou, K.A.; MacDermott, E.J.; Barillas-Arias, L.; Crow, M.K.; Niewold, T.B. Cutting Edge: Autoimmune Disease Risk Variant of STAT4 Confers Increased Sensitivity to IFN-α in Lupus Patients In Vivo. J. Immunol. 2009, 182, 34–38. [Google Scholar] [CrossRef] [PubMed]
- Horita, T.; Atsumi, T.; Yoshida, N.; Nakagawa, H.; Kataoka, H.; Yasuda, S.; Koike, T. STAT4 single nucleotide polymorphism, rs7574865 G/T, as a risk for antiphospholipid syndrome. Ann. Rheum. Dis. 2009, 68, 1366–1367. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Borghi, M.O.; Delgado-Vega, A.M.; Tincani, A.; Meroni, P.L.; Alarcón-Riquelme, M.E. Association of STAT4 and BLK, but not BANK1 or IRF5, with primary antiphospholipid syndrome. Arthritis Rheum. 2009, 60, 2468–2471. [Google Scholar] [CrossRef]
- Fredi, M.; Tincani, A.; Yin, H.; Delgado-Vega, A.M.; Borghi, M.O.; Meroni, P.L.; Alarcón-Riquelme, M.E. IRF5 is associated with primary antiphospholipid syndrome, but is not a major risk factor. Arthritis Rheum. 2010, 62, 1201–1202. [Google Scholar] [CrossRef]
- Ostanek, L.; Ostanek-Pańka, M.; Bobrowska-Snarska, D.; Bińczak-Kuleta, A.; Fischer, K.; Kaczmarczyk, M.; Ciechanowicz, A.; Brzosko, M. PTPN22 1858C>T gene polymorphism in patients with SLE: Association with serological and clinical results. Mol. Biol. Rep. 2014, 41, 6195–6200. [Google Scholar] [CrossRef][Green Version]
- Stanford, S.M.; Bottini, N. PTPN22: The archetypal non-HLA autoimmunity gene. Nat. Rev. Rheumatol. 2014, 10, 602–611. [Google Scholar] [CrossRef]
- Pierangeli, S.S.; Vega-Ostertag, M.E.; Raschi, E.; Liu, X.; Romay-Penabad, Z.; De Micheli, V.; Galli, M.; Moia, M.; Tincani, A.; Borghi, M.O.; et al. Toll-like receptor and antiphospholipid mediated thrombosis: In vivo studies. Ann. Rheum. Dis. 2007, 66, 1327–1333. [Google Scholar] [CrossRef]
- Benhamou, Y.; Bellien, J.; Armengol, G.; Brakenhielm, E.; Adriouch, S.; Iacob, M.; Remy-Jouet, I.; Le Cam-Duchez, V.; Monteil, C.; Renet, S.; et al. Role of toll-like receptors 2 and 4 in mediating endothelial dysfunction and arterial remodeling in primary arterial antiphospholipid syndrome. Arthritis Rheumatol. 2014, 66, 3210–3220. [Google Scholar] [CrossRef]
- Jiménez, S.; Tàssies, D.; Espinosa, G.; García-Criado, Á.; Plaza, J.; Monteagudo, J.; Cervera, R.; Reverter, J.C. Double heterozygosity polymorphisms for platelet glycoproteins Ia/IIa and IIb/IIIa increases arterial thrombosis and arteriosclerosis in patients with the antiphospholipid syndrome or with systemic lupus erythematosus. Ann. Rheum. Dis. 2008, 67, 835–840. [Google Scholar] [CrossRef]
