Monosomy 3 Influences Epithelial-Mesenchymal Transition Gene Expression in Uveal Melanoma Patients; Consequences for Liquid Biopsy
Abstract
:1. Introduction
2. Results
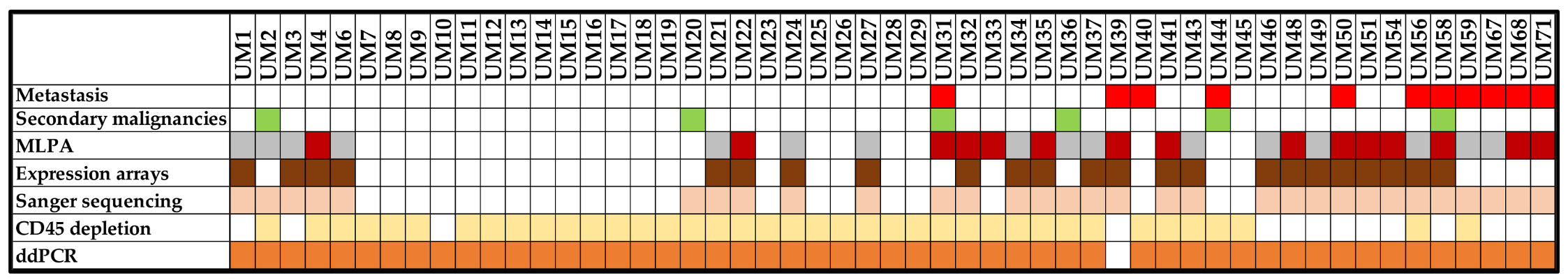
2.1. Clinico-Pathological Characteristics of Patients
2.2. Multiplex Ligation-Dependent Probe Amplification
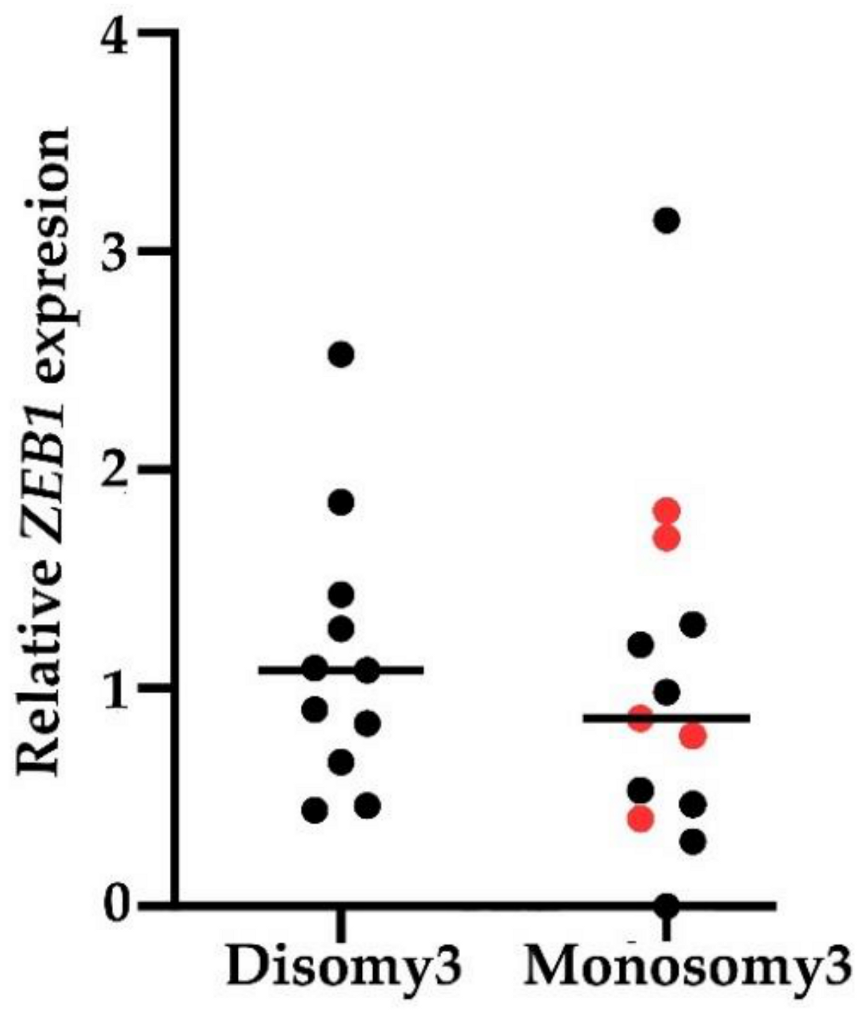
2.3. EMT-Associated TF Expression in Peripheral Blood of UM Patients
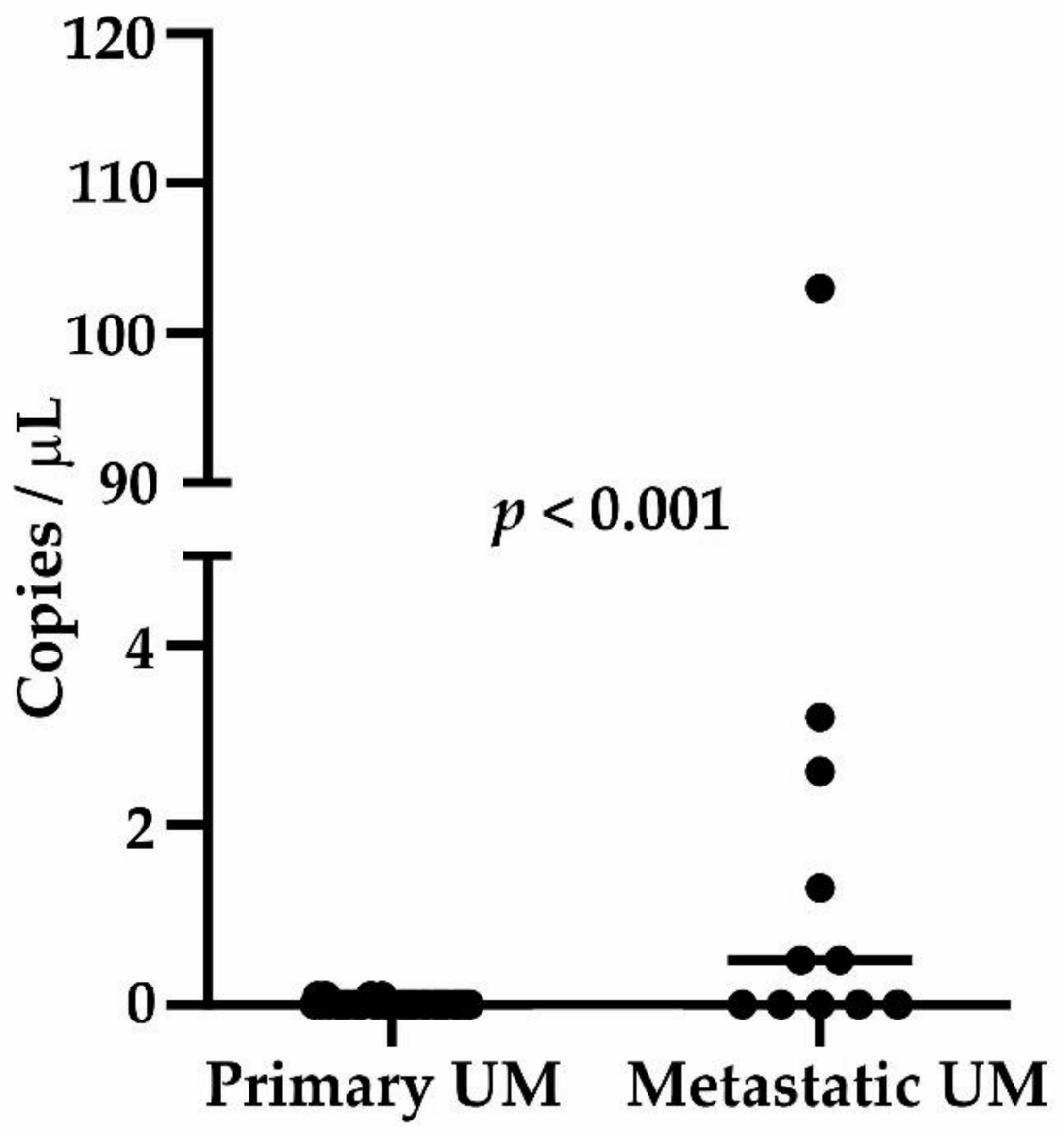
2.4. Circulating Tumor DNA
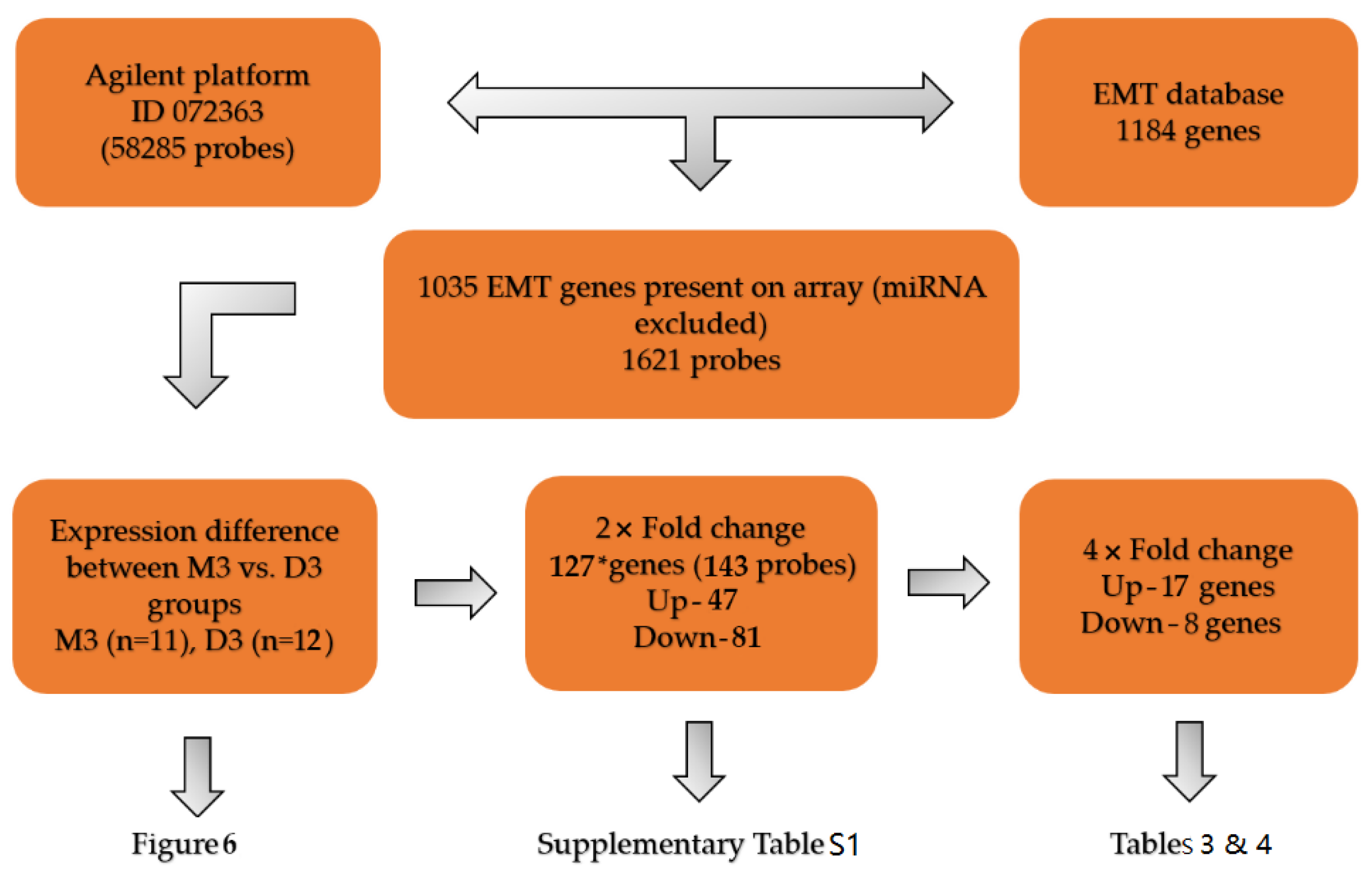
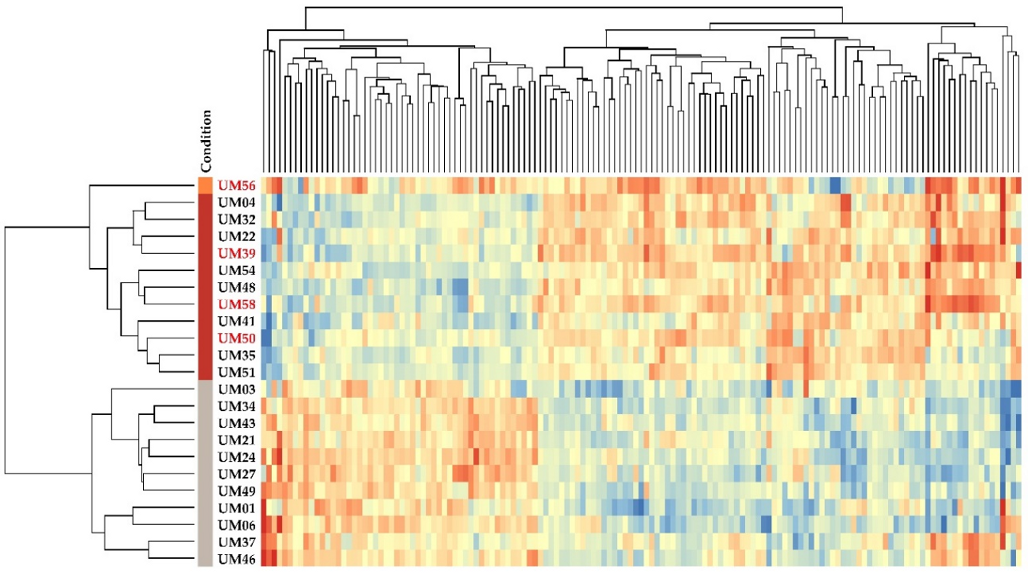
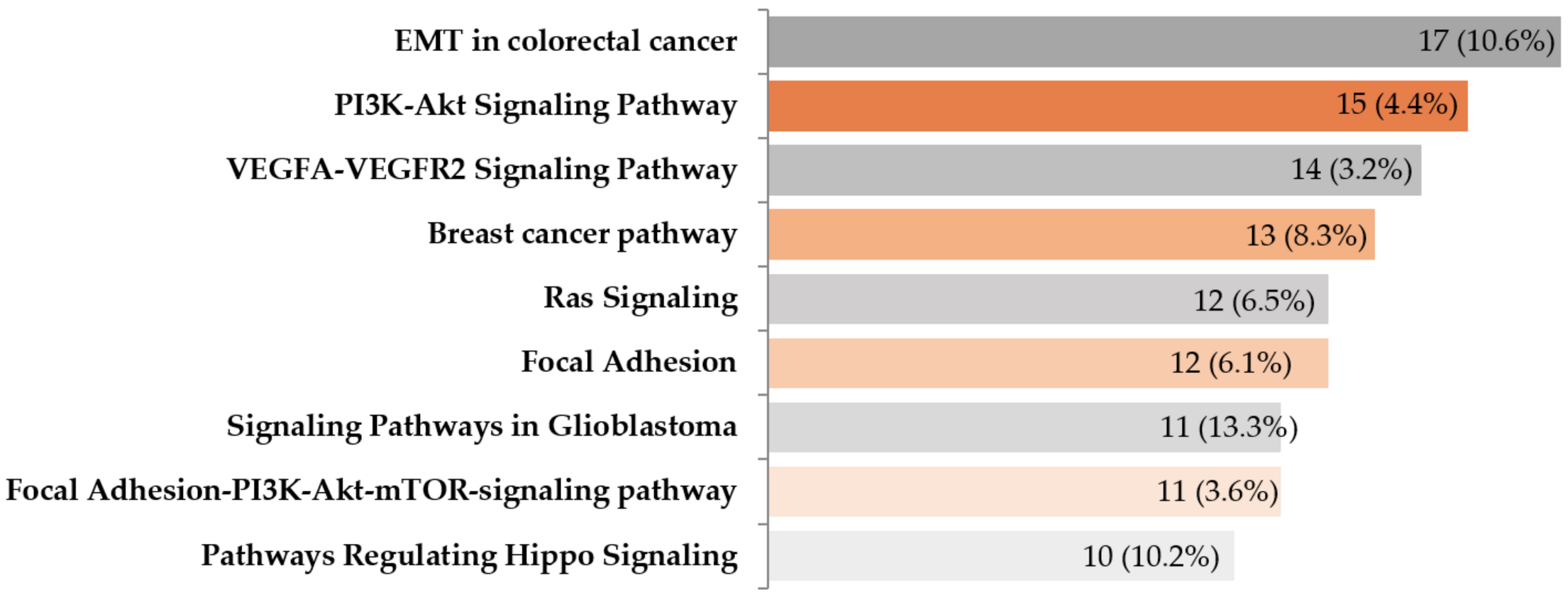
2.5. Gene Expression Profiling of EMT-Associated Genes
3. Discussion
4. Materials and Methods
4.1. Patients and Sample Processing
4.2. Analysis of EMT-Associated TFs Expression in Peripheral Blood
4.2.1. CTCs Enrichment Using CD45 Depletion
4.2.2. Quantitative RT-PCR
4.3. Mutation Detection
4.3.1. DNA Extraction and Quality Assessment
4.3.2. Mutation Detection in Tumor Samples by ddPCR
4.3.3. Sanger Sequencing
4.3.4. Circulating Tumor DNA Detection
4.4. MLPA and Data Analysis
4.5. Microarrays
4.5.1. RNA Extraction and Quality Control
4.5.2. Microarray Assay
4.5.3. Image and Data Analysis
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ALK | Anaplastic lymphoma receptor tyrosine kinase |
| ANG | Angiogenin |
| BAP1 | BRCA1 associated protein |
| CCL18 | C-C motif chemokine ligand 18 |
| CCND2 | Cyclin D2 |
| CDH1 | E-cadherin |
| CNVs | Copy number variations |
| CSPG4 | Chondroitin sulfate proteoglycan 4 |
| CTCs | Circulating tumor cells |
| ctDNA | Circulating tumor DNA |
| CTSZ | Cathepsin Z |
| CYSLTR2 | Cysteinyl leukotriene receptor 2 |
| ddPCR | Droplet digital PCR |
| D3 | Disomy of chromosome 3 |
| EMT | Epithelial-mesenchymal transition |
| ETV1 | ETS variant transcription factor 1 |
| FPR GAPDH | False positive rate Glyceraldehyde-3-phosphate dehydrogenase |
| GDF15 | Growth differentiation factor 15 |
| GJB2 | Gap junction protein beta 2 |
| GNAQ | G protein subunit alfa q |
| GNA11 | G protein subunit alfa 11 |
| HOOK1 | Hook microtubule tethering protein 1 |
| HTN1 | Histatin 1 |
| ID2 | Inhibitor of DNA binding 2 |
| IL | Interleukin |
| JAG1 | Jagged Canonical notch ligand 1 |
| LGALS3 | Galectin |
| KIT | V-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog |
| KRT19 | Epithelial marker keratin 19 |
| MET | Mesenchymal-epithelial transition |
| MITF9 | Melanocyte inducing transcription factor |
| MLPA | Multiplex Ligation-Dependant Probe Amplification |
| MRC2 | Mannose receptor C type 2 |
| M3 | Monosomy of chromosome 3 |
| p | Short arm of chromosome |
| PLCβ4 | Phospholipase C beta 4 |
| POSTN | Periostin |
| PRRX1 | Paired relaxed homeobox 1 |
| PTP4A3 | Protein tyrosine phosphatase type IVA, member 3 |
| RBCs | Red blood cells |
| RORC | RAR related orphan receptor gamma |
| RT-PCR | Real-time PCR |
| q | Long arm of chromosome |
| SATB1 | SATB homeobox 1 |
| SLUG | Snail family transcriptional repressor 2 |
| SPP1 | Secreted phosphoprotein 1 |
| SNAI1 | Snail family transcription repressors 1 |
| SNAI2 | Snail family transcription repressors 2 |
| S100A4 | S100 calcium-binding protein A4 |
| TF | Transcription factor |
| TYR | Tyrosinase |
| TYRP | Tyrosinase-related protein 1 |
| TWIST1 | Twist family bHLH transcription factor 1 |
| TWIST2 | Twist family bHLH transcription factor 2 |
| UM | Uveal melanoma |
| VWCE | von Willebrand factor C and EGF domains |
| WISP2 | Cellular communication network factor 5 |
| ZEB1 | Zinc finger E-box-binding homeobox 1 |
| ZEB2 | Zinc finger E-box-binding homeobox 2 |
References
- McLaughlin, C.C.; Wu, X.C.; Jemal, A.; Martin, H.J.; Roche, L.M.; Chen, V.W. Incidence of noncutaneous melanomas in the US. Cancer Interdiscip. Int. J. Am. Cancer Soc. 2005, 103, 1000–1007. [Google Scholar]
- Murray, T.G.; Boldt, H.C. Ocular Melanoma: Advances in Diagnostic and Therapeutic Strategies; Future Medicine Limited: London, UK, 2014. [Google Scholar]
