Comparative Transcriptome Analysis Provides Insights into the Polyunsaturated Fatty Acid Synthesis Regulation of Fat-1 Transgenic Sheep
Abstract
:1. Introduction
2. Results
2.1. Generation of Fat-1 Transgenic Fetuses and Lambs
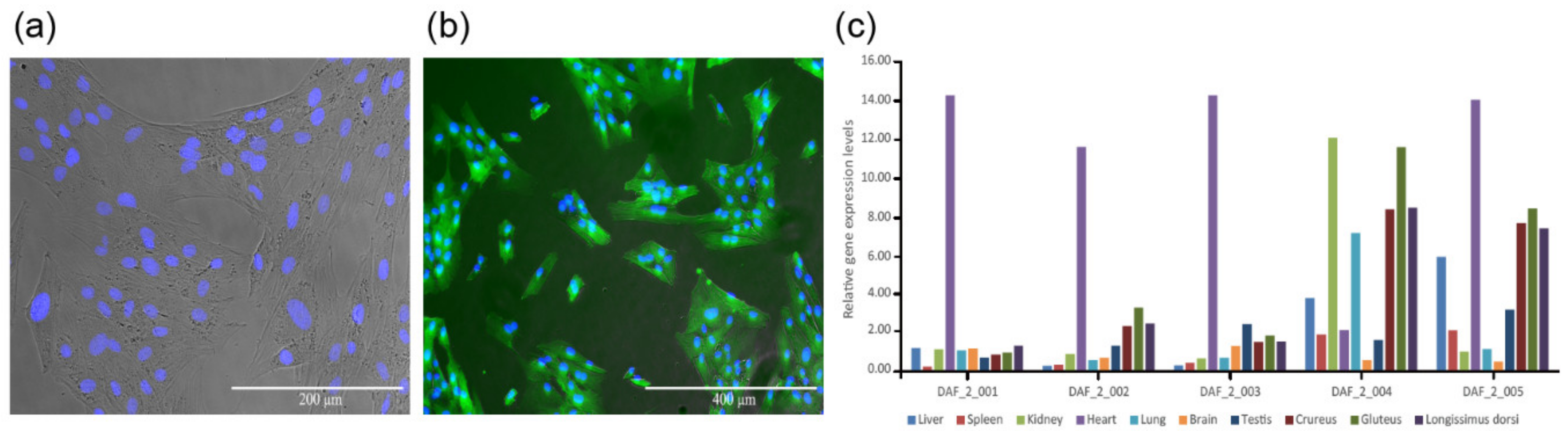
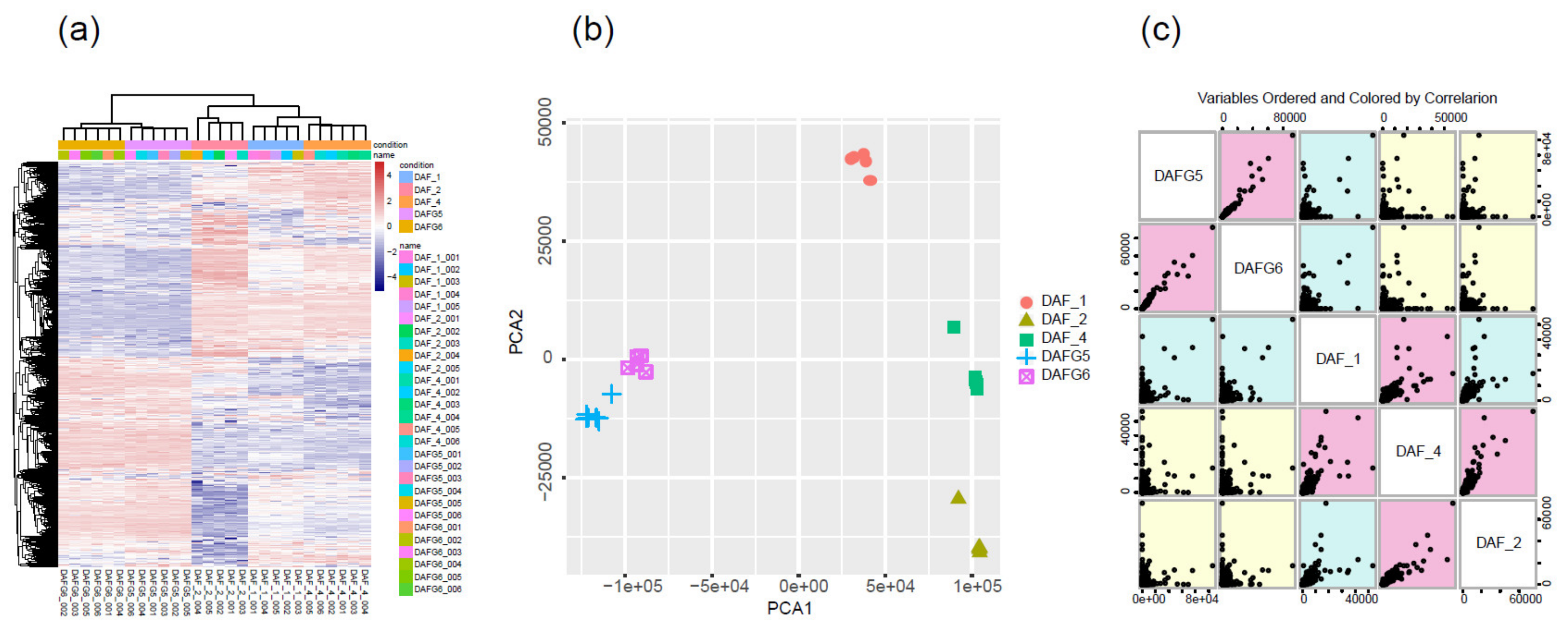
2.2. Global Gene Expression Profiles of Different Tissue and Skin Cell Samples
2.3. Gene Expression Differences between Abnormal and Normal Fetuses
2.4. Gene Expression Differences between Fat-1 Transgenic-Silencing and -Expressing Skin Cells
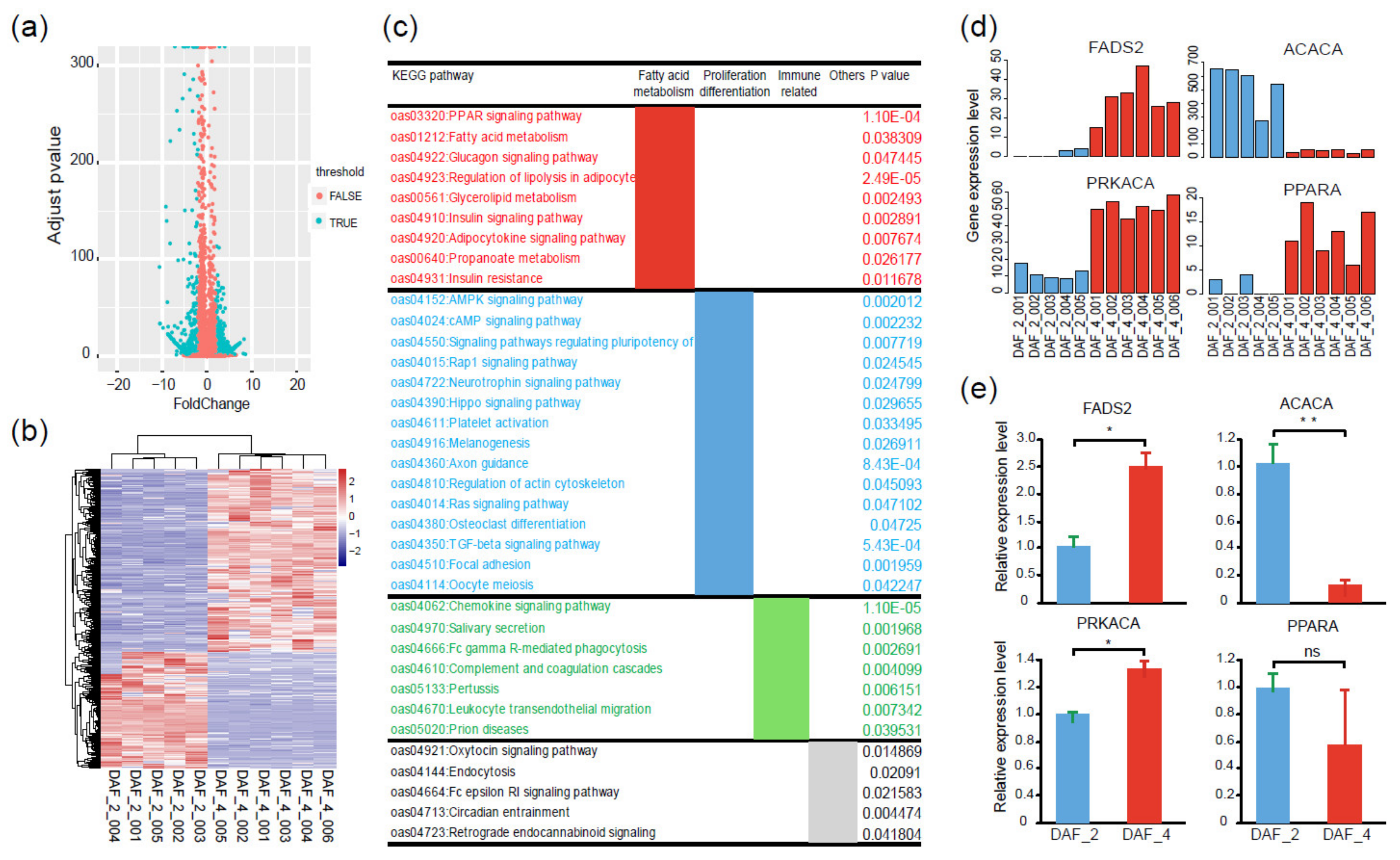
2.5. Gene Expression Differences between Fat-1 Transgenic Fetuses and Postnatal Lambs
2.6. PUFA Contents between 100-day Fat-1 Transgenic Fetuses and Wild Type Fetuses
3. Discussion
4. Materials and Methods
4.1. Ethical Statement
4.2. Generation of Fat-1 Transgenic Lambs and Sample Collection for RNA-seq
4.3. RNA Isolation, Library Construction, and Transcriptome Sequencing
4.4. Read Mapping and Analysis of Differential Gene Expression
4.5. KEGG Pathway Enrichment and PPI Network Analysis
4.6. Quantitative Real-Time PCR (qPCR) Validation
4.7. Determination of Polyunsaturated Fatty Acids
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Data Availability
Conflicts of Interest
Abbreviations
| DAF_1 | Abnormal developed fetus at 100 gestational days |
| DAF_2 | normal fetus at 100 gestational days |
| DAF_4 | postnatal lambs |
| DAFG5 | Fat-1 transgenic-silencing skin cells |
| DAFG6 | Fat-1 transgenic-expressing skin cells |
| PUFAs | Polyunsaturated fatty acids |
| GMO | Genetically modified organism |
| DEG | Differentially expressed gene |
| FPKM | Fragments per kilobase of exon per million mapped reads |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| PCA | Principal components analysis |
| PPI | Protein and protein interaction network |
References
- Lai, D.; Schwantes-An, T.; Corbin, T.J. Skeletal Genetics: From Gene Identification to Murine Models of Disease. In Basic and Applied Bone Biology, 2nd ed.; Burr, D.B., Allen, M.R., Eds.; Academic Press: Cambridge, USA, 2019; pp. 159–185. [Google Scholar]