- Yonal, I.; Hindilerden, F.; Hancer, V.S.; Artim-Esen, B.; Daglar, A.; Akadam, B.; Nalcaci, M.; Diz-Kucukkaya, R. The impact of platelet membrane glycoprotein Ib alpha and Ia/IIa polymorphisms on the risk of thrombosis in the antiphospholipid syndrome. Thromb. Res. 2012, 129, 486–491. [Google Scholar] [CrossRef] [PubMed]
- Plasín-Rodríguez, M.A.; Rodríguez-Pintó, I.; Patricio, P.; Monteagudo, J.; Cervera, R.; Reverter, J.C.; Espinosa, G.; Tàssies, D. The H1 haplotype of the endothelial protein C receptor protects against arterial thrombosis in patients with antiphospholipid syndrome. Thromb. Res. 2018, 169, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Forastiero, R.R.; Martinuzzo, M.E.; Lu, L.; Broze, G.J. Autoimmune antiphospholipid antibodies impairthe inhibition of activated factor X by protein Z/protein Z-dependent protease inhibitor. J. Thromb. Haemost. 2003, 1, 1764–1770. [Google Scholar] [CrossRef] [PubMed]
- Perez-Sanchez, C.; Barbarroja, N.; Messineo, S.; Ruiz-Limon, P.; Rodriguez-Ariza, A.; Jimenez-Gomez, Y.; Khamashta, M.A.; Collantes-Estevez, E.; Cuadrado, M.J.; Aguirre, M.A.; et al. Gene profiling reveals specific molecular pathways in the pathogenesis of atherosclerosis and cardiovascular disease in antiphospholipid syndrome, systemic lupus erythematosus and antiphospholipid syndrome with lupus. Ann. Rheum. Dis. 2015, 74, 1441–1449. [Google Scholar] [CrossRef]
- Islam, M.A.; Khandker, S.S.; Alam, F.; Kamal, M.A.; Gan, S.H. Genetic risk factors in thrombotic primary antiphospholipid syndrome: A systematic review with bioinformatic analyses. Autoimmun. Rev. 2018, 17, 226–243. [Google Scholar] [CrossRef]
- The Relationship between P-Selectin Polymorphisms and Thrombosis in Antiphospholipid Syndrome: A Pilot Case-Control Study. Turk. J. Haematol. 2014, 31, 357–362. [CrossRef]
- Aisina, R.B.; Mukhametova, L.I.; Ostryakova, E.V.; Seredavkina, N.V.; Patrushev, L.I.; Patrusheva, N.L.; Reshetnyak, T.M.; Gulin, D.A.; Gershkovich, K.B.; Nasonov, E.L.; et al. Polymorphism of the plasminogen activator inhibitor type 1 gene, plasminogen level and thromboses in patients with the antiphospholipid syndrome. Biochem. Suppl. Ser. B Biomed. Chem. 2013, 7, 1–15. [Google Scholar] [CrossRef]
- Lincz, L.F.; Adams, M.J.; Scorgie, F.E.; Thom, J.; Baker, R.I.; Seldon, M. Polymorphisms of the tissue factor pathway inhibitor gene are associated with venous thromboembolism in the antiphospholipid syndrome and carriers of factor V Leiden. Blood Coagul. Fibrinolysis 2007, 18, 559–564. [Google Scholar] [CrossRef]
