The Genome-Wide Impact of Nipblb Loss-of-Function on Zebrafish Gene Expression
Abstract
1. Introduction
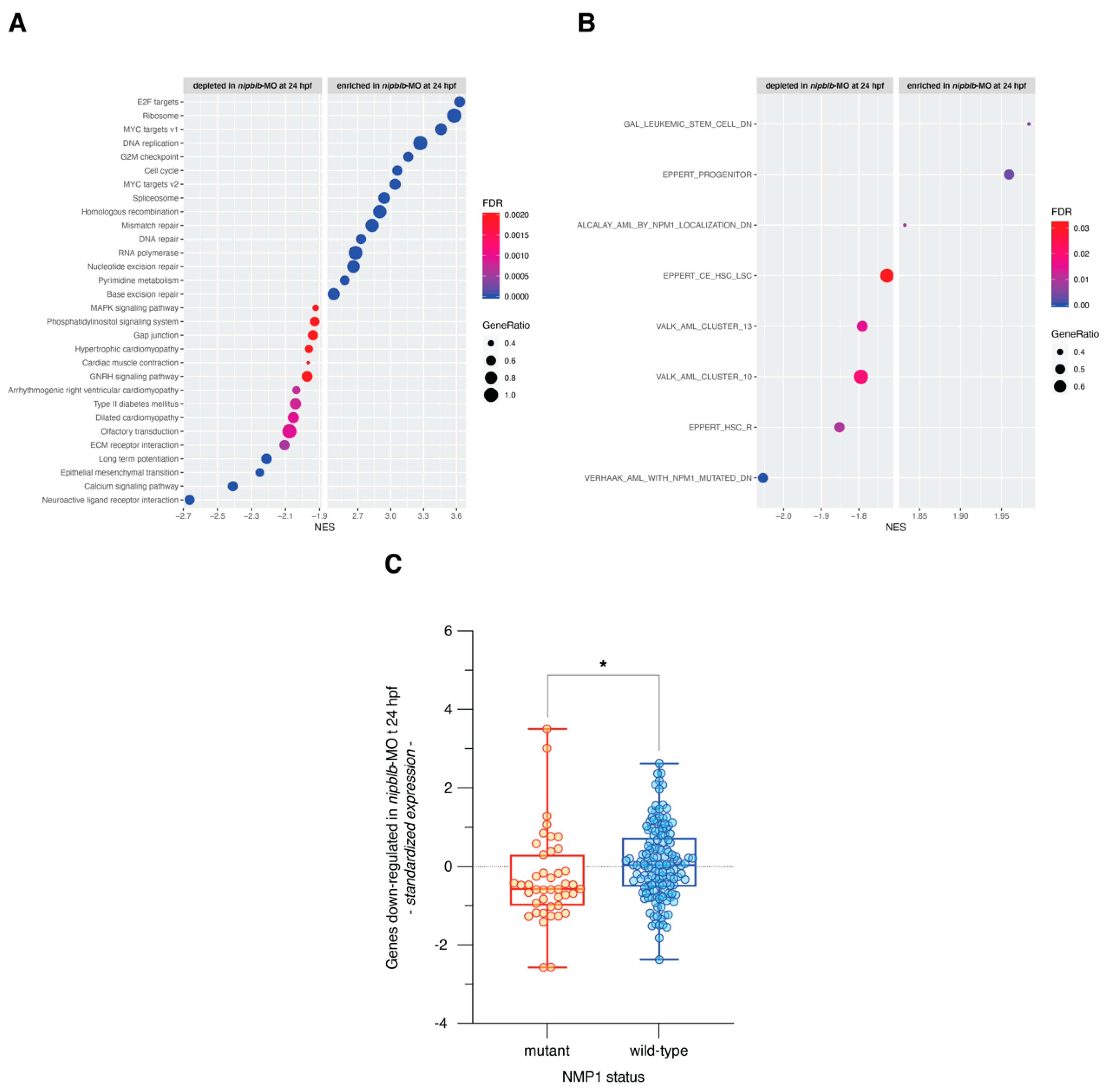
2. Results
3. Discussion
4. Materials and Methods
4.1. Zebrafish Embryos
4.2. Injections
4.3. RNA Extraction
4.4. RNA-Seq and Bioinformatics Analyses
4.5. Data Availability
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| NIPBL | Nipped B-like |
| MO | Morpholino |
| RNA-seq | RNA sequencing |
| EMT | Epithelial–Mesenchymal Transition |
| ECM | Extracellular Matrix |
| AML | Acute Myeloid Leukemia |
| HSC | Hematopoietic Stem Cells |
| LSC | Leukemia Stem Cells |
| FDR | False Discovery Rate |
| GSEA | Gene Set Enrichment Analysis |
| NES | Normalized Enrichment Score |
References
- Michaelis, C.; Ciosk, R.; Nasmyth, K. Cohesins: Chromosomal Proteins that Prevent Premature Separation of Sister Chromatids. Cell 1997, 91, 35–45. [Google Scholar] [CrossRef]
- Watrin, E.; Peters, J.-M. The cohesin complex is required for the DNA damage-induced G2/M checkpoint in mammalian cells. EMBO J. 2009, 28, 2625–2635. [Google Scholar] [CrossRef] [PubMed]
- Gligoris, T.G.; Scheinost, J.C.; Bürmann, F.; Petela, N.; Chan, K.-L.; Uluocak, P.; Beckouët, F.; Gruber, S.; Nasmyth, K.; Löwe, J. Closing the cohesin ring: Structure and function of its Smc3-kleisin interface. Science 2014, 346, 963–967. [Google Scholar] [CrossRef] [PubMed]
- Kagey, M.H.; Newman, J.J.; Bilodeau, S.; Zhan, Y.; Orlando, D.A.; Van Berkum, N.L.; Ebmeier, C.C.; Goossens, J.; Rahl, P.B.; Levine, S.S.; et al. Mediator and cohesin connect gene expression and chromatin architecture. Nat. Cell Biol. 2010, 467, 430–435. [Google Scholar] [CrossRef]
- Seitan, V.C.; Faure, A.J.; Zhan, Y.; Mccord, R.P.; Lajoie, B.R.; Ing-Simmons, E.; Lenhard, B.; Giorgetti, L.; Heard, E.; Fisher, A.G.; et al. Cohesin-based chromatin interactions enable regulated gene expression within preexisting architectural compartments. Genome Res. 2013, 23, 2066–2077. [Google Scholar] [CrossRef]
- Lyu, X.; Rowley, M.J.; Corces, V.G. Architectural Proteins and Pluripotency Factors Cooperate to Orchestrate the Transcriptional Response of hESCs to Temperature Stress. Mol. Cell 2018, 71, 940–955.e7. [Google Scholar] [CrossRef]
- Bernardi, G. The formation of chromatin domains involves a primary step based on the 3-D structure of DNA. Sci. Rep. 2018, 8, 1–7. [Google Scholar] [CrossRef]
- Misulovin, Z.; Schwartz, Y.B.; Li, X.-Y.; Kahn, T.G.; Gause, M.; MacArthur, S.; Fay, J.C.; Eisen, M.B.; Pirrotta, V.; Biggin, M.D.; et al. Association of cohesin and Nipped-B with transcriptionally active regions of the Drosophila melanogaster genome. Chromosoma 2007, 117, 89–102. [Google Scholar] [CrossRef]