- Gordon, J.W.; Scangos, G.A.; Plotkin, D.J.; Barbosa, J.A.; Ruddle, F.H. Genetic transformation of mouse embryos by microinjection of purified DNA. Proc. Natl. Acad. Sci. USA 1980, 77, 7380–7384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilmut, I.; Schnieke, A.E.; McWhir, J.; Kind, A.J.; Campbell, K.H. Viable offspring derived from fetal and adult mammalian cells. Cloning Stem Cells 2007, 9, 3–7. [Google Scholar] [CrossRef] [PubMed]
- Cibelli, J.B.; Stice, S.L.; Golueke, P.J.; Kane, J.J.; Jerry, J.; Blackwell, C.; Ponce, D.L.F.; Robl, J.M. Cloned transgenic calves produced from nonquiescent fetal fibroblasts. Science 1998, 280, 1256–1258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schnieke, A.E.; Kind, A.J.; Ritchie, W.A.; Mycock, K.; Scott, A.R.; Ritchie, M.; Wilmut, I.; Colman, A.; Campbell, K.H. Human factor IX transgenic sheep produced by transfer of nuclei from transfected fetal fibroblasts. Science 1997, 278, 2130–2133. [Google Scholar] [CrossRef] [PubMed]
- Brophy, B.; Smolenski, G.; Wheeler, T.; Wells, D.; L’Huillier, P.; Laible, G. Cloned transgenic cattle produce milk with higher levels of beta-casein and kappa-casein. Nat. Biotechnol. 2003, 21, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Kuroiwa, Y.; Kasinathan, P.; Matsushita, H.; Sathiyaselan, J.; Sullivan, E.J.; Kakitani, M.; Tomizuka, K.; Ishida, I.; Robl, J.M. Sequential targeting of the genes encoding immunoglobulin-mu and prion protein in cattle. Nat. Genet. 2004, 36, 775–780. [Google Scholar] [CrossRef] [Green Version]
- Kyle, D.J.; Arterburn, L.M. Single cell oil sources of docosahexaenoic acid: Clinical studies. World Rev. Nutr. Diet 1998, 83, 116–131. [Google Scholar]
- Watts, J.L.; Browse, J. Genetic dissection of polyunsaturated fatty acid synthesis in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2002, 99, 5854–5859. [Google Scholar] [CrossRef] [Green Version]
- Kang, J.X.; Wang, J.; Wu, L.; Kang, Z.B. Transgenic mice: Fat-1 mice convert n-6 to n-3 fatty acids. Nature 2004, 427, 504. [Google Scholar] [CrossRef]
