Functional Characterization of BRASSINAZOLE-RESISTANT 1 in Panax Ginseng (PgBZR1) and Brassinosteroid Response during Storage Root Formation
Abstract
1. Introduction
2. Results
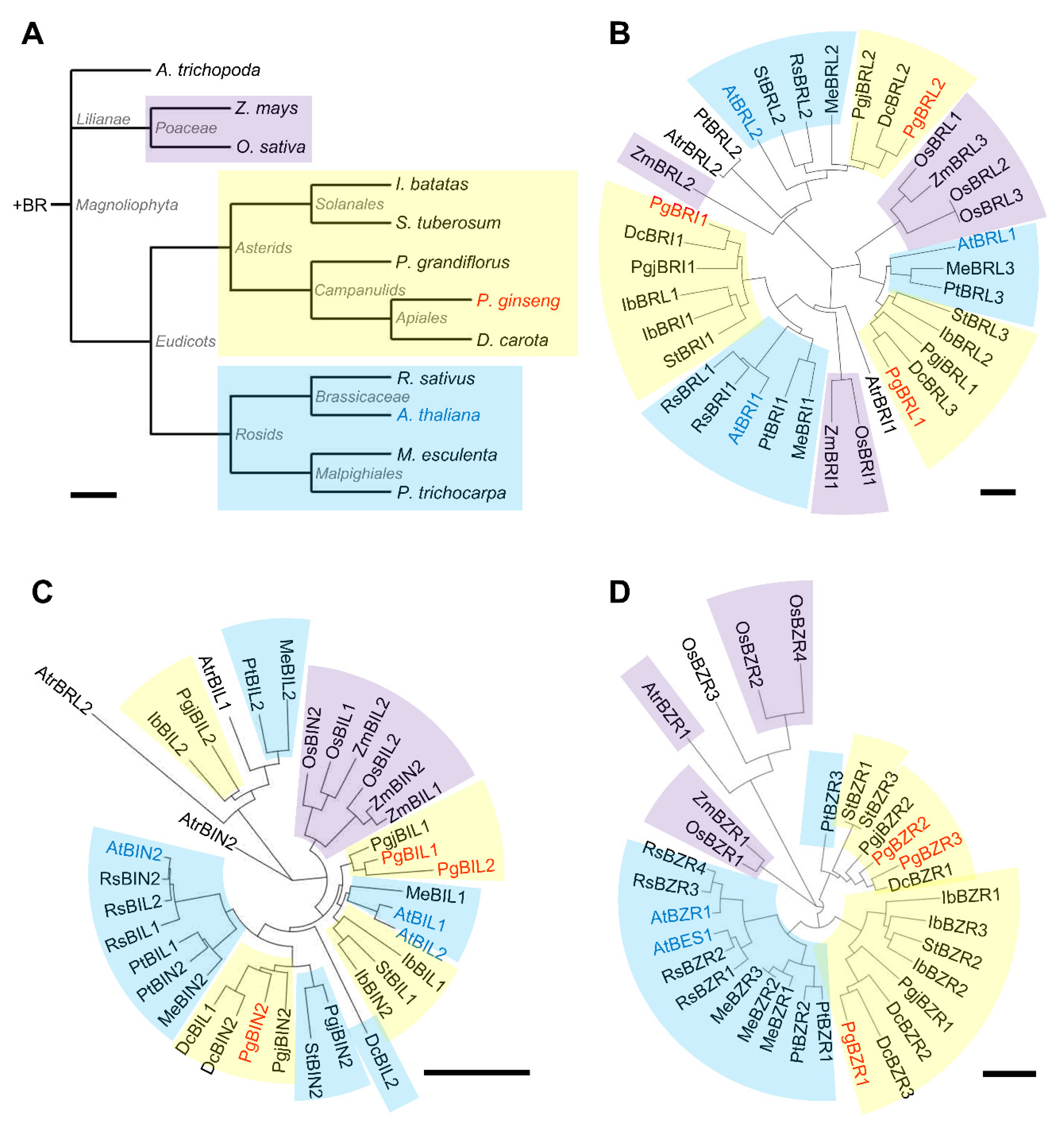
2.1. Phylogenetic Analysis of Brassinosteroid Signaling Components in P. ginseng
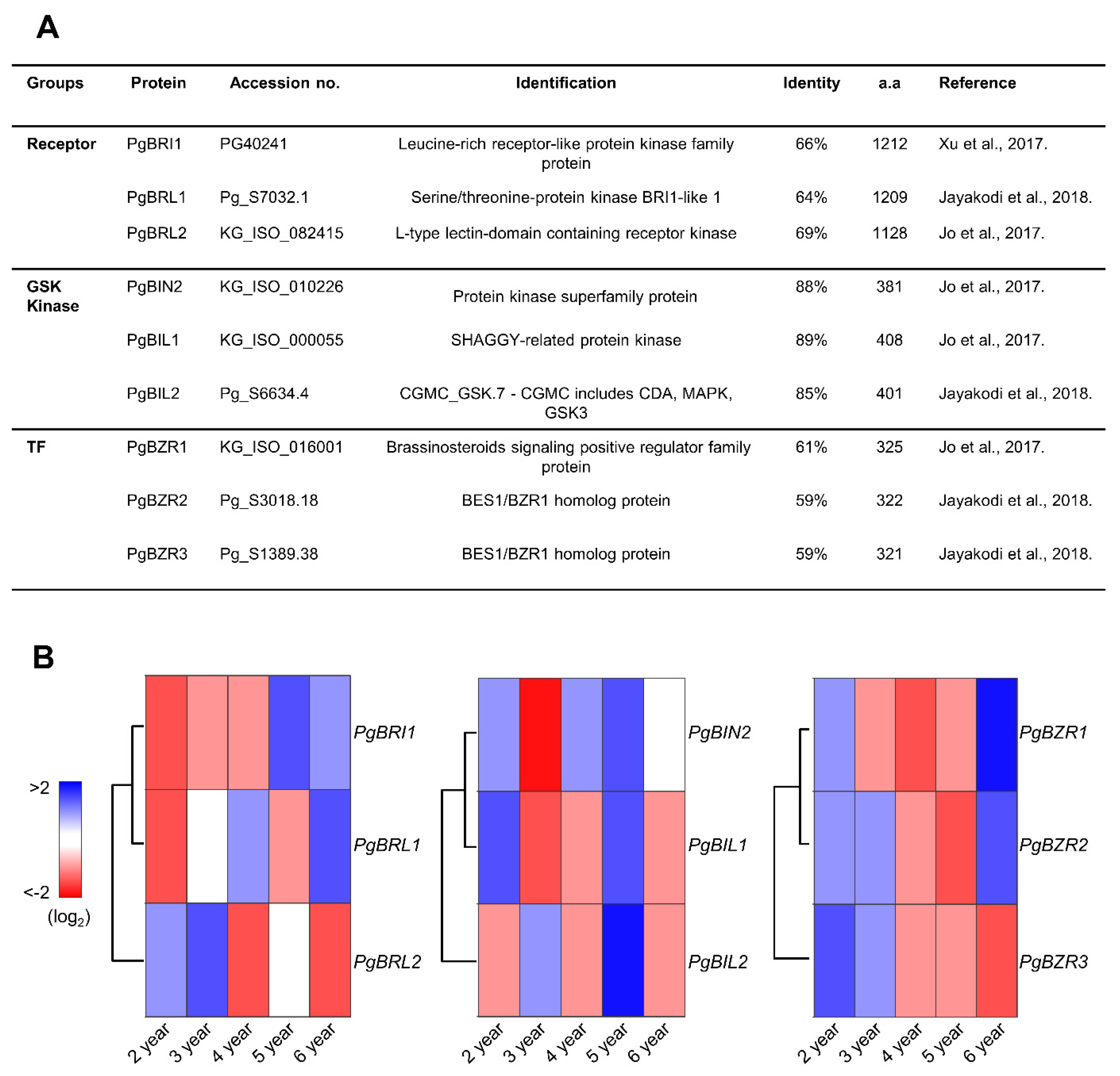
2.2. Identification of Arabidopsis BRI1, BIN2, and BZR1 Homologs in P. ginseng
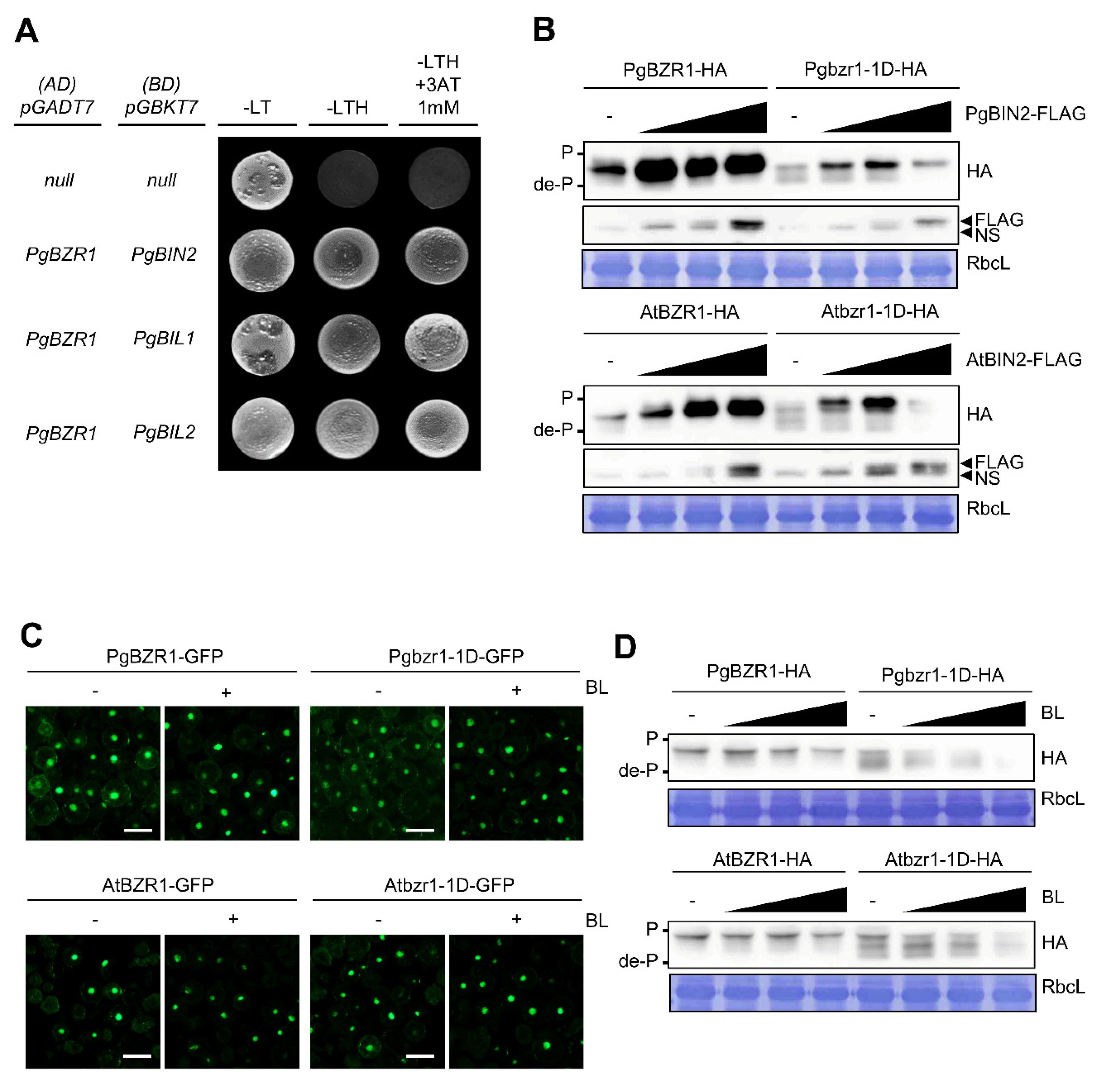
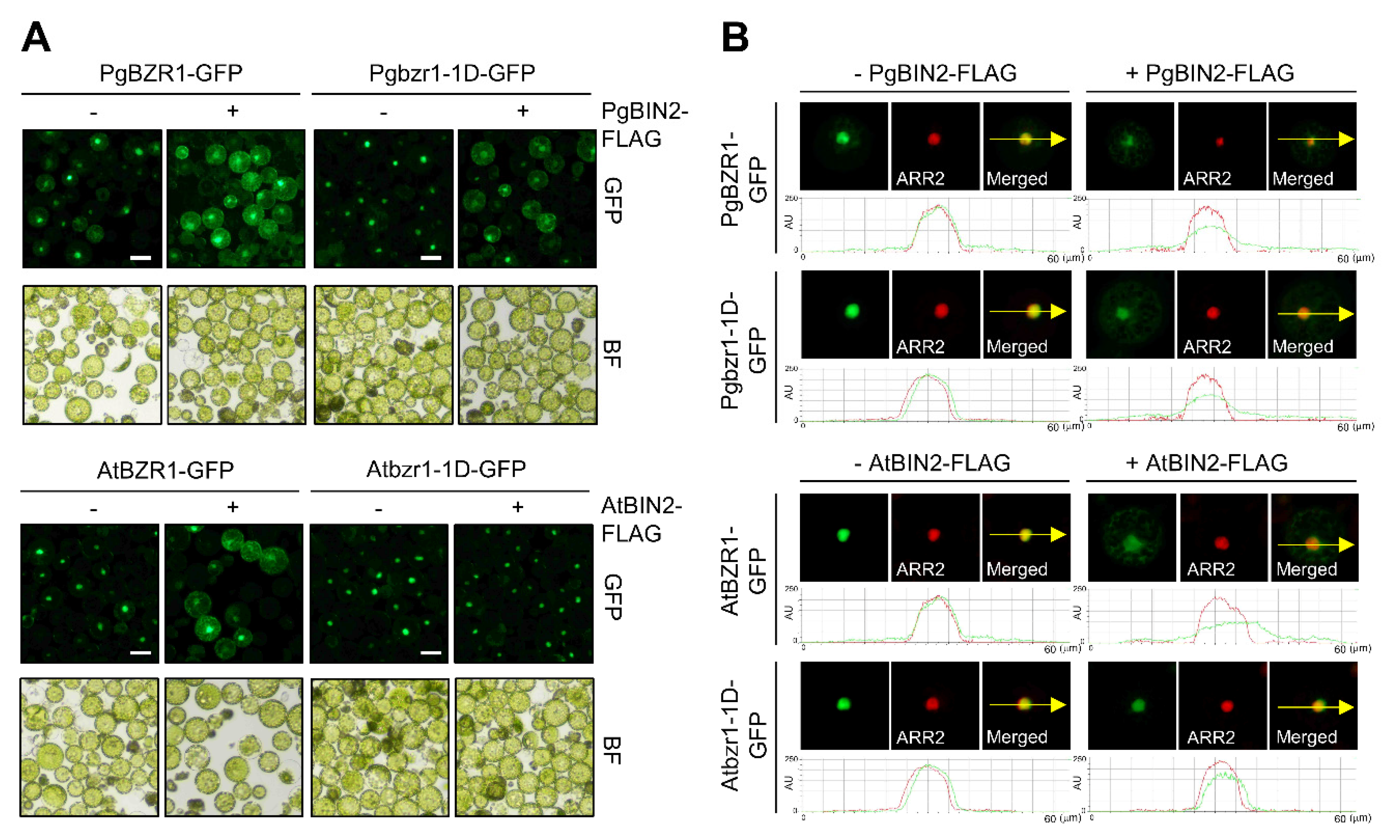
2.3. PgBIN2 Phosphorylates and Leads Nucleocytoplasmic Shuttling of PgBZR1 from the Nucleus to Cytosol
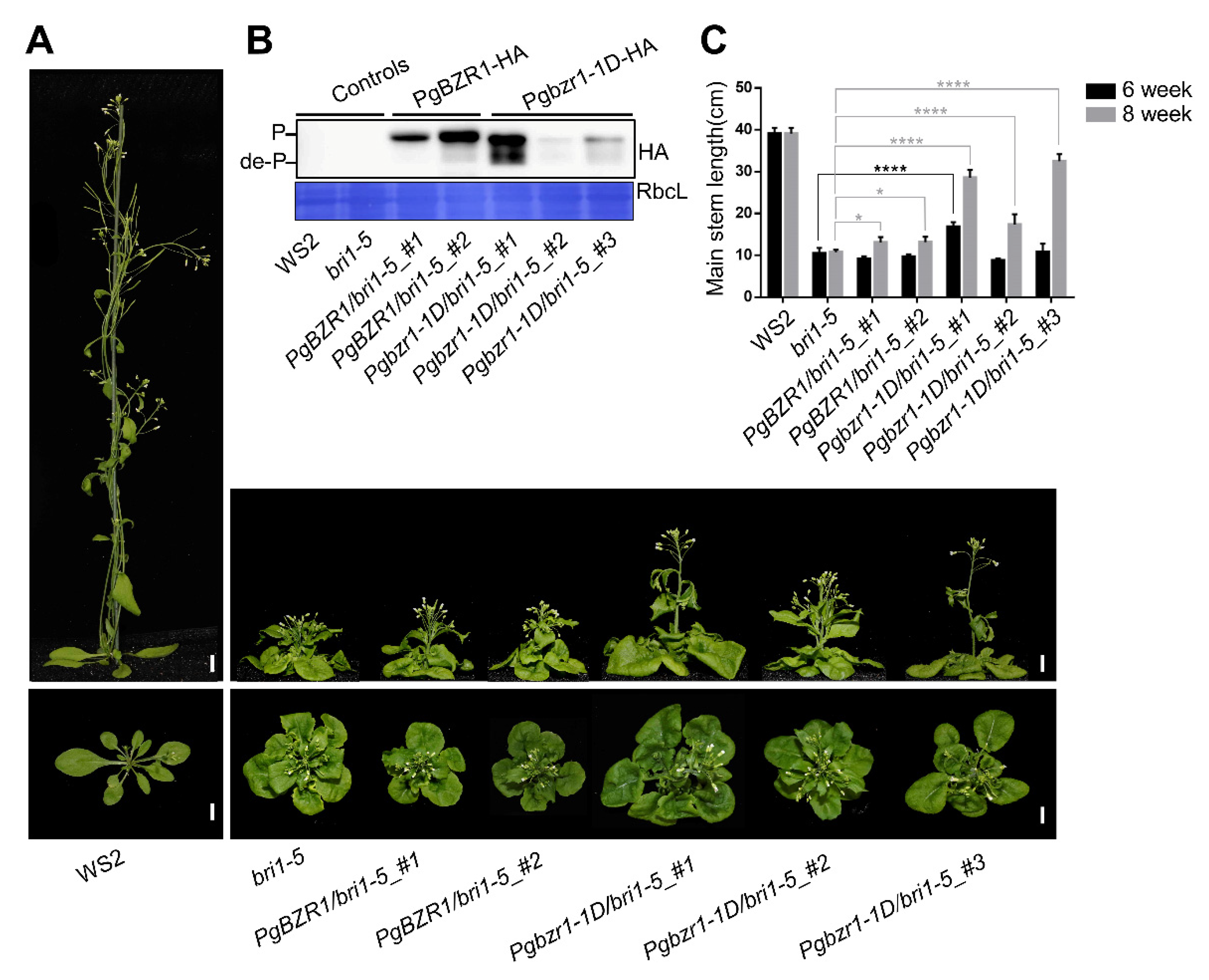
2.4. Ectopic Expression of PgBZR1 and Pgbzr1-1D Alleviates Stem Elongation in BR-Insensitive Bri1-5 Mutant
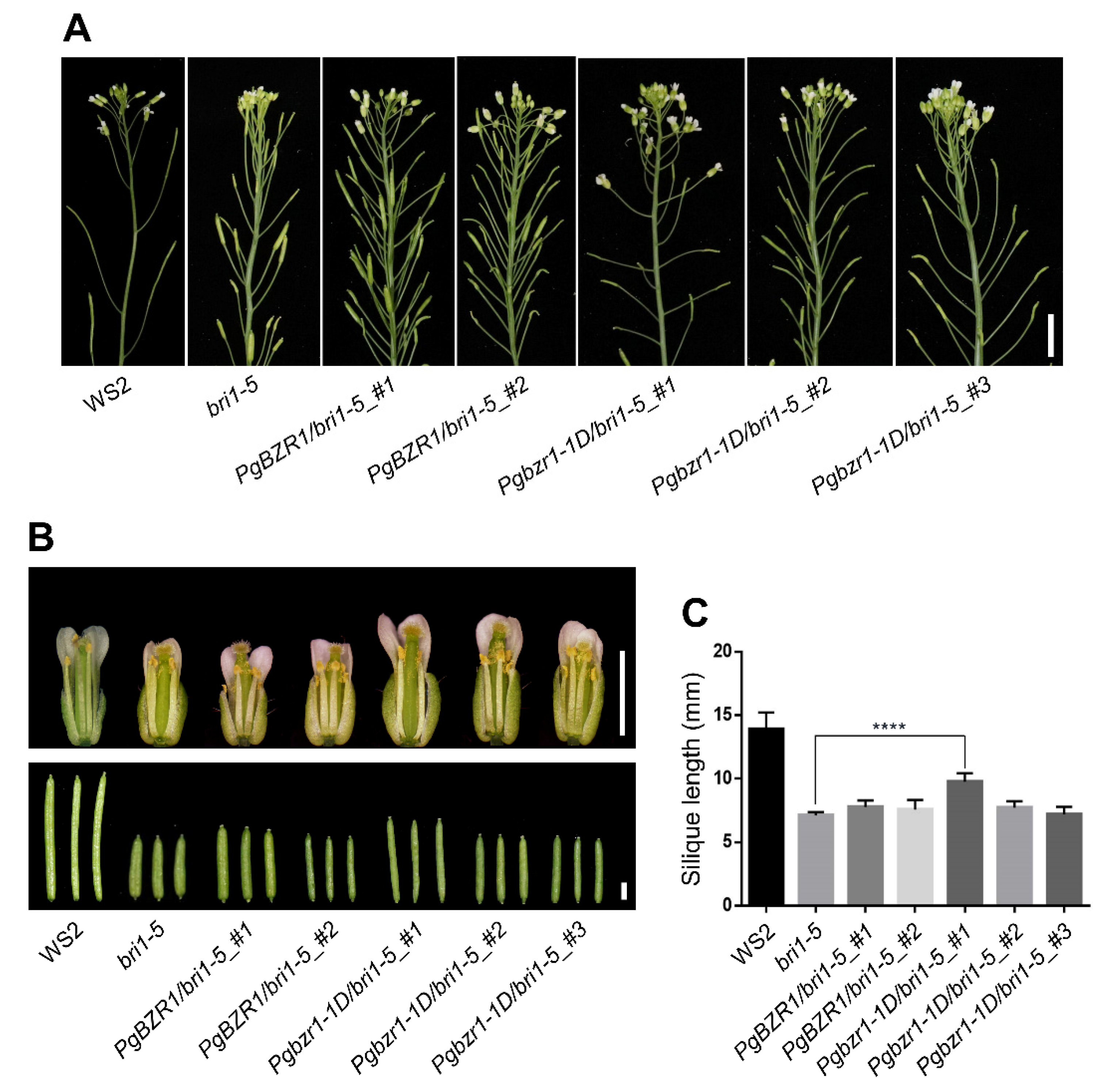
2.5. Ectopic Expression of Pgbzr1-1D Restores Inflorescence Architecture and Reproductive Organ Defects of the bri1-5 Mutant
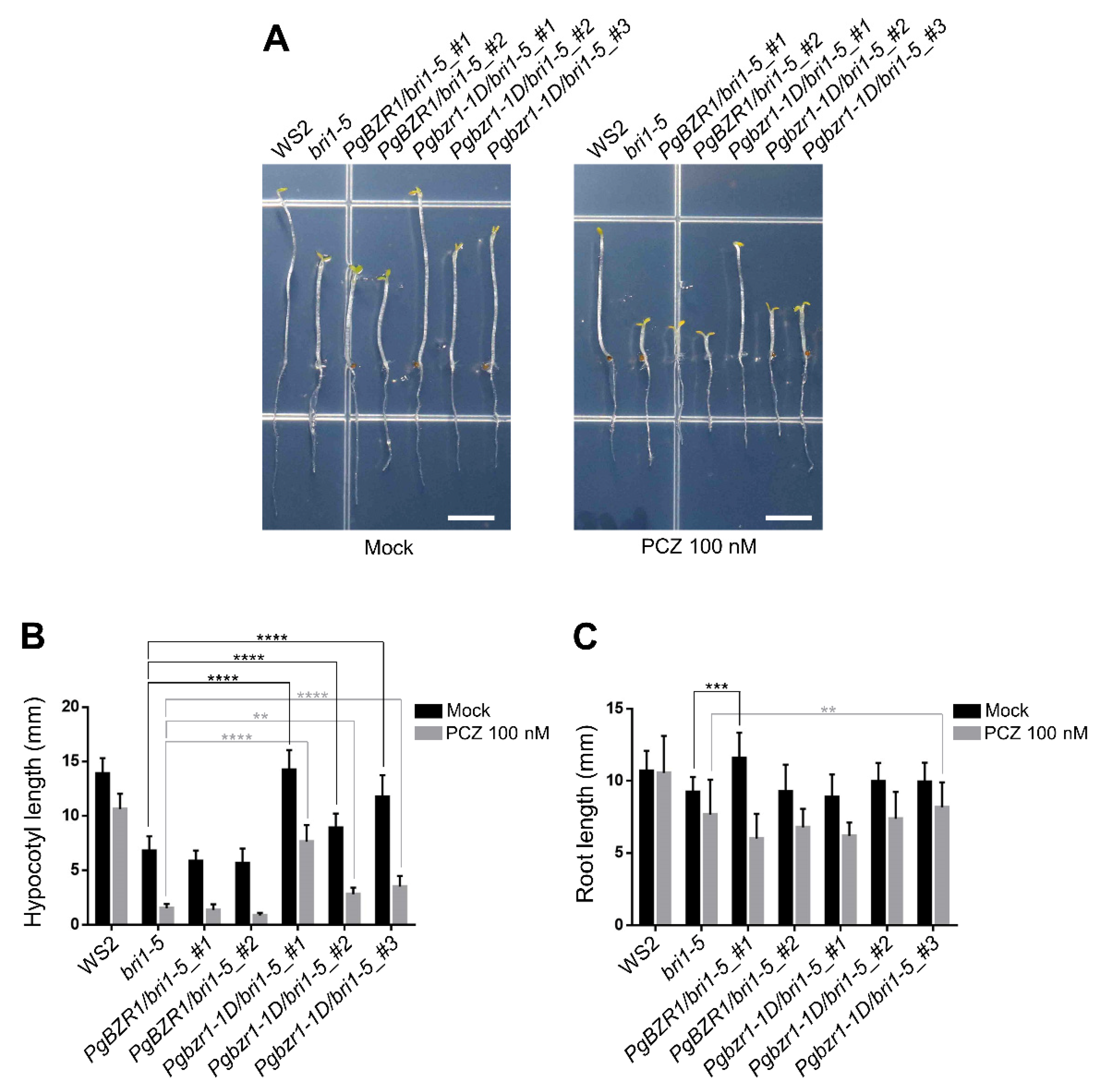
2.6. Ectopic Expression of Pgbzr1-1D Compromises the Deficiency of Dark-Induced Hypocotyl Elongation in bri1-5
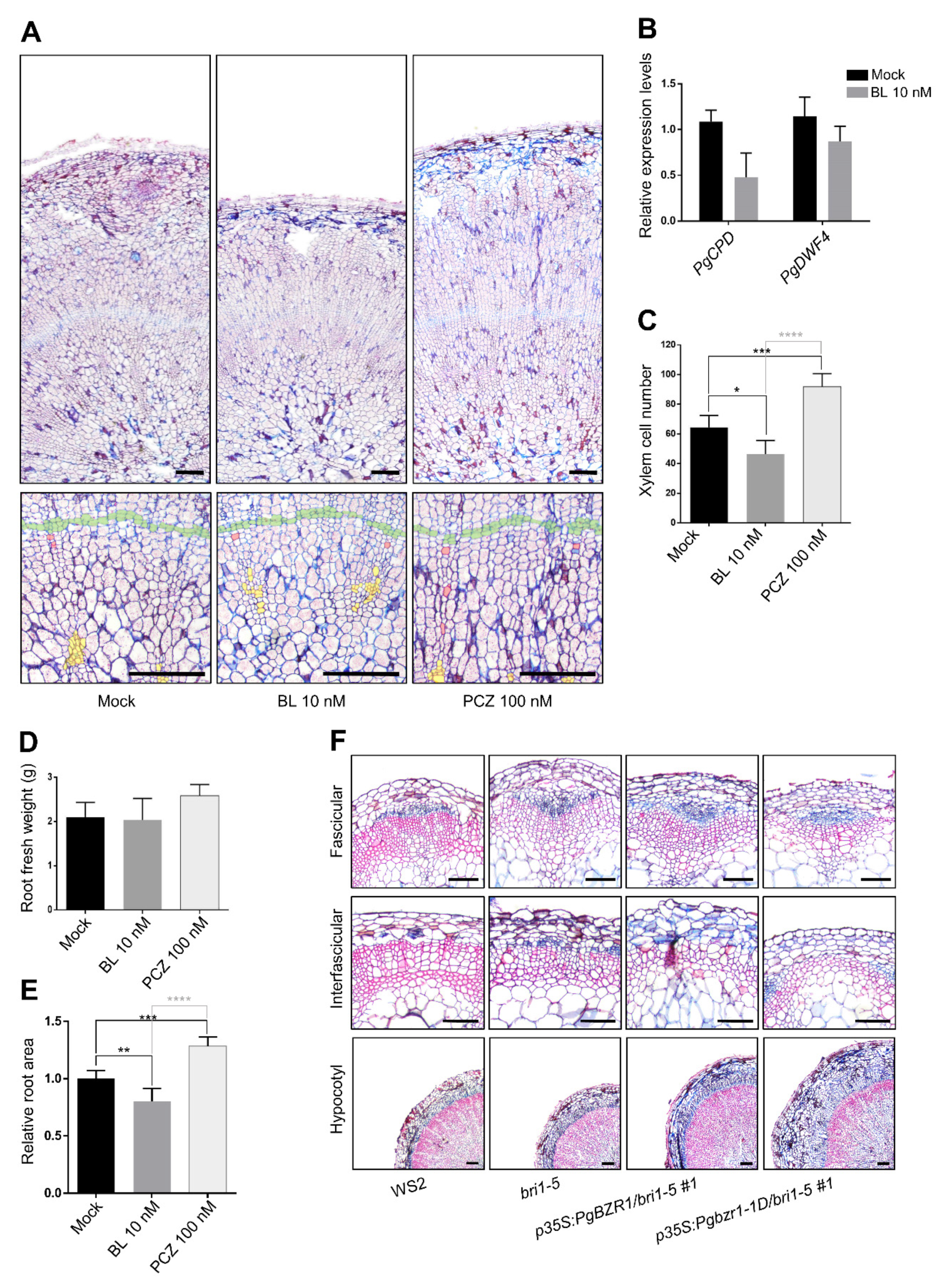
2.7. BR and PgBZR1 Repress Xylem Differentiation during Storage Root Formation of P. ginseng and Secondary Growth of Arabidopsis
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Phylogeny and Heat Map Analysis
4.3. Yeast-Two Hybrid Assay
4.4. Plasmid Construction and Transgenic Plants Analysis
4.5. Transient Expression Assay Using Arabidopsis Mesophyll Protoplast
4.6. Hormone Treatment and Histological Analysis
4.7. Quantitative RT-PCR
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BAK1 | BRI1-ASSOCIATED RECEPTOR KINASE 1 |
| BEH | BES1/BZR1 homolog |
| bHLH | basic helix loop helix |
| BIL1 | BIN2-LIKE 1 |
| BIL2 | BIN2-LIKE 2 |