- Fay, A.; Misulovin, Z.; Li, J.; Schaaf, C.A.; Gause, M.; Gilmour, D.S.; Dorsett, D. Cohesin Selectively Binds and Regulates Genes with Paused RNA Polymerase. Curr. Biol. 2011, 21, 1624–1634. [Google Scholar] [CrossRef]
- Dowen, J.M.; Bilodeau, S.; Orlando, D.A.; Hübner, M.R.; Abraham, B.J.; Spector, D.L.; Young, R.A. Multiple Structural Maintenance of Chromosome Complexes at Transcriptional Regulatory Elements. Stem Cell Rep. 2013, 1, 371–378. [Google Scholar] [CrossRef]
- Zuin, J.; Franke, V.; Van Ijcken, W.F.J.; Van Der Sloot, A.A.; Krantz, I.D.; Van Der Reijden, M.I.J.A.; Nakato, R.; Lenhard, B.; Wendt, K.S. A Cohesin-Independent Role for NIPBL at Promoters Provides Insights in CdLS. PLoS Genet. 2014, 10, e1004153. [Google Scholar] [CrossRef] [PubMed]
- Rollins, R.A.; Morcillo, P.; Dorsett, D. Nipped-B, a Drosophila homologue of chromosomal adherins, participates in activation by remote enhancers in the cut and Ultrabithorax genes. Genetics 1999, 152, 577–593. [Google Scholar] [PubMed]
- Kawauchi, S.; Calof, A.L.; Santos, R.; Lopez-Burks, M.E.; Young, C.M.; Hoang, M.P.; Chua, A.; Lao, T.; Lechner, M.S.; Daniel, J.A.; et al. Multiple Organ System Defects and Transcriptional Dysregulation in the Nipbl+/− Mouse, a Model of Cornelia de Lange Syndrome. PLoS Genet. 2009, 5, e1000650. [Google Scholar] [CrossRef] [PubMed]
- Muto, A.; Ikeda, S.; Lopez-Burks, M.E.; Kikuchi, Y.; Calof, A.L.; Lander, A.D.; Schilling, T.F. Nipbl and Mediator Cooperatively Regulate Gene Expression to Control Limb Development. PLoS Genet. 2014, 10, e1004671. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, Z.; Bando, M.; Itoh, T.; Deardorff, M.A.; Clark, D.; Kaur, M.; Tandy, S.; Kondoh, T.; Rappaport, E.; et al. Transcriptional Dysregulation in NIPBL and Cohesin Mutant Human Cells. PLoS Biol. 2009, 7, e1000119. [Google Scholar] [CrossRef]
- Mills, J.A.; Herrera, P.S.; Kaur, M.; Leo, L.; McEldrew, D.; Tintos-Hernández, J.A.; Rajagopalan, R.; Gagne, A.; Zhang, Z.; Ortiz-Gonzalez, X.R.; et al. NIPBL+/− haploinsufficiency reveals a constellation of transcriptome disruptions in the pluripotent and cardiac states. Sci. Rep. 2018, 8, 1056. [Google Scholar] [CrossRef]
- Yuen, K.C.; Xu, B.; Krantz, I.D.; Gerton, J.L. NIPBL Controls RNA Biogenesis to Prevent Activation of the Stress Kinase PKR. Cell Rep. 2016, 14, 93–102. [Google Scholar] [CrossRef]
- Liu, T.; Han, Y.; Yu, C.; Ji, Y.; Wang, C.; Chen, X.; Wang, X.; Shen, J.; Zhang, Y.; Lang, J.-Y. MYC predetermines the sensitivity of gastrointestinal cancer to antifolate drugs through regulating TYMS transcription. EBioMedicine 2019, 48, 289–300. [Google Scholar] [CrossRef]
