Mitochondrial Localization of the Yeast Forkhead Factor Hcm1
Abstract
:1. Introduction
2. Results and Discussion
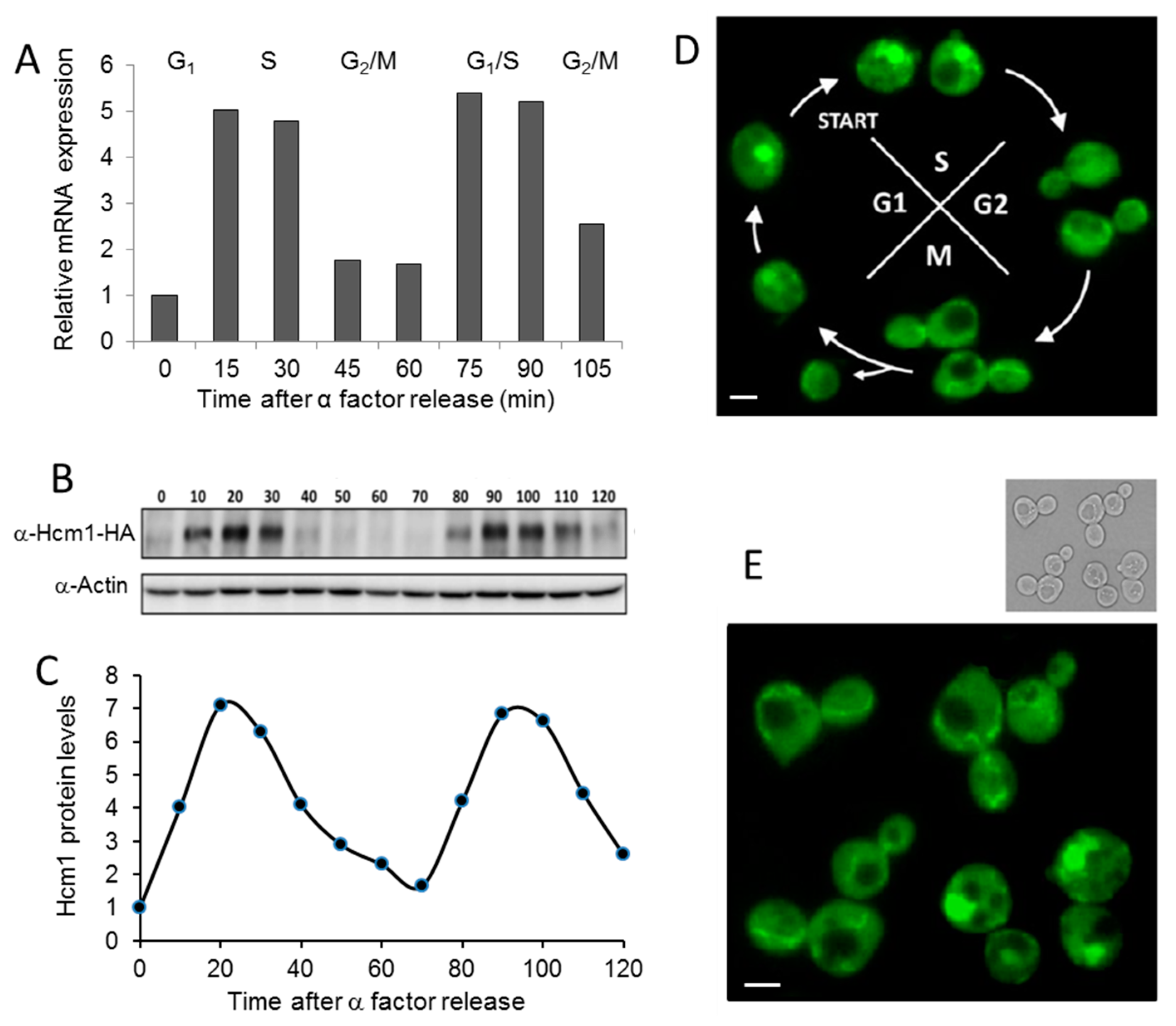
2.1. Cell Cycle Regulates Hcm1 Levels and Localization
2.2. Mitochondrial Localization of Hcm1
2.3. Hcm1, mtDNA Copy Number, and Mitochondrial Gene Activation
3. Materials and Methods
3.1. Yeast Strains and Growth Conditions
3.2. Cell Extracts Fractionation and Mitochondria Purification
3.3. Yeast Cell Synchronization
3.4. Gene Expression Analysis
3.5. Western Blot Analysis
3.6. Microscopy Studies
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| FKH-TF | Forkhead transcription factor |
| ER | Endoplasmic reticulum |
| mtDNA | Mitochondrial DNA |
| nDNA | Nuclear DNA |
References
- Martins, R.S.T.; Lithgow, G.J.; Link, W. Long live FOXO: Unraveling the role of FOXO proteins in aging and longevity. Aging Cell 2016, 15, 196–207. [Google Scholar] [CrossRef] [PubMed]
- Hagenbuchner, J.; Ausserlechner, M.J. Mitochondria and FOXO3: Breath or die. Front. Physiol. 2013, 4, 147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calnan, D.R.; Brunet, A. The FoxO code. Oncogene 2008, 27, 2276–2288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Link, W. Introduction to FOXO Biology. Methods Mol. Biol. 2019, 1890, 1–9. [Google Scholar] [CrossRef]
- Brown, A.K.; Webb, A.E. Regulation of FOXO Factors in Mammalian Cells. Curr. Top. Dev. Biol. 2018, 127, 165–192. [Google Scholar] [CrossRef]
- Kaestner, K.H.; Knochel, W.; Martinez, D.E. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 2000, 14, 142–146. [Google Scholar]
- Zhu, G.; Muller, E.G.; Amacher, S.L.; Northrop, J.L.; Davis, T.N. A dosage-dependent suppressor of a temperature-sensitive calmodulin mutant encodes a protein related to the fork head family of DNA-binding proteins. Mol. Cell. Biol. 1993, 13, 1779–1787. [Google Scholar] [CrossRef] [Green Version]
- Horak, C.E.; Luscombe, N.M.; Qian, J.; Bertone, P.; Piccirrillo, S.; Gerstein, M.; Snyder, M. Complex transcriptional circuitry at the G1/S transition in Saccharomyces cerevisiae. Genes Dev. 2002, 16, 3017–3033. [Google Scholar] [CrossRef] [Green Version]
- Pramila, T. The Forkhead transcription factor Hcm1 regulates chromosome segregation genes and fills the S-phase gap in the transcriptional circuitry of the cell cycle. Genes Dev. 2006, 20, 2266–2278. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-Colman, M.J.; Reverter-Branchat, G.; Sorolla, M.A.; Tamarit, J.; Ros, J.; Cabiscol, E. The Forkhead Transcription Factor Hcm1 Promotes Mitochondrial Biogenesis and Stress Resistance in Yeast. J. Biol. Chem. 2010, 285, 37092–37101. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-Colman, M.J.; Sorolla, M.A.; Vall-Llaura, N.; Tamarit, J.; Ros, J.; Cabiscol, E. The FOX transcription factor Hcm1 regulates oxidative metabolism in response to early nutrient limitation in yeast. Role of Snf1 and Tor1/Sch9 kinases. Biochim. Biophys. Acta (BBA) Bioenerg. 