| BIN2 | BRASSINOSTEROIDS INSENSITIVE 2 |
| BKI1 | BRI1 KINASE INHIBITOR 1 |
| BL | Brassinolide |
| BR | Brassinosteroid |
| BRI1 | BRASSINOSTEROIDS INSENSITIVE 1 |
| BSU1 | BRI1 SUPPRESSOR 1 |
| BZR1/BES1 | BRASSINAZOLE RESISTANCE 1/BRI1-EMS-SUPPRESSOR 1 |
| CPD | CONSTITUTIVE PHOTOMORPHOGENESIS AND DWARFISM |
| DWF4 | DWARF 4 |
| ECD | Extracellular domain |
| ETH | Ethylene |
| GA | Gibberellic acid |
| GSK3 | Glycogen synthase kinase 3 |
| HA | Hemagglutinin |
| ICD | Intracellular domain |
| JA | Jasmonic acid |
| PCZ | propiconazole |
| PXY/TDR | PHLOEM INTERCALATED WITH XYLEM/TDIF RECEPTOR |
| TDIF | TRACHEARY ELEMENT DIFFERENTIATION INHIBITORY FACTOR |
| TF | Transcription factor |
| WOX4 | WUSCHEL RELATED HOMEOBOX 4 |
| WT | Wild type |
| Y2H | Yeast two hybrid |
References
- Kim, T.-W.; Wang, Z.-Y. Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu. Rev. Plant Biol. 2010, 61, 681–704. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-Y.; Bai, M.-Y.; Oh, E.; Zhu, J.-Y. Brassinosteroid signaling network and regulation of photomorphogenesis. Annu. Rev. Genet. 2012, 46, 701–724. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Zhu, W.; Chen, Y.; Ito, S.; Asami, T.; Wang, X. Brassinosteroids control root epidermal cell fate via direct regulation of a MYB-bHLH-WD40 complex by GSK3-like kinases. Elife 2014, 3, e02525. [Google Scholar] [CrossRef] [PubMed]
- González-García, M.-P.; Vilarrasa-Blasi, J.; Zhiponova, M.; Divol, F.; Mora-García, S.; Russinova, E.; Caño-Delgado, A.I. Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development 2011, 138, 849–859. [Google Scholar] [CrossRef] [PubMed]