- Shields, C.L.; Ganguly, A.; Bianciotto, C.G.; Turaka, K.; Tavallali, A.; Shields, J.A. Prognosis of uveal melanoma in 500 cases using genetic testing of fine-needle aspiration biopsy specimens. Ophthalmology 2011, 118, 396–401. [Google Scholar] [CrossRef]
- Blum, E.S.; Yang, J.; Komatsubara, K.M.; Carvajal, R.D. Clinical management of uveal and conjunctival melanoma. Oncol. (Williston Park) 2016, 30, 29–43. [Google Scholar]
- Kaliki, S.; Shields, C.L.; Shields, J.A. Uveal melanoma: Estimating prognosis. Indian J. Ophthalmol. 2015, 63, 93. [Google Scholar] [CrossRef]
- Versluis, M.; de Lange, M.J.; van Pelt, S.I.; Ruivenkamp, C.A.; Kroes, W.G.; Cao, J.; Jager, M.J.; Luyten, G.P.; van der Velden, P.A. Digital PCR validates 8q dosage as prognostic tool in uveal melanoma. PLoS ONE 2015, 10, e0116371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalirai, H.; Dodson, A.; Faqir, S.; Damato, B.; Coupland, S. Lack of BAP1 protein expression in uveal melanoma is associated with increased metastatic risk and has utility in routine prognostic testing. Br. J. Cancer 2014, 111, 1373–1380. [Google Scholar] [CrossRef] [Green Version]
- Onken, M.D.; Worley, L.A.; Char, D.H.; Augsburger, J.J.; Correa, Z.M.; Nudleman, E.; Aaberg Jr, T.M.; Altaweel, M.M.; Bardenstein, D.S.; Finger, P.T. Collaborative Ocular Oncology Group report number 1: Prospective validation of a multi-gene prognostic assay in uveal melanoma. Ophthalmology 2012, 119, 1596–1603. [Google Scholar] [CrossRef] [Green Version]
- Shain, A.H.; Bagger, M.M.; Yu, R.; Chang, D.; Liu, S.; Vemula, S.; Weier, J.F.; Wadt, K.; Heegaard, S.; Bastian, B.C. The genetic evolution of metastatic uveal melanoma. Nat. Genet. 2019, 51, 1123–1130. [Google Scholar] [CrossRef]
- Batlle, E.; Sancho, E.; Francí, C.; Domínguez, D.; Monfar, M.; Baulida, J.; De Herreros, A.G. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat. Cell Biol. 2000, 2, 84–89. [Google Scholar] [CrossRef]
- Eger, A.; Aigner, K.; Sonderegger, S.; Dampier, B.; Oehler, S.; Schreiber, M.; Berx, G.; Cano, A.; Beug, H.; Foisner, R. DeltaEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene 2005, 24, 2375–2385. [Google Scholar] [CrossRef] [Green Version]
- Comijn, J.; Berx, G.; Vermassen, P.; Verschueren, K.; van Grunsven, L.; Bruyneel, E.; Mareel, M.; Huylebroeck, D.; Van Roy, F. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol. Cell 2001, 7, 1267–1278. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Mani, S.A.; Donaher, J.L.; Ramaswamy, S.; Itzykson, R.A.; Come, C.; Savagner, P.; Gitelman, I.; Richardson, A.; Weinberg, R.A. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 2004, 117, 927–939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asnaghi, L.; Gezgin, G.; Tripathy, A.; Handa, J.T.; Merbs, S.L.; van der Velden, P.A.; Jager, M.J.; Harbour, J.W.; Eberhart, C.G. EMT-associated factors promote invasive properties of uveal melanoma cells. Mol. Vis. 2015, 21, 919. [Google Scholar]
- Eskelin, S.; Pyrhönen, S.; Summanen, P.; Hahka-Kemppinen, M.; Kivelä, T. Tumor doubling times in metastatic malignant melanoma of the uvea: Tumor progression before and after treatment. Ophthalmology 2000, 107, 1443–1449. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; Hoon, D.S.; Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 2011, 11, 426–437. [Google Scholar] [CrossRef]
- Park, J.J.; Diefenbach, R.J.; Byrne, N.; Kefford, R.; Long, G.V.; Scolyer, R.A.; Gray, E.; Carlino, M.S.; Rizos, H. Circulating Tumor DNA (ctDNA) in Patients (pts) with Metastatic Uveal Melanoma (UM) Treated with Protein Kinase C Inhibitor (PKCi); American Society of Clinical Oncology: Alexandria, VA, USA, 2020. [Google Scholar]
- Ambrosini, G.; Pratilas, C.A.; Qin, L.-X.; Tadi, M.; Surriga, O.; Carvajal, R.D.; Schwartz, G.K. Identification of unique MEK-dependent genes in GNAQ mutant uveal melanoma involved in cell growth, tumor cell invasion, and MEK resistance. Clin. Cancer Res. 2012, 18, 3552–3561. [Google Scholar] [CrossRef] [Green Version]