- Bertolaccini, M.L.; Atsumi, T.; Lanchbury, J.S.; Caliz, A.R.; Katsumata, K.; Vaughan, R.W.; Kondeatis, E.; Khamashta, M.A.; Koike, T.; Hughes, G.R.V. Plasma tumor necrosis factor α levels and the -238* A promoter polymorphism in patients with antiphospholipid syndrome. Thromb. Haemost. 2001, 85, 198–203. [Google Scholar] [CrossRef]
- Forastiero, R.; Martinuzzo, M.; Adamczuk, Y.; Iglesias Varela, M.L.; Pombo, G.; Carreras, L.O. The combination of thrombophilic genotypes is associated with definite antiphospholipid syndrome. Haematologica 2001, 86, 735–741. [Google Scholar]
- Pablos, J.L.; Caliz, R.A.; Carreira, P.E.; Atsumi, T.; Serrano, L.; Amengual, O.; Santiago, B.; Khamashta, M.A.; Hughes, G.R.V.; Gomez-Reino, J.J. Risk of thrombosis in patients with antiphospholipid antibodies and factor V Leiden mutation. J. Rheumatol. 1999, 26, 588–590. [Google Scholar]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Kopanos, C.; Tsiolkas, V.; Kouris, A.; Chapple, C.E.; Albarca Aguilera, M.; Meyer, R.; Massouras, A. VarSome: The human genomic variant search engine. Bioinformatics 2019, 35, 1978–1980. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Merisov, J. P Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef] [PubMed]
- Ramanadham, S.; Tomader, A.; Ashley, J.W.; Bone, R.N.; Hancock, W.D.; Lei, X. Calcium-independent phospholipases A2 and their roles in biological processes and diseases. J. Lipid Res. 2015, 56, 1643–1668. [Google Scholar] [CrossRef]
- Song, H.; Wohltmann, M.; Tan, M.; Ladenson, J.H.; Turk, J. Group VIA phospholipase A2 mitigates palmitate-induced β-cell mitochondrial injury and apoptosis. J. Biol. Chem. 2014, 289, 14194–14210. [Google Scholar] [CrossRef]
- Cohen, D.; Papillon, J.; Aoudjit, L.; Li, H.; Cybulsky, A.V.; Takano, T. Role of calcium-independent phospholipase A2 in complement-mediated glomerular epithelial cell injury. Am. J. Physiol.-Ren. Physiol. 2008, 294, 469–479. [Google Scholar] [CrossRef]
- Malley, K.R.; Koroleva, O.; Miller, I.; Sanishvili, R.; Jenkins, C.M.; Gross, R.W.; Korolev, S. The structure of iPLA2β reveals dimeric active sites and suggests mechanisms of regulation and localization. Nat. Commun. 2018, 9, 1–11. [Google Scholar] [CrossRef]
- Murdock, A.; Dodge, G.; Cohen, I.; Tuan, R.; Iozzo, R. Primary structure of the human heparan sulfate proteoglycan from basement membrane (HSPG2/perlecan). J. Biol. Chem. 1992, 267, 8544–8557. [Google Scholar]
- Segev, A.; Nili, N.; Strauss, B.H. The role of perlecan in arterial injury and angiogenesis. Cardiovasc. Res. 2004, 63, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Nugent, M.A.; Nugent, H.M.; Iozzo, R.V.; Sanchack, K.; Edelman, E.R. Perlecan is required to inhibit thrombosis after deep vascular injury and contributes to endothelial cell-mediated inhibition of intimal hyperplasia. Proc. Natl. Acad. Sci. USA 2000, 97, 6722–6727. [Google Scholar] [CrossRef] [PubMed]