- Ibañez, C.; Delker, C.; Martinez, C.; Bürstenbinder, K.; Janitza, P.; Lippmann, R.; Ludwig, W.; Sun, H.; James, G.V.; Klecker, M. Brassinosteroids dominate hormonal regulation of plant thermomorphogenesis via BZR1. Curr. Biol. 2018, 28, 303–310.e3. [Google Scholar]
- Jiang, H.; Tang, B.; Xie, Z.; Nolan, T.; Ye, H.; Song, G.Y.; Walley, J.; Yin, Y. GSK 3-like kinase BIN 2 phosphorylates RD 26 to potentiate drought signaling in Arabidopsis. Plant J. 2019, 100, 923–937. [Google Scholar] [CrossRef]
- Kim, T.-W.; Michniewicz, M.; Bergmann, D.C.; Wang, Z.-Y. Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 2012, 482, 419–422. [Google Scholar] [CrossRef]
- Li, H.; Ye, K.; Shi, Y.; Cheng, J.; Zhang, X.; Yang, S. BZR1 positively regulates freezing tolerance via CBF-dependent and CBF-independent pathways in Arabidopsis. Mol. Plant 2017, 10, 545–559. [Google Scholar] [CrossRef]
- Li, J.; Nagpal, P.; Vitart, V.; McMorris, T.C.; Chory, J. A role for brassinosteroids in light-dependent development of Arabidopsis. Science 1996, 272, 398–401. [Google Scholar] [CrossRef]
- Martínez, C.; Espinosa-Ruíz, A.; de Lucas, M.; Bernardo-García, S.; Franco-Zorrilla, J.M.; Prat, S. PIF 4-induced BR synthesis is critical to diurnal and thermomorphogenic growth. Embo J. 2018, 37, e99552. [Google Scholar] [CrossRef]
- Ryu, H.; Cho, H.; Bae, W.; Hwang, I. Control of early seedling development by BES1/TPL/HDA19-mediated epigenetic regulation of ABI3. Nat. Commun. 2014, 5, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Steber, C.M.; McCourt, P. A role for brassinosteroids in germination in Arabidopsis. Plant Physiol. 2001, 125, 763–769. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Liu, S.; Tang, B.; Chen, J.; Xie, Z.; Nolan, T.M.; Jiang, H.; Guo, H.; Lin, H.-Y.; Li, L. RD26 mediates crosstalk between drought and brassinosteroid signalling pathways. Nat. Commun. 2017, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chory, J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 1997, 90, 929–938. [Google Scholar] [CrossRef]