- Lai, L.; Kang, J.X.; Li, R.; Wang, J.; Witt, W.T.; Yong, H.Y.; Hao, Y.; Wax, D.M.; Murphy, C.N.; Rieke, A.; et al. Generation of cloned transgenic pigs rich in omega-3 fatty acids. Nat. Biotechnol. 2006, 24, 435–436. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Ouyang, H.; Duan, B.; Pang, D.; Zhang, L.; Yuan, T.; Xue, L.; Ni, D.; Cheng, L.; Dong, S.; et al. Production of cloned transgenic cow expressing omega-3 fatty acids. Transgenic Res 2012, 21, 537–543. [Google Scholar] [CrossRef]
- Turkmen, S.; Zamorano, M.J.; Fernández-Palacios, H.; Hernández-Cruz, C.M.; Montero, D.; Robaina, L.; Izquierdo, M. Parental nutritional programming and a reminder during juvenile stage affect growth, lipid metabolism and utilisation in later developmental stages of a marine teleost, the gilthead sea bream (Sparus aurata). Brit J. Nutr. 2017, 118, 500–512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simopoulos, A.P. The importance of the omega-6/omega-3 fatty acid ratio in cardiovascular disease and other chronic diseases. Exp. Biol. Med. (Maywood) 2008, 233, 674–688. [Google Scholar] [CrossRef] [PubMed]
- Connor, W.E. Importance of n-3 fatty acids in health and disease. Am. J. Clin. Nutr. 2000, 71 (Suppl. S1), 171S–175S. [Google Scholar] [CrossRef] [PubMed]
- Turkmen, S.; Hernández-Cruz, C.M.; Zamorano, M.J.; Fernández-Palacios, H.; Montero, D.; Afonso, J.M.; Izquierdo, M. Long-chain PUFA profiles in parental diets induce long-term effects on growth, fatty acid profiles, expression of fatty acid desaturase 2 and selected immune system-related genes in the offspring of gilthead seabream. Brit J. Nutr. 2019, 122, 25–38. [Google Scholar] [CrossRef] [PubMed]
- Chilton, F.; Dutta, R.; Reynolds, L.; Sergeant, S.; Mathias, R.; Seeds, M. Precision Nutrition and Omega-3 Polyunsaturated Fatty Acids: A Case for Personalized Supplementation Approaches for the Prevention and Management of Human Diseases. Nutrients 2017, 9, 1165. [Google Scholar] [CrossRef]
- Lands, B. Dietary omega-3 and omega-6 fatty acids compete in producing tissue compositions and tissue responses. Mil. Med. 2014, 179 (Suppl. S11), 76–81. [Google Scholar] [CrossRef] [Green Version]
- Lands, W.E.; Libelt, B.; Morris, A.; Kramer, N.C.; Prewitt, T.E.; Bowen, P.; Schmeisser, D.; Davidson, M.H.; Burns, J.H. Maintenance of lower proportions of (n-6) eicosanoid precursors in phospholipids of human plasma in response to added dietary (n-3) fatty acids. Biochim. Biophys. Acta 1992, 2, 147–162. [Google Scholar] [CrossRef]