- Nonaka, R.; Iesaki, T.; de Vega, S.; Daida, H.; Okada, T.; Sasaki, T.; Arikawa-Hirasawa, E. Perlecan deficiency causes endothelial dysfunction by reducing the expression of endothelial nitric oxide synthase. Physiol. Rep. 2015, 3, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Dieudé, M.; Cardinal, H.; Hébert, M.J. Injury derived autoimmunity: Anti-perlecan/LG3 antibodies in transplantation. Hum. Immunol. 2019, 80, 608–613. [Google Scholar] [CrossRef] [PubMed]
- Pilon, E.A.; Dieudé, M.; Qi, S.; Hamelin, K.; Pomerleau, L.; Beillevaire, D.; Durocher, Y.; Zutter, M.; Coutu, D.; Perreault, C.; et al. The perlecan fragment lg3 regulates homing of mesenchymal stem cells and neointima formation during vascular rejection. Am. J. Transplant. 2015, 15, 1205–1218. [Google Scholar] [CrossRef]
- Herrington, F.; Nibbs, R. Regulation of the Adaptive Immune Response by the IκB Family Protein Bcl-3. Cells 2016, 5, 14. [Google Scholar] [CrossRef]
- Weyrich, A.S.; Denis, M.M.; Schwertz, H.; Tolley, N.D.; Foulks, J.; Spencer, E.; Kraiss, L.W.; Albertine, K.H.; McIntyre, T.M.; Zimmerman, G.A. mTOR-dependent synthesis of Bcl-3 controls the retraction of fibrin clots by activated human platelets. Blood 2007, 109, 1975–1983. [Google Scholar] [CrossRef]
- Koyanagi, M.; Nakabayashi, K.; Fujimoto, T.; Gu, N.; Baba, I.; Takashima, Y.; Doi, K.; Harada, H.; Kato, N.; Sasazuki, T.; et al. ZFAT expression in B and T lymphocytes and identification of ZFAT-regulated genes. Genomics 2008, 91, 451–457. [Google Scholar] [CrossRef]
- Yoshida, Y.; Tsunoda, T.; Takashima, Y.; Fujimoto, T.; Doi, K.; Sasazuki, T.; Kuroki, M.; Iwasaki, A.; Shirasawa, S. ZFAT is essential for endothelial cell assembly and the branch point formation of capillary-like structures in an angiogenesis model. Cell Mol. Biol. Lett. 2010, 15, 541–550. [Google Scholar] [CrossRef]
- Ogawa, M.; Okamura, T.; Ishikura, S.; Doi, K.; Matsuzaki, H.; Tanaka, Y.; Ota, T.; Hayakawa, K.; Suzuki, H.; Tsunoda, T. Zfat-Deficiency Results in a Loss of CD3ζ Phosphorylation with Dysregulation of ERK and Egr Activities Leading to Impaired Positive Selection. PLoS ONE 2013, 8, e76254. [Google Scholar] [CrossRef]
- Brini, M.; Di Leva, F.; Ortega, C.K.; Domi, T.; Ottolini, D.; Leonardi, E.; Tosatto, S.C.E.; Carafoli, E. Deletions and mutations in the acidic lipid-binding region of the plasma membrane Ca2+ pump: A study on different splicing variants of isoform 2. J. Biol. Chem. 2010, 285, 30779–30791. [Google Scholar] [CrossRef] [PubMed]