- Nam, K.H.; Li, J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 2002, 110, 203–212. [Google Scholar] [CrossRef]
- Li, J.; Nam, K.H. Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 2002, 295, 1299–1301. [Google Scholar]
- Mora-García, S.; Vert, G.; Yin, Y.; Caño-Delgado, A.; Cheong, H.; Chory, J. Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 2004, 18, 448–460. [Google Scholar] [CrossRef]
- Wang, X.; Chory, J. Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science 2006, 313, 1118–1122. [Google Scholar] [CrossRef]
- Wang, Z.-Y.; Nakano, T.; Gendron, J.; He, J.; Chen, M.; Vafeados, D.; Yang, Y.; Fujioka, S.; Yoshida, S.; Asami, T. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2002, 2, 505–513. [Google Scholar] [CrossRef]
- Yin, Y.; Wang, Z.-Y.; Mora-Garcia, S.; Li, J.; Yoshida, S.; Asami, T.; Chory, J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 2002, 109, 181–191. [Google Scholar] [CrossRef]
- Yu, X.; Li, L.; Zola, J.; Aluru, M.; Ye, H.; Foudree, A.; Guo, H.; Anderson, S.; Aluru, S.; Liu, P.; et al. A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 2011, 65, 634–646. [Google Scholar] [CrossRef] [PubMed]
- Gampala, S.S.; Kim, T.-W.; He, J.-X.; Tang, W.; Deng, Z.; Bai, M.-Y.; Guan, S.; Lalonde, S.; Sun, Y.; Gendron, J.M. An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev. Cell 2007, 13, 177–189. [Google Scholar] [CrossRef] [PubMed]
- He, J.-X.; Gendron, J.M.; Yang, Y.; Li, J.; Wang, Z.-Y. The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2002, 99, 10185–10190. [Google Scholar] [CrossRef] [PubMed]