- Yoshida, K.; Toki, T.; Okuno, Y.; Kanezaki, R.; Shiraishi, Y.; Sato-Otsubo, A.; Sanada, M.; Park, M.-J.; Terui, K.; Suzuki, H.; et al. The landscape of somatic mutations in Down syndrome–related myeloid disorders. Nat. Genet. 2013, 45, 1293–1299. [Google Scholar] [CrossRef]
- Pistocchi, A.; Fazio, G.; Cereda, A.; Ferrari, L.; Bettini, L.R.; Messina, G.; Cotelli, F.; Biondi, A.; Selicorni, A.; Massa, V. Cornelia de Lange Syndrome: NIPBL haploinsufficiency downregulates canonical Wnt pathway in zebrafish embryos and patients fibroblasts. Cell Death Dis. 2013, 4, e866. [Google Scholar] [CrossRef]
- Mazzola, M.; Deflorian, G.; Pezzotta, A.; Ferrari, L.; Fazio, G.; Bresciani, E.; Saitta, C.; Ferrari, L.; Fumagalli, M.; Parma, M.; et al. NIPBL: A new player in myeloid cell differentiation. Haematologica 2019, 104, 1332–1341. [Google Scholar] [CrossRef] [PubMed]
- Dang, J.; Nance, S.; Ma, J.; Cheng, J.; Walsh, M.P.; Vogel, P.; Easton, J.; Song, G.; Rusch, M.; Gedman, A.L.; et al. AMKL chimeric transcription factors are potent inducers of leukemia. Leukemia 2017, 31, 2228–2234. [Google Scholar] [CrossRef] [PubMed]
- Mazzola, M.; Pezzotta, A.; Fazio, G.; Rigamonti, A.; Bresciani, E.; Gaudenzi, G.; Pelleri, M.C.; Saitta, C.; Ferrari, L.; Parma, M.; et al. Dysregulation of NIPBL leads to impaired RUNX1 expression and haematopoietic defects. J. Cell. Mol. Med. 2020, 24, 6272–6282. [Google Scholar] [CrossRef] [PubMed]
- Tomasson, M.H.; Xiang, Z.; Walgren, R.; Zhao, Y.; Kasai, Y.; Miner, T.; Ries, R.E.; Lubman, O.; Fremont, D.H.; McLellan, M.D.; et al. Somatic mutations and germline sequence variants in the expressed tyrosine kinase genes of patients with de novo acute myeloid leukemia. Blood 2008, 111, 4797–4808. [Google Scholar] [CrossRef]
- Nasmyth, K.; Haering, C.H. Cohesin: Its Roles and Mechanisms. Annu. Rev. Genet. 2009, 43, 525–558. [Google Scholar] [CrossRef]
- Zuin, J.; Dixon, J.R.; van der Reijden, M.I.; Ye, Z.; Kolovos, P.; Brouwer, R.W.; van de Corput, M.P.; van de Werken, H.J.; Knoch, T.A.; van IJcken, W.F.; et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc. Natl. Acad. Sci. USA 2014, 111, 996–1001. [Google Scholar] [CrossRef]
- Sofueva, S.; Yaffe, E.; Chan, W.-C.; Georgopoulou, D.; Rudan, M.V.; Mira-Bontenbal, H.; Pollard, S.M.; Schroth, G.P.; Tanay, A.; Hadjur, S. Cohesin-mediated interactions organize chromosomal domain architecture. EMBO J. 2013, 32, 3119–3129. [Google Scholar] [CrossRef]
- Rollins, R.A.; Korom, M.; Aulner, N.; Martens, A.; Dorsett, D. Drosophila Nipped-B Protein Supports Sister Chromatid Cohesion and Opposes the Stromalin/Scc3 Cohesion Factor To Facilitate Long-Range Activation of the cut Gene. Mol. Cell. Biol. 2004, 24, 3100–3111. [Google Scholar] [CrossRef]
- Muto, A.; Calof, A.L.; Lander, A.D.; Schilling, T.F. Multifactorial Origins of Heart and Gut Defects in nipbl-Deficient Zebrafish, a Model of Cornelia de Lange Syndrome. PLoS Biol. 2011, 9, e1001181. [Google Scholar] [CrossRef]