- Okuyama, H.; Kobayashi, T.; Watanabe, S. Dietary fatty acids--the N-6/N-3 balance and chronic elderly diseases. Excess linoleic acid and relative N-3 deficiency syndrome seen in Japan. Prog. Lipid Res. 1996, 35, 409–457. [Google Scholar] [CrossRef]
- Cleland, L.G.; James, M.J.; Neumann, M.A.; D’Angelo, M.; Gibson, R.A. Linoleate inhibits EPA incorporation from dietary fish-oil supplements in human subjects. Am. J. Clin. Nutr. 1992, 55, 395–399. [Google Scholar] [CrossRef]
- Duan, B.; Cheng, L.; Gao, Y.; Yin, F.X.; Su, G.H.; Shen, Q.Y.; Liu, K.; Hu, X.; Liu, X.; Li, G.P. Silencing of fat-1 transgene expression in sheep may result from hypermethylation of its driven cytomegalovirus (CMV) promoter. theriogenology 2012, 78, 793–802. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Shang, X.; Cheng, L.; Yang, L.; Liu, X.; Bai, C.; Wei, Z.; Hua, J.; Li, G. DNMT 1 maintains hypermethylation of CAG promoter specific region and prevents expression of exogenous gene in fat-1 transgenic sheep. PLoS ONE 2017, 12, e0171442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuijpers, T.W.; Nguyen, M.; Hopman, C.T.; Nieuwenhuys, E.; Dewald, G.; Lankester, A.C.; Roos, A.; van der Ende, A.; Fijen, C.; de Boer, M. Complement factor 7 gene mutations in relation to meningococcal infection and clinical recurrence of meningococcal disease. Mol. Immunol. 2010, 47, 671–677. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.J.; Im, E.; Chung, H.K.; Pothoulakis, C.; Rhee, S.H. TRIF mediates Toll-like receptor 5-induced signaling in intestinal epithelial cells. J. Biol. Chem. 2010, 284, 37570–37578. [Google Scholar] [CrossRef] [Green Version]
- Blohmke, C.J.; Victor, R.E.; Hirschfeld, A.F.; Elias, I.M.; Hancock, D.G.; Lane, C.R.; Davidson, A.G.; Wilcox, P.G.; Smith, K.D.; Overhage, J.; et al. Innate immunity mediated by TLR5 as a novel antiinflammatory target for cystic fibrosis lung disease. J. Immunol. 2008, 180, 7764–7773. [Google Scholar] [CrossRef] [Green Version]