- Ryu, H.; Kim, K.; Cho, H.; Park, J.; Choe, S.; Hwang, I. Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. Plant. Cell 2007, 19, 2749–2762. [Google Scholar] [CrossRef]
- Yin, Y.; Vafeados, D.; Tao, Y.; Yoshida, S.; Asami, T.; Chory, J. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 2005, 120, 249–259. [Google Scholar] [CrossRef]
- He, J.-X.; Gendron, J.M.; Sun, Y.; Gampala, S.S.; Gendron, N.; Sun, C.Q.; Wang, Z.-Y. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 2005, 307, 1634–1638. [Google Scholar] [CrossRef]
- Hu, Y.; Yu, D. BRASSINOSTEROID INSENSITIVE2 interacts with ABSCISIC ACID INSENSITIVE5 to mediate the antagonism of brassinosteroids to abscisic acid during seed germination in Arabidopsis. Plant. Cell 2014, 26, 4394–4408. [Google Scholar] [CrossRef]
- Li, L.; Ye, H.; Guo, H.; Yin, Y.; Estelle, M. Arabidopsis IWS1 interacts with transcription factor BES1 and is involved in plant steroid hormone brassinosteroid regulated gene expression. Proc. Natl. Acad. Sci. USA 2010, 107, 3918–3923. [Google Scholar] [CrossRef]
- Belkhadir, Y.; Jaillais, Y. The molecular circuitry of brassinosteroid signaling. New Phytol. 2015, 206, 522–540. [Google Scholar] [CrossRef]
- Jouannet, V.; Brackmann, K.; Greb, T. (Pro) cambium formation and proliferation: Two sides of the same coin? Curr. Opin. Plant Biol. 2015, 23, 54–60. [Google Scholar] [CrossRef]
- Shi, D.; Lebovka, I.; López-Salmerón, V.; Sanchez, P.; Greb, T. Bifacial cambium stem cells generate xylem and phloem during radial plant growth. Development 2019, 146, dev171355. [Google Scholar] [CrossRef] [PubMed]
- Fisher, K.; Turner, S. PXY, a receptor-like kinase essential for maintaining polarity during plant vascular-tissue development. Curr. Biol. 2007, 17, 1061–1066. [Google Scholar] [CrossRef] [PubMed]