- Di Leva, F.; Domi, T.; Fedrizzi, L.; Lim, D.; Carafoli, E. The plasma membrane Ca2+ ATPase of animal cells: Structure, function and regulation. Arch. Biochem. Biophys. 2008, 476, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Holton, M.; Mohamed, T.M.A.; Oceandy, D.; Wang, W.; Lamas, S.; Emerson, M.; Neyses, L.; Armesilla, A.L. Endothelial nitric oxide synthase activity is inhibited by the plasma membrane calcium ATPase in human endothelial cells. Cardiovasc. Res. 2010, 87, 440–448. [Google Scholar] [CrossRef] [PubMed]
- Hudson, C.; Kimura, T.E.; Duggirala, A.; Sala-Newby, G.B.; Newby, A.C.; Bond, M. Dual Role of CREB in The Regulation of VSMC Proliferation: Mode of Activation Determines Pro- or Anti-Mitogenic Function. Sci. Rep. 2018, 8, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Chava, K.R.; Tauseef, M.; Sharma, T.; Mehta, D. Cyclic AMP response element-binding protein prevents endothelial permeability increase through transcriptional controlling p190RhoGAP expression. Blood 2012, 119, 308–319. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; He, B.; Abdel-Halim, S.M.; Tibell, A.; Brendel, M.D.; Bretzel, R.G.; Efendic, S.; Hillert, J. Molecular cloning of a full-length cDNA for human type 3 adenylyl cyclase and its expression in human islets. Biochem. Biophys. Res. Commun. 1999, 254, 548–551. [Google Scholar] [CrossRef]
- Tong, T.; Shen, Y.; Lee, H.W.; Yu, R.; Park, T. Adenylyl cyclase 3 haploinsufficiency confers susceptibility to diet-induced obesity and insulin resistance in mice. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef]
- Van Cott, E.M.; Khor, B.; Zehnder, J.L. Factor VLeiden. Am. J. Hematol. 2016, 91, 46–49. [Google Scholar] [CrossRef]
- Hotoleanu, C. Genetic risk factors in venous thromboembolism. Adv. Exp. Med. Biol. 2017, 906. [Google Scholar] [CrossRef]
- Bagoly, Z.; Koncz, Z.; Hársfalvi, J.; Muszbek, L. Factor XIII, clot structure, thrombosis. Thromb. Res. 2012, 129, 382–387. [Google Scholar] [CrossRef]
- Luo, H.; Li, X.; Jiang, A.; Zhang, B.; Bi, P.; Dong, Y.; Guo, Y. Associations of β-Fibrinogen Polymorphisms with the Risk of Ischemic Stroke: A Meta-analysis. J. Stroke Cerebrovasc. Dis. 2019, 28, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Sartori, M.T.; Danesin, C.; Saggiorato, G.; Tormene, D.; Simioni, P.; Spiezia, L.; Patrassi, G.M.; Girolami, A. The PAI-1 gene 4G/5G Polymorphism and Deep Vein Thrombosis in Patients with Inherited Thrombophilia. Clin. Appl. Thromb. 2003, 9, 299–307. [Google Scholar] [CrossRef]
- Barabási, A.L.; Gulbahce, N.; Loscalzo, J. Network medicine: A network-based approach to human disease. Nat. Rev. Genet. 2011, 12, 56–68. [Google Scholar] [CrossRef] [PubMed]