- Kawai, T.; Sato, S.; Ishii, K.J.; Coban, C.; Hemmi, H.; Yamamoto, M.; Terai, K.; Matsuda, M.; Inoue, J.; Uematsu, S.; et al. Interferon-alpha induction through Toll-like receptors involves a direct interaction of IRF7 with MyD88 and TRAF6. Nat. Immunol. 2004, 5, 1061–1068. [Google Scholar] [CrossRef]
- Hayashi, F.; Smith, K.D.; Ozinsky, A.; Hawn, T.R.; Yi, E.C.; Goodlett, D.R.; Eng, J.K.; Akira, S.; Underhill, D.M.; Aderem, A. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 2001, 410, 1099–1103. [Google Scholar] [CrossRef]
- Nicklin, M.J.; Weith, A.; Duff, G.W. A physical map of the region encompassing the human interleukin-1 alpha, interleukin-1 beta, and interleukin-1 receptor antagonist genes. Genomics 1994, 19, 382–384. [Google Scholar] [CrossRef]
- Enserink, J.M.; Kolodner, R.D. An overview of Cdk1-controlled targets and processes. Cell. Div. 2010, 5, 11. [Google Scholar] [CrossRef] [Green Version]
- Saltiel, A.R.; Olefsky, J.M. Inflammatory mechanisms linking obesity and metabolic disease. J. Clin. Invest. 2017, 127, 1–4. [Google Scholar] [CrossRef]
- Ge, L.; Gordon, J.S.; Hsuan, C.; Stenn, K.; Prouty, S.M. Identification of the delta-6 desaturase of human sebaceous glands: Expression and enzyme activity. J. Invest. Dermatol. 2003, 120, 707–714. [Google Scholar] [CrossRef] [PubMed]
- Sher, T.; Yi, H.F.; McBride, O.W.; Gonzalez, F.J. cDNA cloning, chromosomal mapping, and functional characterization of the human peroxisome proliferator activated receptor. Biochemistry-US 1993, 32, 5598–5604. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, D.; Quintanilla, R.; Varona, L.; Díaz, I.; Ramírez, O.; Pena, R.N.; Amills, M. Polymorphism of the pigacetyl-coenzyme A carboxylaseαgene is associated with fatty acid composition in a Duroc commercial line. Anim. Genet. 2009, 40, 410–417. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Lu, D.; Xue, J.; Huang, Y.; Huang, S. Construction of mammary gland-specific expression vectors for human clotting factor IX and its secretory expression in goat milk. Chin. J. Biotechnol. 1997, 13, 271–276. [Google Scholar]
- Hammer, R.E.; Pursel, V.G.; Rexroad, C.J.; Wall, R.J.; Bolt, D.J.; Ebert, K.M.; Palmiter, R.D.; Brinster, R.L. Production of transgenic rabbits, sheep and pigs by microinjection. Nature 1985, 315, 680–683. [Google Scholar] [CrossRef] [Green Version]