- Hirakawa, Y.; Shinohara, H.; Kondo, Y.; Inoue, A.; Nakanomyo, I.; Ogawa, M.; Sawa, S.; Ohashi-Ito, K.; Matsubayashi, Y.; Fukuda, H. Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc. Natl. Acad. Sci. USA 2008, 105, 15208–15213. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Strable, J.; Shimizu, R.; Koenig, D.; Sinha, N.; Scanlon, M.J. WOX4 promotes procambial development. Plant Physiol. 2010, 152, 1346–1356. [Google Scholar] [CrossRef]
- Kondo, Y.; Ito, T.; Nakagami, H.; Hirakawa, Y.; Saito, M.; Tamaki, T.; Shirasu, K.; Fukuda, H. Plant GSK3 proteins regulate xylem cell differentiation downstream of TDIF–TDR signalling. Nat. Commun. 2014, 5, 1–11. [Google Scholar] [CrossRef]
- Cho, H.; Ryu, H.; Rho, S.; Hill, K.; Smith, S.; Audenaert, D.; Park, J.; Han, S.; Beeckman, T.; Bennett, M.J.; et al. A secreted peptide acts on BIN2-mediated phosphorylation of ARFs to potentiate auxin response during lateral root development. Nat. Cell Biol. 2014, 16, 66–76. [Google Scholar] [CrossRef]
- Ibañes, M.; Fàbregas, N.; Chory, J.; Caño-Delgado, A.I. Brassinosteroid signaling and auxin transport are required to establish the periodic pattern of Arabidopsis shoot vascular bundles. Proc. Natl. Acad. Sci. USA 2009, 106, 13630–13635. [Google Scholar] [CrossRef]
- Oh, E.; Zhu, J.-Y.; Bai, M.-Y.; Arenhart, R.A.; Sun, Y.; Wang, Z.-Y. Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. Elife 2014, 3, e03031. [Google Scholar] [CrossRef]
- Suer, S.; Agusti, J.; Sanchez, P.; Schwarz, M.; Greb, T. WOX4 imparts auxin responsiveness to cambium cells in Arabidopsis. Plant. Cell 2011, 23, 3247–3259. [Google Scholar] [CrossRef]
- Vert, G.; Walcher, C.L.; Chory, J.; Nemhauser, J.L. Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proc. Natl. Acad. Sci. USA 2008, 105, 9829–9834. [Google Scholar] [CrossRef]