- Mego, M.; Cholujova, D.; Minarik, G.; Sedlackova, T.; Gronesova, P.; Karaba, M.; Benca, J.; Cingelova, S.; Cierna, Z.; Manasova, D. CXCR4-SDF-1 interaction potentially mediates trafficking of circulating tumor cells in primary breast cancer. Bmc Cancer 2016, 16, 127. [Google Scholar] [CrossRef] [Green Version]
- Anfuso, C.D.; Longo, A.; Distefano, A.; Amorini, A.M.; Salmeri, M.; Zanghì, G.; Giallongo, C.; Giurdanella, G.; Lupo, G. Uveal Melanoma Cells Elicit Retinal Pericyte Phenotypical and Biochemical Changes in an in Vitro Model of Coculture. Int. J. Mol. Sci. 2020, 21, 5557. [Google Scholar] [CrossRef]
- Fei, F.; Qu, J.; Zhang, M.; Li, Y.; Zhang, S. S100A4 in cancer progression and metastasis: A systematic review. Oncotarget 2017, 8, 73219. [Google Scholar] [CrossRef] [Green Version]
- Onken, M.D.; Ehlers, J.P.; Worley, L.A.; Makita, J.; Yokota, Y.; Harbour, J.W. Functional gene expression analysis uncovers phenotypic switch in aggressive uveal melanomas. Cancer Res. 2006, 66, 4602–4609. [Google Scholar] [CrossRef] [Green Version]
- Laura, D.; Océane, A.; Kalpana, M.; Laurent, C.; Nathalie, P.; Coquelle, F.M.; Gentien, D.; Jean-Baptiste, M.; Saule, S. Protein tyrosine phosphatase 4A3 (PTP4A3/PRL-3) promotes the aggressiveness of human uveal melanoma through dephosphorylation of CRMP2. Sci. Rep. 2019, 9, 1–13. [Google Scholar]
- Laurent, C.; Valet, F.; Planque, N.; Silveri, L.; Maacha, S.; Anezo, O.; Hupe, P.; Plancher, C.; Reyes, C.; Albaud, B. High PTP4A3 phosphatase expression correlates with metastatic risk in uveal melanoma patients. Cancer Res. 2011, 71, 666–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Dijk, I.A.; Laura Ferrando, M.; van der Wijk, A.E.; Hoebe, R.A.; Nazmi, K.; de Jonge, W.J.; Krawczyk, P.M.; Bolscher, J.G.; Veerman, E.C.; Stap, J. Human salivary peptide histatin-1 stimulates epithelial and endothelial cell adhesion and barrier function. Faseb J. 2017, 31, 3922–3933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Proença, R.P.; Fonseca, C.; Goyeneche, A.A.; Ito, H.; Dias, A.; Marques-Neves, C.; Burnier, M.N. The expression of ALK, RET, ROS1, c-MET, EGFR and IGF-R1 in uveal melanoma and its association to clinicopathological characteristics and prognosis. Investig. Ophthalmol. Vis. Sci. 2020, 61, 2833. [Google Scholar]
- Willecke, K.; Eiberger, J.; Degen, J.; Eckardt, D.; Romualdi, A.; Güldenagel, M.; Deutsch, U.; Söhl, G. Structural and functional diversity of connexin genes in the mouse and human genome. Biol. Chem. 2002, 383, 725–737. [Google Scholar] [CrossRef]
- Melander, M.C.; Jürgensen, H.J.; Madsen, D.H.; Engelholm, L.H.; Behrendt, N. The collagen receptor uPARAP/Endo180 in tissue degradation and cancer. Int. J. Oncol. 2015, 47, 1177–1188. [Google Scholar] [CrossRef] [Green Version]
- Suesskind, D.; Schatz, A.; Schnichels, S.; Coupland, S.E.; Lake, S.L.; Wissinger, B.; Bartz-Schmidt, K.U.; Henke-Fahle, S. GDF-15: A novel serum marker for metastases in uveal melanoma patients. Graefe’s Arch. Clin. Exp. Ophthalmol. 2012, 250, 887–895. [Google Scholar] [CrossRef]
- Pache, M.; Glatz, K.; Bösch, D.; Dirnhofer, S.; Mirlacher, M.; Simon, R.; Schraml, P.; Rufle, A.; Flammer, J.; Sauter, G. Sequence analysis and high-throughput immunhistochemical profiling of KIT (CD 117) expression in uveal melanoma using tissue microarrays. Virchows Arch. 2003, 443, 741–744. [Google Scholar] [CrossRef]
- Figueiredo, C.R.; Kalirai, H.; Sacco, J.J.; Azevedo, R.A.; Duckworth, A.; Slupsky, J.R.; Coulson, J.M.; Coupland, S.E. Loss of BAP1 expression is associated with an immunosuppressive microenvironment in uveal melanoma, with implications for immunotherapy development. J. Pathol. 2020, 250, 420–439. [Google Scholar] [CrossRef] [Green Version]
- Krishna, Y.; Acha-Sagredo, A.; Sabat-Pośpiech, D.; Kipling, N.; Clarke, K.; Figueiredo, C.R.; Kalirai, H.; Coupland, S.E. Transcriptome Profiling Reveals New Insights into the Immune Microenvironment and Upregulation of Novel Biomarkers in Metastatic Uveal Melanoma. Cancers 2020, 12. [Google Scholar] [CrossRef]