- Nitsch, D.; Tranchevent, L.-C.; Thienpont, B.; Thorrez, L.; Van Esch, H.; Devriendt, K.; Moreau, Y. Network Analysis of Differential Expression for the Identification of Disease-Causing Genes. PLoS ONE 2009, 4, e5526. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Itan, Y. Biological network approaches and applications in rare disease studies. Genes 2019, 10, 797. [Google Scholar] [CrossRef] [PubMed]
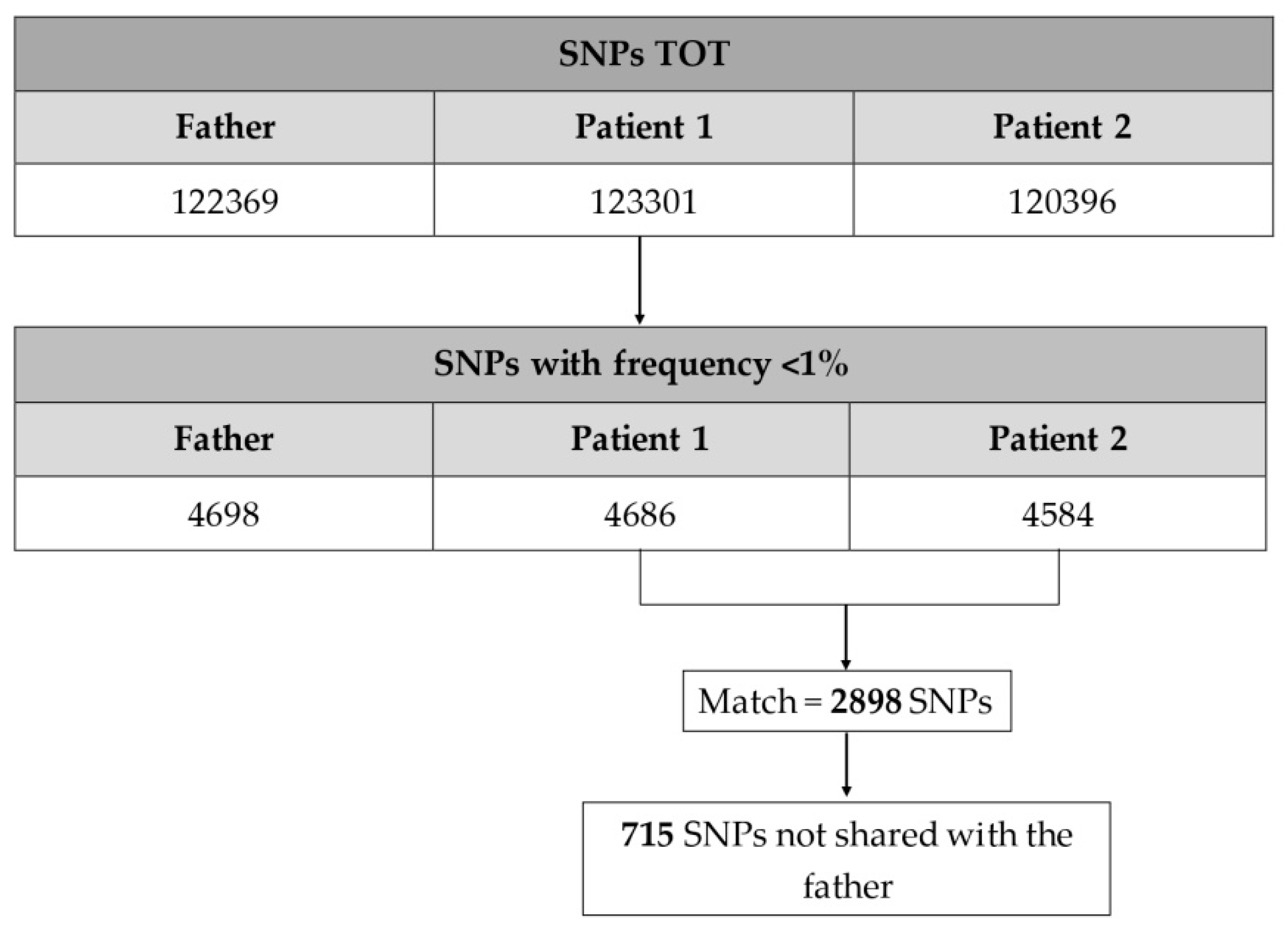
| Gene | Full Name | SNP | Chr | Coordinate | Transcript | Variant Effect | Ref |
|---|---|---|---|---|---|---|---|
| BLK | BLK proto-oncogene, Src family tyrosine kinase | rs2736340 | 8 | 11343973 | NM_001715.3 | 5′ UTR variant | [34] |
| B2GP1/APOH | Beta-2-glycoprotein I, Apolipoprotein H | Position 247 | 17 | 64210757 | NM_000042.3 | Missense variant (p.Val247Leu) | [27,28] |
| F2 | Coagulation factor II, Thrombin | rs1799963 | 11 | 46761055 | NM_000506.5 | 3′ UTR variant | [50] |
| F5 | Coagulation factor V | rs6025 | 1 | 169519049 | NM_000130.5 | Missense variant (p.Gln534Arg) | [50,51] |
| GPIa/ITGA2 | Integrin subunit alpha 2 | rs1126643 | 5 | 52347369 | NM_002203.4 | Synonymous variant (p.Phe253 =) | [40,41] |
| GPIIIa/ITGB3 | Integrin subunit beta 3 | rs5918 | 17 | 45360730 | NM_000212.3 | Missense variant (p.Leu59Pro) | [40,41] |
| GP1BA | Glycoprotein Ib platelet subunit alpha | rs2243093 | 17 | 4835895 | NM_000173.7 | 5′ UTR variant | [41] |
| IRF5 | Interferon regulatory factor 5 | rs2070197 | 7 | 128589000 | NM_001098629.3 | 3′ UTR variant | [34,35] |
| rs10954213 | 7 | 128589427 | NM_001098629.3 | 3′ UTR variant | [34] | ||
| PROCR | Protein C receptor | H1 haplotype | 20 | NM_006404.5 | [42] | ||
| PTPN22 | Protein tyrosine phosphatase non-receptor type 22 | rs2476601 | 1 | 114377568 | NM_015967.7 | Missense variant (p.Arg620Trp) | [36,37] |
| SELP | Selectin P | rs6127 | 1 | 169566313 | NM_003005.4 | Missense variant (p.Asp603Asn) | [46] |
| SERPINE1 | Serpin family E member 1 | c.-817dupG | 7 | 101126426 | NM_000602.3 | [47] | |
| STAT4 | Signal transducer and activator of transcription 4 | rs7574865 rs3821236 rs3024866 | 2 | 191964633 191,902,758 191922841 | NM_00124385.2 NM_00124385.2 NM_00124385.2 | Intronic Intronic Intronic | [30,31,32,33,34] |
| TFPI | Tissue factor pathway inhibitor | intron 7 -33T>C | 2 | 188385299 | NM_006287.6 | Intronic | [48] |
| TLR4 | Toll-like receptor 4 | rs4986790 | 9 | 12047502 | NM_138554.5 | Missense variant (p.Asp299Gly) | [38,39] |
| TNF | Tumor necrosis factor | rs361525 | 6 | 31543101 | NM_000594.4 | Upstream Transcript Variant | [49] |