- Wang, C.; Liu, Y.; Li, S.-S.; Han, G.-Z. Insights into the origin and evolution of the plant hormone signaling machinery. Plant Physiol. 2015, 167, 872–886. [Google Scholar] [CrossRef] [PubMed]
- Jayakodi, M.; Choi, B.-S.; Lee, S.-C.; Kim, N.-H.; Park, J.Y.; Jang, W.; Lakshmanan, M.; Mohan, S.V.; Lee, D.-Y.; Yang, T.-J. Ginseng Genome Database: An open-access platform for genomics of Panax ginseng. BMC Plant Biol. 2018, 18, 62. [Google Scholar] [CrossRef] [PubMed]
- Jo, I.-H.; Lee, J.; Hong, C.E.; Lee, D.J.; Bae, W.; Park, S.-G.; Ahn, Y.J.; Kim, Y.C.; Kim, J.U.; Lee, J.W. Isoform sequencing provides a more comprehensive view of the Panax ginseng transcriptome. Genes 2017, 8, 228. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Chu, Y.; Liao, B.; Xiao, S.; Yin, Q.; Bai, R.; Su, H.; Dong, L.; Li, X.; Qian, J. Panax ginseng genome examination for ginsenoside biosynthesis. Gigascience 2017, 6, gix093. [Google Scholar] [CrossRef]
- De Rybel, B.; Audenaert, D.; Vert, G.; Rozhon, W.; Mayerhofer, J.; Peelman, F.; Coutuer, S.; Denayer, T.; Jansen, L.; Nguyen, L. Chemical inhibition of a subset of Arabidopsis thaliana GSK3-like kinases activates brassinosteroid signaling. Chem. Biol. 2009, 16, 594–604. [Google Scholar] [CrossRef]
- Espinosa-Ruiz, A.; Martínez, C.; de Lucas, M.; Fàbregas, N.; Bosch, N.; Caño-Delgado, A.I.; Prat, S. TOPLESS mediates brassinosteroid control of shoot boundaries and root meristem development in Arabidopsis thaliana. Development 2017, 144, 1619–1628. [Google Scholar] [CrossRef]
- Huang, H.-Y.; Jiang, W.-B.; Hu, Y.-W.; Wu, P.; Zhu, J.-Y.; Liang, W.-Q.; Wang, Z.-Y.; Lin, W.-H. BR signal influences Arabidopsis ovule and seed number through regulating related genes expression by BZR1. Mol. Plant 2013, 6, 456–469. [Google Scholar] [CrossRef]