- Fazilaty, H.; Rago, L.; Kass Youssef, K.; Ocaña, O.H.; Garcia-Asencio, F.; Arcas, A.; Galceran, J.; Nieto, M.A. A gene regulatory network to control EMT programs in development and disease. Nat. Commun. 2019, 10, 5115. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.; Zhang, J.; Liu, J. URG11 predicts poor prognosis of pancreatic cancer by enhancing epithelial–mesenchymal transition-driven invasion. Med. Oncol. 2014, 31, 64. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.Y.; Li, R.; Zhang, Q.J.; Wang, Y.; Jiang, Y.; Meng, Q.Y.; Xi, Q.L.; Wu, G.H. Prognostic role of cyclin D2/D3 in multiple human malignant neoplasms: A systematic review and meta-analysis. Cancer Med. 2019, 8, 2717–2729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, F.; Lord, M.S.; Stallcup, W.B.; Whitelock, J.M. Cell surface chondroitin sulphate proteoglycan 4 (CSPG4) binds to the basement membrane heparan sulphate proteoglycan, perlecan, and is involved in cell adhesion. J. Biochem. 2018, 163, 399–412. [Google Scholar] [CrossRef] [PubMed]
- Asnaghi, L.; Lin, M.H.; Lim, K.S.; Lim, K.J.; Tripathy, A.; Wendeborn, M.; Merbs, S.L.; Handa, J.T.; Sodhi, A.; Bar, E.E. Hypoxia promotes uveal melanoma invasion through enhanced Notch and MAPK activation. PLoS ONE 2014, 9, e105372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Lei, C.; Long, K.; Yang, X.; Zhu, Z.; Zhang, L.; Liu, J. Mutant GNAQ promotes cell viability and migration of uveal melanoma cells through the activation of Notch signaling. Oncol. Rep. 2015, 34, 295–301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Triozzi, P.L.; Schoenfield, L.; Plesec, T.; Saunthararajah, Y.; Tubbs, R.R.; Singh, A.D. Molecular profiling of primary uveal melanomas with tumor-infiltrating lymphocytes. Oncoimmunology 2019, 8, e947169. [Google Scholar] [CrossRef]
- Souri, Z.; Wierenga, A.; van Weeghel, C.; van Der Velden, P.A.; Kroes, W.G.; Luyten, G.P.; van Der Burg, S.H.; Jochemsen, A.G.; Jager, M.J. Loss of BAP1 Is Associated with Upregulation of the NFkB Pathway and Increased HLA Class I Expression in Uveal Melanoma. Cancers 2019, 11, 1102. [Google Scholar] [CrossRef] [Green Version]
- Kadkol, S.S.; Lin, A.Y.; Barak, V.; Kalickman, I.; Leach, L.; Valyi-Nagy, K.; Majumdar, D.; Setty, S.; Maniotis, A.J.; Folberg, R. Osteopontin expression and serum levels in metastatic uveal melanoma: A pilot study. Investig. Ophthalmol. Vis. Sci. 2006, 47, 802–806. [Google Scholar] [CrossRef]
- Das, A.; Dhar, K.; Maity, G.; Sarkar, S.; Ghosh, A.; Haque, I.; Dhar, G.; Banerjee, S.; Banerjee, S.K. Deficiency of CCN5/WISP-2-Driven Program in breast cancer Promotes Cancer Epithelial cells to mesenchymal stem cells and Breast Cancer growth. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Shi, X.; Chen, M.; Xu, N.; Sun, D.; Bai, R.; Chen, H.; Ding, K.; Sheng, J.; Xu, Z. Angiogenin promotes colorectal cancer metastasis via tiRNA production. Int. J. Cancer 2019, 145, 1395–1407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, X.; Zhang, Q.; Chen, W.; Hu, Q.; Lou, Y.; Fu, Q.-H.; Zhang, J.-Y.; Chen, Y.-W.; Ye, L.-Y.; Wang, Y. Hook1 inhibits malignancy and epithelial–mesenchymal transition in hepatocellular carcinoma. Tumor Biol. 2017, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harbour, J.W. A prognostic test to predict the risk of metastasis in uveal melanoma based on a 15-gene expression profile. Methods Mol. Biol. 2014, 1102, 427–440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, Y.; Zhang, D.; Hu, T.; Zhao, H.; Zhao, X.; Lou, Z.; He, Y.; Qin, W.; Xia, J.; Zhang, X. KMT2A histone methyltransferase contributes to colorectal cancer development by promoting cathepsin Z transcriptional activation. Cancer Med. 2019, 8, 3544–3552. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, L.; Ma, Z.; Yang, M.; Tang, J.; Fu, Y.; Mao, Y.; Hong, X.; Zhang, Y. ETV1 induces epithelial to mesenchymal transition in human gastric cancer cells through the upregulation of Snail expression. Oncol. Rep. 2013, 30, 2859–2863. [Google Scholar] [CrossRef]