| Patient | Age | aPL Profile | Relevant Clinical History |
|---|---|---|---|
| 1 (F) | 51 | Triple positive (LA, aCL IgG, aβ2GPI IgG) | Two episodes of ischemic stroke, one episode of CAPS (renal thrombotic microangiopathy, visual impairment, ischemic stroke) |
| 2 (M) | 47 | Triple positive (LA, aCL IgG, aβ2GPI IgG) | Three episodes of deep vein thrombosis, regardless ongoing well conducted therapy vitamin k antagonist and additional retinal vein thrombosis |
| Gene | Full Name | SNP | Chr | Coordinate | Transcript | Variant Effect |
|---|---|---|---|---|---|---|
| ADCY3 | Adenylate cyclase 3 | rs754839662 (C/T) Het | 2 | 25050928 | NM_004036.3 | Missense variant (p.Val759Met) |
| ATP2B2 | ATPase plasma membrane Ca2+ transporting 2 | rs751257556 (G/A) Het | 3 | 10382352 | NM_001001331.2 | Missense variant (p.Ala985Val) |
| BCL3 | BCL3 transcription coactivator | rs747655476 (C/A) Het | 19 | 45262063 | NM_005178.4 | Missense variant (p.Thr381Asn) |
| CC2D1A | Coiled-coil and C2 domain containing 1A | c.794G>T Het | 19 | 14028928 | NM_017721.4 | Missense variant (p.Arg265Leu) |
| CROCC | Ciliary rootlet coiled-coil/Rootletin | rs1444279934 (A/T) Het | 1 | 17270657 | NM_014675.3 | Missense variant (p.Gln624Leu) |
| CRTC3 | CREB regulated transcription coactivator 3 | c.163C>T Het | 15 | 91083301 | NM_022769.4 | Missense variant (p.Leu55Phe) |
| FAT2 | FAT atypical cadherin 2 | rs761199516 (C/A) Het | 5 | 150945648 | NM_001447.2 | Missense variant (p.Ala949Ser) |
| GRM7 | Glutamate metabotropic receptor 7 | rs1480175679 (A/C) Het | 3 | 7494339 | NM_181874.2 | Missense variant (p.Lys407Thr) |
| HSPG2 | Heparan sulfate proteoglycan 2 | rs766963773 (C/T) Het | 1 | 22207015 | NM_005529.5 | Missense variant (p.Arg679His) |
| IFNA17 | Interferon alpha 17 | c.338A>T Het | 9 | 21227835 | NM_021268.2 | Missense variant (p.Tyr113Phe) |
| IMPA2 | Inositol monophosphatase 2 | rs1423846345 (T/C) Het | 18 | 12009980 | NM_014214.2 | Missense variant (p.Val110Ala) |
| KIF14 | Kinesin family member 14 | rs373895990 (C/T) Het | 1 | 200573037 | NM_014875.2 | Missense variant (p.Arg598Gln) |
| LTBP2 | Latent transforming growth factor beta binding protein 2 | rs1310944162 (G/A) Het | 14 | 74995323 | NM_000428.2 | Missense variant (p.Ala744Val) |
| MAP3K10 | Mitogen-activated protein kinase kinase kinase 10 | c.358G>A Het | 19 | 40698296 | NM_002446.3 | Missense variant (p.Glu120Lys) |
| MCM8 | Minichromosome maintenance 8 homologous recombination repair factor | rs768426546 (G/A) Het | 20 | 5953350 | NM_001281521.1 | Missense variant (p.Arg451His) |