- Choe, S.; Dilkes, B.P.; Fujioka, S.; Takatsuto, S.; Sakurai, A.; Feldmann, K.A. The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22α-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell 1998, 10, 231–243. [Google Scholar] [CrossRef]
- Kauschmann, A.; Jessop, A.; Koncz, C.; Szekeres, M.; Willmitzer, L.; Altmann, T. Genetic evidence for an essential role of brassinosteroids in plant development. Plant J. 1996, 9, 701–713. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hwang, H.; Lee, H.-Y.; Ryu, H.; Cho, H. Functional Characterization of BRASSINAZOLE-RESISTANT 1 in Panax Ginseng (PgBZR1) and Brassinosteroid Response during Storage Root Formation. Int. J. Mol. Sci. 2020, 21, 9666. https://doi.org/10.3390/ijms21249666
Hwang H, Lee H-Y, Ryu H, Cho H. Functional Characterization of BRASSINAZOLE-RESISTANT 1 in Panax Ginseng (PgBZR1) and Brassinosteroid Response during Storage Root Formation. International Journal of Molecular Sciences. 2020; 21(24):9666. https://doi.org/10.3390/ijms21249666
Chicago/Turabian StyleHwang, Hyeona, Hwa-Yong Lee, Hojin Ryu, and Hyunwoo Cho. 2020. "Functional Characterization of BRASSINAZOLE-RESISTANT 1 in Panax Ginseng (PgBZR1) and Brassinosteroid Response during Storage Root Formation" International Journal of Molecular Sciences 21, no. 24: 9666. https://doi.org/10.3390/ijms21249666
APA StyleHwang, H., Lee, H.-Y., Ryu, H., & Cho, H. (2020). Functional Characterization of BRASSINAZOLE-RESISTANT 1 in Panax Ginseng (PgBZR1) and Brassinosteroid Response during Storage Root Formation. International Journal of Molecular Sciences, 21(24), 9666. https://doi.org/10.3390/ijms21249666