- Pereira, P.R.; Odashiro, A.N.; Lim, L.-A.; Miyamoto, C.; Blanco, P.L.; Odashiro, M.; Maloney, S.; De Souza, D.F.; Burnier Jr, M.N. Current and emerging treatment options for uveal melanoma. Clin. Ophthalmol. 2013, 7, 1669. [Google Scholar] [CrossRef] [Green Version]
- Palmirotta, R.; Lovero, D.; Cafforio, P.; Felici, C.; Mannavola, F.; Pellè, E.; Quaresmini, D.; Tucci, M.; Silvestris, F. Liquid biopsy of cancer: A multimodal diagnostic tool in clinical oncology. Ther. Adv. Med. Oncol. 2018, 10, 1758835918794630. [Google Scholar] [CrossRef]
- Mattox, A.K.; Bettegowda, C.; Zhou, S.; Papadopoulos, N.; Kinzler, K.W.; Vogelstein, B. Applications of liquid biopsies for cancer. Sci. Transl. Med. 2019, 11, eaay1984. [Google Scholar] [CrossRef] [Green Version]
- Tura, A.; Lueke, J.; Grisanti, S. Liquid biopsy for uveal melanoma. Exon Publ. 2018, 37–48. [Google Scholar] [CrossRef] [Green Version]
- Ly, L.V.; Odish, O.F.; de Wolff-Rouendaal, D.; Missotten, G.S.; Luyten, G.P.; Jager, M.J. Intravascular presence of tumor cells as prognostic parameter in uveal melanoma: A 35-year survey. Investig. Ophthalmol. Vis. Sci. 2010, 51, 658–665. [Google Scholar] [CrossRef] [Green Version]
- Callejo, S.A.; Antecka, E.; Blanco, P.L.; Edelstein, C.; Burnier, M.N., Jr. Identification of circulating malignant cells and its correlation with prognostic factors and treatment in uveal melanoma. A prospective longitudinal study. Eye 2007, 21, 752–759. [Google Scholar] [CrossRef] [PubMed]
- Bidard, F.C.; Madic, J.; Mariani, P.; Piperno-Neumann, S.; Rampanou, A.; Servois, V.; Cassoux, N.; Desjardins, L.; Milder, M.; Vaucher, I. Detection rate and prognostic value of circulating tumor cells and circulating tumor DNA in metastatic uveal melanoma. Int. J. Cancer 2014, 134, 1207–1213. [Google Scholar] [CrossRef] [PubMed]
- Terai, M.; Mu, Z.; Eschelman, D.J.; Gonsalves, C.F.; Kageyama, K.; Chervoneva, I.; Orloff, M.; Weight, R.; Mastrangelo, M.J.; Cristofanilli, M.; et al. Arterial Blood, Rather Than Venous Blood, is a Better Source for Circulating Melanoma Cells. EBioMedicine 2015, 2, 1821–1826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roszik, J.; Gombos, D.; Upshaw, J.; Sarli, V.; Meas, S.; Lucci, A.; Hall, C.; Patel, S.; Anand, K. Abstract A12: Presence of Circulating Tumor Cells is an Adverse Risk Factor for Early-Stage Uveal Melanoma; AACR: Philadelphia, PA, USA, 2020. [Google Scholar]
- Beasley, A.; Isaacs, T.; Khattak, M.A.; Freeman, J.B.; Allcock, R.; Chen, F.K.; Pereira, M.R.; Yau, K.; Bentel, J.; Vermeulen, T. Clinical application of circulating tumor cells and circulating tumor DNA in uveal melanoma. Jco Precis. Oncol. 2018, 2, 1–12. [Google Scholar] [CrossRef]
- Eide, N.; Faye, R.S.; Høifødt, H.K.; Sandstad, B.; Qvale, G.; Faber, R.; Jebsen, P.; Kvalheim, G.; Fodstad, Ø. Immunomagnetic detection of micrometastatic cells in bone marrow of uveal melanoma patients: A paradox. Acta Ophthalmol. 2015, 93, 59–66. [Google Scholar] [CrossRef]
- Mego, M.; Mani, S.A.; Lee, B.N.; Li, C.; Evans, K.W.; Cohen, E.N.; Gao, H.; Jackson, S.A.; Giordano, A.; Hortobagyi, G.N. Expression of epithelial–mesenchymal transition-inducing transcription factors in primary breast cancer: The effect of neoadjuvant therapy. Int. J. Cancer 2012, 130, 808–816. [Google Scholar] [CrossRef]
- Mego, M.; Karaba, M.; Minarik, G.; Benca, J.; Silvia, J.; Sedlackova, T.; Manasova, D.; Kalavska, K.; Pindak, D.; Cristofanilli, M.; et al. Circulating Tumor Cells With Epithelial-to-mesenchymal Transition Phenotypes Associated With Inferior Outcomes in Primary Breast Cancer. Anticancer Res. 2019, 39, 1829–1837. [Google Scholar] [CrossRef]
- Madic, J.; Piperno-Neumann, S.; Servois, V.; Rampanou, A.; Milder, M.; Trouiller, B.; Gentien, D.; Saada, S.; Assayag, F.; Thuleau, A. Pyrophosphorolysis-activated polymerization detects circulating tumor DNA in metastatic uveal melanoma. Clin. Cancer Res. 2012, 18, 3934–3941. [Google Scholar] [CrossRef] [Green Version]