2013, 1833, 2004–2015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maoz, N.; Gabay, O.; Ben-Asher, H.W.; Cohen, H.Y. The Yeast Forkhead HCM1 Controls Life Span Independent of Calorie Restriction. J. Gerontol. Ser. A Boil. Sci. Med. Sci. 2014, 70, 444–453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghavidel, A.; Baxi, K.; Prusinkiewicz, M.; Swan, C.; Belak, Z.R.; Eskiw, C.H.; Carvalho, C.E.; Harkness, T.A. Rapid Nuclear Exclusion of Hcm1 in AgingSaccharomyces cerevisiaeLeads to Vacuolar Alkalization and Replicative Senescence. G3 Genes Genomes Genet. 2018, 8, 1579–1592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Negishi, T.; Veis, J.; Hollenstein, D.M.; Sekiya, M.; Ammerer, G.; Ohya, Y. The Late S-Phase Transcription Factor Hcm1 Is Regulated through Phosphorylation by the Cell Wall Integrity Checkpoint. Mol. Cell. Biol. 2016, 36, 941–953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arsenault, H.E.; Roy, J.; Mapa, C.E.; Cyert, M.S.; Benanti, J.A. Hcm1 integrates signals from Cdk1 and calcineurin to control cell proliferation. Mol. Biol. Cell 2015, 26, 3570–3577. [Google Scholar] [CrossRef] [PubMed]
- Linke, C.; Klipp, E.; Lehrach, H.; Barberis, M.; Krobitsch, S. Fkh1 and Fkh2 associate with Sir2 to control CLB2 transcription under normal and oxidative stress conditions. Front. Physiol. 2013, 4, 173. [Google Scholar] [CrossRef] [Green Version]
- Peserico, A.; Chiacchiera, F.; Grossi, V.; Matrone, A.; Latorre, D.; Simonatto, M.; Fusella, A.; Ryall, J.G.; Finley, L.W.S.; Haigis, M.C.; et al. A novel AMPK-dependent FoxO3A-SIRT3 intramitochondrial complex sensing glucose levels. Cell. Mol. Life Sci. 2013, 70, 2015–2029. [Google Scholar] [CrossRef]
- Celestini, V.; Tezil, T.; Russo, L.; Fasano, C.; Sanese, P.; Forte, G.; Peserico, A.; Signorile, M.L.; Longo, G.; De Rasmo, D.; et al. Uncoupling FoxO3A mitochondrial and nuclear functions in cancer cells undergoing metabolic stress and chemotherapy. Cell Death Dis. 2018, 9, 231. [Google Scholar] [CrossRef]
- Ghaemmaghami, S.; Huh, W.-K.; Bower, K.; Howson, R.W.; Belle, A.; Dephoure, N.; O’Shea, E.K.; Weissman, J.S. Global analysis of protein expression in yeast. Nat. Cell Biol. 2003, 425, 737–741. [Google Scholar] [CrossRef]
- Forgac, M. Structure and properties of the clathrin-coated vesicle and yeast vacuolar V-ATPases. J. Bioenerg. Biomembr. 1999, 31, 57–65. [Google Scholar] [CrossRef]
- Kane, P.M.; Kuehn, M.C.; Howald-Stevenson, I.; Stevens, T.H. Assembly and targeting of peripheral and integral membrane subunits of the yeast vacuolar H(+)-ATPase. J. Biol. Chem. 1992, 267, 447–454. [Google Scholar] [PubMed]