- Jahnke, P.; Xu, W.; Wülling, M.; Albrecht, M.; Gabriel, H.; Gillessen-Kaesbach, G.; Kaiser, F.J. The Cohesin loading factor NIPBL recruits histone deacetylases to mediate local chromatin modifications. Nucleic Acids Res. 2008, 36, 6450–6458. [Google Scholar] [CrossRef]
- Lechner, M.S.; Schultz, D.C.; Negorev, D.; Maul, G.G.; Rauscher, F.J. The mammalian heterochromatin protein 1 binds diverse nuclear proteins through a common motif that targets the chromoshadow domain. Biochem. Biophys. Res. Commun. 2005, 331, 929–937. [Google Scholar] [CrossRef] [PubMed]
- Muto, A.; Schilling, T.F. Zebrafish as a model to study cohesin and cohesinopathies. In Methods in Molecular Biology; Humana Press: New York, NY, USA, 2017. [Google Scholar]
- Rossi, A.; Kontarakis, Z.; Gerri, C.; Nolte, H.; Hölper, S.; Krüger, M.; Stainier, D.Y.R. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nat. Cell Biol. 2015, 524, 230–233. [Google Scholar] [CrossRef] [PubMed]
- Kimmel, C.B.; Ballard, W.W.; Kimmel, S.R.; Ullmann, B.; Schilling, T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995, 203, 253–310. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef]
- Irizarry, R.A.; Hobbs, B.; Collin, F.; Beazer-Barclay, Y.D.; Antonellis, K.J.; Scherf, U.; Speed, T.P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003, 4, 249–264. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spreafico, M.; Mangano, E.; Mazzola, M.; Consolandi, C.; Bordoni, R.; Battaglia, C.; Bicciato, S.; Marozzi, A.; Pistocchi, A. The Genome-Wide Impact of Nipblb Loss-of-Function on Zebrafish Gene Expression. Int. J. Mol. Sci. 2020, 21, 9719. https://doi.org/10.3390/ijms21249719
Spreafico M, Mangano E, Mazzola M, Consolandi C, Bordoni R, Battaglia C, Bicciato S, Marozzi A, Pistocchi A. The Genome-Wide Impact of Nipblb Loss-of-Function on Zebrafish Gene Expression. International Journal of Molecular Sciences. 2020; 21(24):9719. https://doi.org/10.3390/ijms21249719
Chicago/Turabian StyleSpreafico, Marco, Eleonora Mangano, Mara Mazzola, Clarissa Consolandi, Roberta Bordoni, Cristina Battaglia, Silvio Bicciato, Anna Marozzi, and Anna Pistocchi. 2020. "The Genome-Wide Impact of Nipblb Loss-of-Function on Zebrafish Gene Expression" International Journal of Molecular Sciences 21, no. 24: 9719. https://doi.org/10.3390/ijms21249719
APA StyleSpreafico, M., Mangano, E., Mazzola, M., Consolandi, C., Bordoni, R., Battaglia, C., Bicciato, S., Marozzi, A., & Pistocchi, A. (2020). The Genome-Wide Impact of Nipblb Loss-of-Function on Zebrafish Gene Expression. International Journal of Molecular Sciences, 21(24), 9719. https://doi.org/10.3390/ijms21249719