- Metz, C.H.; Scheulen, M.; Bornfeld, N.; Lohmann, D.; Zeschnigk, M. Ultradeep sequencing detects GNAQ and GNA11 mutations in cell-free DNA from plasma of patients with uveal melanoma. Cancer Med. 2013, 2, 208–215. [Google Scholar] [CrossRef]
- Chen, Y.; Lu, X.; Montoya-Durango, D.E.; Liu, Y.-H.; Dean, K.C.; Darling, D.S.; Kaplan, H.J.; Dean, D.C.; Gao, L.; Liu, Y. ZEB1 regulates multiple oncogenic components involved in uveal melanoma progression. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Tsantoulis, P.; Delorenzi, M.; Bièche, I.; Vacher, S.; Mariani, P.; Cassoux, N.; Houy, A.; Stern, M.-H.; Roman-Roman, S.; Dietrich, P.-Y. Prospective validation in epithelial tumors of a gene expression predictor of liver metastasis derived from uveal melanoma. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Tao, Y.; Ni, N.; Zhou, X.; Xiong, J.; Zeng, X.; Xu, X.; Qi, J.; Sun, J. miR-128 targets the CC chemokine ligand 18 gene (CCL18) in cutaneous malignant melanoma progression. J. Dermatol. Sci. 2018, 91, 317–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, F.; Huang, Q.-M.; Hu, D.-N.; Jonas, J.B.; Wei, W.-B. Immune oppression array elucidating immune escape and survival mechanisms in uveal melanoma. Int. J. Ophthalmol. 2016, 9, 1701. [Google Scholar]
- Wang, X.; Wang, Y.; Yu, L.; Sakakura, K.; Visus, C.; Schwab, J.; Ferrone, C.; Favoino, E.; Koya, Y.; Campoli, M. CSPG4 in cancer: Multiple roles. Curr. Mol. Med. 2010, 10, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Madigan, M.; Lai, K.; Conway, R.; Billson, F.; Crouch, R.; Allen, B. Human uveal melanoma expresses NG2 immunoreactivity. Br. J. Ophthalmol. 2003, 87, 629–632. [Google Scholar] [CrossRef] [PubMed]
- Ilieva, K.M.; Cheung, A.; Mele, S.; Chiaruttini, G.; Crescioli, S.; Griffin, M.; Nakamura, M.; Spicer, J.F.; Tsoka, S.; Lacy, K.E. Chondroitin sulfate proteoglycan 4 and its potential as an antibody immunotherapy target across different tumor types. Front. Immunol. 2018, 8, 1911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sturge, J. Endo180 at the cutting edge of bone cancer treatment and beyond. J. Pathol. 2016, 238, 485–488. [Google Scholar] [CrossRef] [Green Version]
- Pan, B.; Ye, Y.; Liu, H.; Zhen, J.; Zhou, H.; Li, Y.; Qu, L.; Wu, Y.; Zeng, C.; Zhong, W. URG11 Regulates Prostate Cancer Cell Proliferation, Migration, and Invasion. Biomed. Res. Int. 2018, 2018, 4060728. [Google Scholar] [CrossRef]
- Liu, Z.L.; Wu, J.; Wang, L.X.; Yang, J.F.; Xiao, G.M.; Sun, H.P.; Chen, Y.J. Knockdown of downregulated Gene 11 (URG11) Inhibits Proliferation, Invasion, and β-Catenin Expression in Non-Small Cell Lung Cancer Cells. Oncol. Res. 2016, 24, 197–204. [Google Scholar] [CrossRef]
- Xie, H.; Liu, J. Increased expression URG11 in hepatocellular carcinoma tissues promotes the growth of hepatocellular carcinoma cells. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 2015, 31, 1523–1527. [Google Scholar]
- Kotini, M.; Mayor, R. Connexins in migration during development and cancer. Dev. Biol. 2015, 401, 143–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nieto, M.A.; Huang, R.Y.-J.; Jackson, R.A.; Thiery, J.P. EMT: 2016. Cell 2016, 166, 21–45. [Google Scholar] [CrossRef] [Green Version]
- Santarlasci, V.; Maggi, L.; Capone, M.; Querci, V.; Beltrame, L.; Cavalieri, D.; D’Aiuto, E.; Cimaz, R.; Nebbioso, A.; Liotta, F.; et al. Rarity of human T helper 17 cells is due to retinoic acid orphan receptor-dependent mechanisms that limit their expansion. Immunity 2012, 36, 201–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Onken, M.D.; Worley, L.A.; Ehlers, J.P.; Harbour, J.W. Gene expression profiling in uveal melanoma reveals two molecular classes and predicts metastatic death. Cancer Res 2004, 64, 7205–7209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, P.P.; Purbey, P.K.; Sinha, C.K.; Notani, D.; Limaye, A.; Jayani, R.S.; Galande, S. Phosphorylation of SATB1, a global gene regulator, acts as a molecular switch regulating its transcriptional activity in vivo. Mol. Cell 2006, 22, 231–243. [Google Scholar] [CrossRef]


| All n (%) | Metastasis Absent n (%) | Metastasis Present n (%) | p-Value | |