| MRPL48 | Mitochondrial ribosomal protein L48 | rs745995390 (G/T) Het | 11 | 73555903 | NM_016055.5 | Missense variant (p.Asp85Tyr) |
| MUC16 | Mucin 16 | rs200972932 (C/T) Het | 19 | 9067504 | NM_024690.2 | Missense variant (p.Glu6648Lys) |
| OR2F1 | Olfactory receptor family 2 subfamily F member 1 | rs777034277 (T/C) Het | 7 | 143657370 | NM_012369.2 | Missense variant (p.Phe103Leu) |
| OXNAD1 | Oxidoreductase NAD binding domain containing 1 | rs1456594626 (T/G) Het rs1159857217 (T/A) Het rs1390159575 (C/G) Het | 3 | 163431751634317616343177 | NM_138381.3 | Missense variant (p.Phe159Glu) |
| PDZD2 | PDZ domain containing 2 | rs1345334581 (C/G) Het | 5 | 32098731 | NM_178140.2 | Missense variant (p.Pro2737Ala) |
| PLA2G6 | Phospholipase A2 group VI | rs780423461 (C/G) Het | 22 | 38528920 | NM_003560.2 | Missense variant (p.Cys332Ser) |
| SRGAP3 | SLIT-ROBO Rho GTPase activating protein 3 | rs764134718 (C/T) Het | 3 | 9146464 | NM_014850.3 | Missense variant (p.Arg108Gln) |
| USP32 | Ubiquitin specific peptidase 32 | c.1169T>C Het | 17 | 58313569 | NM_032582.3 | Missense variant (p.Leu390Pro) |
| ZFAT | Zinc finger and AT-hook domain containing | rs748138009 (G/A) Het | 8 | 135596174 | NM_020863.3 | Missense variant (p.Arg930Cys) |
| ZNF462 | Zinc finger protein 462 | c.226A>T Het | 9 | 109686419 | NM_021224.4 | Missense variant (p.Asn76Tyr) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barinotti, A.; Radin, M.; Cecchi, I.; Foddai, S.G.; Rubini, E.; Roccatello, D.; Sciascia, S.; Menegatti, E. Genetic Factors in Antiphospholipid Syndrome: Preliminary Experience with Whole Exome Sequencing. Int. J. Mol. Sci. 2020, 21, 9551. https://doi.org/10.3390/ijms21249551
Barinotti A, Radin M, Cecchi I, Foddai SG, Rubini E, Roccatello D, Sciascia S, Menegatti E. Genetic Factors in Antiphospholipid Syndrome: Preliminary Experience with Whole Exome Sequencing. International Journal of Molecular Sciences. 2020; 21(24):9551. https://doi.org/10.3390/ijms21249551
Chicago/Turabian StyleBarinotti, Alice, Massimo Radin, Irene Cecchi, Silvia Grazietta Foddai, Elena Rubini, Dario Roccatello, Savino Sciascia, and Elisa Menegatti. 2020. "Genetic Factors in Antiphospholipid Syndrome: Preliminary Experience with Whole Exome Sequencing" International Journal of Molecular Sciences 21, no. 24: 9551. https://doi.org/10.3390/ijms21249551
APA StyleBarinotti, A., Radin, M., Cecchi, I., Foddai, S. G., Rubini, E., Roccatello, D., Sciascia, S., & Menegatti, E. (2020). Genetic Factors in Antiphospholipid Syndrome: Preliminary Experience with Whole Exome Sequencing. International Journal of Molecular Sciences, 21(24), 9551. https://doi.org/10.3390/ijms21249551