- Orlean, P. Dolichol phosphate mannose synthase is required in vivo for glycosyl phosphatidylinositol membrane anchoring, O mannosylation, and N glycosylation of protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 1990, 10, 5796–5805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, A.; Xu, X.; Blachly-Dyson, E.; Forte, M.; Colombini, M. The role of yeast VDAC genes on the permeability of the mitochondrial outer membrane. J. Membr. Biol. 1998, 161, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, N.S.; Pearce, D.A.; Cardillo, T.S.; Uribe, S.; Sherman, F. Requirements of Cyc2p and the Porin, Por1p, for Ionic Stability and Mitochondrial Integrity in Saccharomyces cerevisiae. Arch. Biochem. Biophys. 2001, 392, 326–332. [Google Scholar] [CrossRef]
- Grissom, J.H.; Segarra, V.A.; Chi, R.J. New Perspectives on SNARE Function in the Yeast Minimal Endomembrane System. Genes 2020, 11, 899. [Google Scholar] [CrossRef] [PubMed]
- Lis, M.; Walther, D. The orientation of transcription factor binding site motifs in gene promoter regions: Does it matter? BMC Genom. 2016, 17, 185. [Google Scholar] [CrossRef] [Green Version]
- Gallego, C.; Garí, E.; Colomina, N.; Herrero, E.; Aldea, M. The Cln3 cyclin is down-regulated by translational repression and degradation during the G1 arrest caused by nitrogen deprivation in budding yeast. EMBO J. 1997, 16, 7196–7206. [Google Scholar] [CrossRef] [Green Version]
- Goldstein, A.L.; McCusker, J.H. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 1999, 15, 1541–1553. [Google Scholar] [CrossRef]
- Meisinger, C.; Pfanner, N.; Truscott, K.N.; Xiao, W. Isolation of Yeast Mitochondria; Humana Press: New Jersey, NJ, USA, 2006; pp. 33–40. [Google Scholar] [CrossRef]
- Vall-Llaura, N.; Mir, N.; Garrido, L.; Vived, C.; Cabiscol, E. Redox control of yeast Sir2 activity is involved in acetic acid resistance and longevity. Redox Biol. 2019, 24, 101229. [Google Scholar] [CrossRef]
- Reverter-Branchat, G.; Cabiscol, E.; Tamarit, J.; Sorolla, M.A.; De La Torre-Ruiz, M.A.; Ros, J. Chronological and replicative life-span extension in Saccharomyces cerevisiae by increased dosage of alcohol dehydrogenase 1. Microbiology 2007, 153, 3667–3676. [Google Scholar] [CrossRef] [PubMed] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez Colman, M.J.; Ros, J.; Cabiscol, E. Mitochondrial Localization of the Yeast Forkhead Factor Hcm1. Int. J. Mol. Sci. 2020, 21, 9574. https://doi.org/10.3390/ijms21249574
Rodríguez Colman MJ, Ros J, Cabiscol E. Mitochondrial Localization of the Yeast Forkhead Factor Hcm1. International Journal of Molecular Sciences. 2020; 21(24):9574. https://doi.org/10.3390/ijms21249574
Chicago/Turabian StyleRodríguez Colman, María José, Joaquim Ros, and Elisa Cabiscol. 2020. "Mitochondrial Localization of the Yeast Forkhead Factor Hcm1" International Journal of Molecular Sciences 21, no. 24: 9574. https://doi.org/10.3390/ijms21249574
APA StyleRodríguez Colman, M. J., Ros, J., & Cabiscol, E. (2020). Mitochondrial Localization of the Yeast Forkhead Factor Hcm1. International Journal of Molecular Sciences, 21(24), 9574. https://doi.org/10.3390/ijms21249574