|---|---|---|---|---|
| Gender | ||||
| Male | 25 (47.2) | 22 (52.4) | 3 (27.3) | 0.138 |
| Female | 28 (52.8) | 20 (47.6) | 8 (72.7) | |
| Eye | ||||
| Right | 26 (49.1) | 21 (50.0) | 5 (45.5) | 0.788 |
| Left | 27 (50.9) | 21 (50.0) | 6 (54.5) | |
| Median age (range) | 67 years (33–87) | 67 | 65 | |
| Median tumor volume (range) | 1.1 cm3 (0.2–2.6) | 1.0 cm3 (0.2–2.6) | 1.4 cm3 (0.3–2.6) | 0.414 |
| <1.55 cm3 | 38 (71.7) | 30 (71.4) | 8 (72.7) | 0.932 |
| ≥1.55 cm3 | 15 (28.3) | 12 (28.6) | 3 (27.3) | |
| Diagnosis | ||||
| C69.3 | 43 (81.1) | 36 (85.7) | 7 (63.6) | 0.096 |
| C69.4 | 10 (18.9) | 6 (14.3) | 4 (36.4) | |
| Cell type | ||||
| Epitheloid | 10 (25.6) | 5 (16.7) | 5 (55.6) | 0.060 |
| Spindle | 19 (48.7) | 16 (53.3) | 3 (33.3) | |
| Mixed | 10 (25.6) | 9 (30.0) | 1 (11.1) | |
| Melan-A | ||||
| Positive | 37 (94.9) | 28 (93.3) | 9 (100) | 0.426 |
| Negative | 2 (5.1) | 2 (6.7) | 0 (0) | |
| S100 | ||||
| Positive | 36 (92.3) | 28 (93.3) | 8 (88.9) | 0.661 |
| Negative | 3 (7.7) | 2 (6.7) | 1 (11.1) | |
| HMB45 | ||||
| Positive | 36 (92.3) | 28 (93.3) | 8 (88.9) | 0.661 |
| Negative | 3 (7.7) | 2 (6.7) | 1 (11.1) | |
| Therapy | ||||
| Enucleation | 32 (60.4) * | 22 (52.4) | 9 (81.1) | 0.031 |
| Enucleation after radiosurgery in the past | 8 (15.1) | 7 (16.7) | 1 (9.1) | |
| Stereotactic radiosurgery | 13 (24.5) | 13 (31.0) | 0 (0.0) | |
| Vascular invasion | ||||
| Present | 5 (12.8) | 2 (6.9) | 3 (30.0) | 0.060 |
| Absent | 37 (87.2) | 27 (93.1) | 7 (70.0) | |
| Lymphogenic invasion | ||||
| Present | 10 (25.6) | 5 (17.2) | 5 (50.0) | 0.041 |
| Absent | 29 (74.4) | 24 (82.2) | 5 (50.0) | |
| Perineural invasion | ||||
| Present | 9 (23.1) | 6 (20.7) | 3 (30.0) | 0.547 |
| Absent | 30 (76.9) | 27 (79.0) | 7 (70.0) | |
| Extrabulbar overgrowth | ||||
| Present | 30 (76.9) | 23 (76.7) | 5 (55.6) | 0.217 |
| Absent | 9 (23.1) | 7 (23.3) | 4 (44.4) | |
| TNM staging | ||||
| I-IIB | 30 (56.6) | 27 (62.8) | 3 (30.0) | 0.059 |
| IIIA-IIIC | 23 (43.4) | 16 (37.2) | 7 (70.0) | |
| MLPA status $ | ||||
| Monosomy 3 | 16 (51.6) | 10 (43.5) | 6 (75.0) | 0.124 |
| Disomy 3 | 15 (48.4) | 13 (56.5) | 2 (25.0) |
| Mutations | All Tumor Tissues n (%) | All Plasma n (%) | Primary UM Plasma n (%) | Metastatic UM Plasma n (%) |
|---|---|---|---|---|
| Number * | 31 (100) | 42 (100) | 32 (100) | 11 (100) |
| GNA11 p.Q209L | 17 (54.8) | 4 (9.5) | 1 (3.1) | 3 (27.3) |
| GNA11 p.Q209P | 1 (3.2) | 0 | ||
| GNA11 p.R183C | 1 (3.2) | 1 (2.4) | 1 (9.1) | |
| GNAQ p.Q209L | 1 (3.2) | 0 | ||
| GNAQ p.Q209P | 10 (32.3) | 4 (9.5) | 2 (6.3) | 2 (18.2) |
| GNAQ p.Q209R | 1 (3.2) | 0 | ||
| GNAQ p.R183Q | 0 | 0 | ||
| CYSLTR2 p.L129Q | 0 | 0 | ||
| PLCB4 p.D630Y | 0 | 0 | ||
| Total | 31 (100) | 9 (21.4) | 3 (9.4) | 6 (54.5) |
| Gene (n = 17) | Chromosome Location | p-Value | Fold Change | Function | Reference |
|---|---|---|---|---|---|
| CCL18 | 17q12 | 1.44 × 10−6 | 22.7 | Expressed in retinal pericyte UM cells, maintain blood-retinal barrier integrity in UM | [20] |
| S100A4 | 1q21.3 | 2.19 × 10−5 | 7.9 | Role in motility, invasion, tubulin polymerization, its chromosomal rearrangements and expression changes implicated in tumor metastasis | [21] |
| CDH1 | 16q22.1 | 1.05 × 10−5 | 7.7 | ↑ in Class 2 UM tumors, associated with transcriptional up-regulation, and plasma membrane colocalization | [22] |
| PTP4A3 | 8q24.3 | 1.45 × 10−6 | 6.4 | ↑ the aggressiveness of human UM, ↑metastatic risk in UM patients | [23,24] |
| HTN1 | 4q13.3 | 0.031 | 7.1 | ↑ epithelial and endothelial cell adhesion and barrier function | [25] |
| ALK | 2p23.2-p23.1 | 0.001 | 6.0 | Role in cellular communication and the normal development and function of the nervous system | [26] |
| GJB2 | 13q12.11 | 0.006 | 5.6 | Gap junction channel protein having a role in direct communication between cells | [27] |
| MRC2 | 17q23.2 | 8.44 × 10−5 | 5.4 | Role in extracellular matrix remodeling by mediating the internalization and lysosomal degradation of collagen ligands | [28] |
| GDF15 | 19p13.11 | 0.003 | 5.2 | A serum marker for metastases in UM | [29] |
| KIT | 4q12 | 9.62 × 10−6 | 5.1 | ↑ in UM, role in cell survival, proliferation, melanogenesis, and hematopoiesis | [30] |
| TWIST2 | 2q37.3 | 3.31 × 10−7 | 5.1 | Role in the progression of non-epithelium-derived tumors, including melanomas | [14] |
| LGALS3 | 14q22.3 | 6.61 × 10−5 | 4.9 | ↑ in BAP1 loss UM tumors and in metastatic liver of UM patients | [31,32] |
| PRRX1 | 1q24.2 | 2.77 × 10−5 | 4.7 | Regulates differentiation of mesenchymal precursors | [33] |
| VWCE | 11q12.2 | 1.39 × 10−9 | 4.5 | Correlated with poor prognosis of pancreatic cancer by ↑EMT-driven invasion | [34] |
| CCND2 | 12p13.32 | 0.002 | 4.4 | Drives cell cycle progression, associated with tumorigenesis | [35] |
| CSPG4 | 15q24.2 | 8.44 × 10−8 | 4.3 | ↑ expression in tumor, perivascular, and oligodendrocyte cells, involved in cell adhesion and migration | [36] |
| JAG1 | 20p12.2 | 8.10 × 10−9 | 4.1 | ↑ expression as response to hypoxia in metastatic spread of UM | [37,38] |
| Gene (n = 8) | Chromosome Location | p-Value | Fold Change | Function | Reference |
|---|---|---|---|---|---|
| RORC | 1q21.3 | 4.23 ×10−7 | −28.5 | Th17-associated transcription factor | [39] |
| SPP1 | 4q22.1 | 2.37 × 10−6 | −15.4 | ↓ in M3/BAP1-negative tumors, expressed in hepatic metastases from UM, ↑ serum levels correlate with metastatic melanoma to the liver | [40,41] |
| WISP2 | 20q13.12 | 1.26 × 10−4 | −8.1 | Regulates diverse cellular functions, including cell adhesion, migration, proliferation, differentiation | [42] |
| ANG | 14q11.2 | 2.04 × 10−9 | −7.5 | Induce tumor angiogenesis, ↑ cell survival, proliferation and/or migration | [43] |
| HOOK1 | 1p32.1 | 1.23 × 10−8 | −7.2 | Involved in endocytic membrane trafficking to the microtubule cytoskeleton | [44] |
| SATB1 | 3p24.3 | 9.28 × 10−9 | −5.1 | One of the 12 gene expression profile predictive of primary UM metastasis | [45] |
| CTSZ | 20q13.32 | 0.013 | −4.6 | Specific functions in cancer invasion and metastasis | [46] |
| ETV1 | 7p21.2 | 3.56 × 10−5 | −4.3 | Role in cell growth, angiogenesis, migration, proliferation, and differentiation | [47] |
| Genes | Strand | Sequence (5′-3′) |
|---|---|---|
| GNA11 p.Q209P/L | Forward | TCTCTGAGAGCGTCCTTGC |
| Reverse | GACTTGGTCGTATTCGCTGAG | |
| GNA11 p.R183C | Forward | TGGTTTGGGTGCTGTGTC |
| Reverse | CGGAAGATGATGTTCTCC | |
| GNAQ p.Q209P/L/R | Forward | CATTGCCTGTCTAAAGAACAC |
| Reverse | TGCAGAATGGTCGATGTAGG | |
| GNAQ p.R183Q | Forward | ATTCGATGATCCCTGTGGTG |
| Reverse | ACAGCTTTGGTGTGATGGTG | |
| PLCB4 p.D630Y | Forward | CAGTGAACTGTGATCTTAAGC |
| Reverse | ATCTGGCAGCCAGCGTTC | |
| CYSLTR2 p.L129Q | Forward | ATCTCCTGTTCATAAGCACGC |
| Reverse | AACGCACAACACTCAGCACG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soltysova, A.; Sedlackova, T.; Dvorska, D.; Jasek, K.; Chokhachi Baradaran, P.; Horvathova Kajabova, V.; Demkova, L.; Buocikova, V.; Kurucova, T.; Lyskova, D.; et al. Monosomy 3 Influences Epithelial-Mesenchymal Transition Gene Expression in Uveal Melanoma Patients; Consequences for Liquid Biopsy. Int. J. Mol. Sci. 2020, 21, 9651. https://doi.org/10.3390/ijms21249651
Soltysova A, Sedlackova T, Dvorska D, Jasek K, Chokhachi Baradaran P, Horvathova Kajabova V, Demkova L, Buocikova V, Kurucova T, Lyskova D, et al. Monosomy 3 Influences Epithelial-Mesenchymal Transition Gene Expression in Uveal Melanoma Patients; Consequences for Liquid Biopsy. International Journal of Molecular Sciences. 2020; 21(24):9651. https://doi.org/10.3390/ijms21249651
Chicago/Turabian StyleSoltysova, Andrea, Tatiana Sedlackova, Dana Dvorska, Karin Jasek, Pooneh Chokhachi Baradaran, Viera Horvathova Kajabova, Lucia Demkova, Verona Buocikova, Terezia Kurucova, Darina Lyskova, and et al. 2020. "Monosomy 3 Influences Epithelial-Mesenchymal Transition Gene Expression in Uveal Melanoma Patients; Consequences for Liquid Biopsy" International Journal of Molecular Sciences 21, no. 24: 9651. https://doi.org/10.3390/ijms21249651