- Chan, A.W.; Homan, E.J.; Ballou, L.U.; Burns, J.C.; Bremel, R.D. Transgenic cattle produced by reverse-transcribed gene transfer in oocytes. Proc. Natl. Acad. Sci. USA 1998, 95, 14028–14033. [Google Scholar] [CrossRef] [Green Version]
- Van Cott, K.E.; Butler, S.P.; Russell, C.G.; Subramanian, A.; Lubon, H.; Gwazdauskas, F.C.; Knight, J.; Drohan, W.N.; Velander, W.H. Transgenic pigs as bioreactors: A comparison of gamma-carboxylation of glutamic acid in recombinant human protein C and factor IX by the mammary gland. Genet. Anal. 1999, 15, 155–160. [Google Scholar] [CrossRef]
- Tian, X.C.; Kubota, C.; Enright, B.; Yang, X. Cloning animals by somatic cell nuclear transfer-biological factors. Reprod. Biol. Endocrinol. 2003, 1, 98. [Google Scholar] [CrossRef] [Green Version]
- Lou, Y.; Schwender, J.; Shanklin, J. FAD2 and FAD3 desaturases form heterodimers that facilitate metabolic channeling in vivo. J. Biol. Chem. 2014, 289, 17996–18007. [Google Scholar] [CrossRef] [Green Version]
- Wei, Z.; Li, D.; Zhu, L.; Yang, L.; Chen, C.; Bai, C.; Li, G. Omega 3 polyunsaturated fatty acids inhibit cell proliferation by regulating cell cycle in fad3b transgenic mouse embryonic stem cells. LipidS Health Dis. 2018, 17, 210. [Google Scholar] [CrossRef] [Green Version]
- Newell, M.; Brun, M.; Field, C.J. Treatment with DHA Modifies the Response of MDA-MB-231 Breast Cancer Cells and Tumors from nu/nu Mice to Doxorubicin through Apoptosis and Cell Cycle Arrest. J. Nutr. 2019, 149, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Zhou, S.; Xiang, R.; Zhao, Z.; Liu, S.; Ding, N.; Gong, S.; Lin, Y.; Li, X.; Bai, X.; et al. An Omega-3 fatty acid desaturase-expressing gene attenuates prostate cancer proliferation by cell cycle regulation. Oncol. Lett. 2017, 13, 3717–3721. [Google Scholar] [CrossRef] [Green Version]
- So, W.W.; Liu, W.N.; Leung, K.N. Omega-3 Polyunsaturated Fatty Acids Trigger Cell Cycle Arrest and Induce Apoptosis in Human Neuroblastoma LA-N-1 Cells. Nutrients 2015, 7, 6956–6973. [Google Scholar] [CrossRef] [PubMed]
- Yao, Q.H.; Zhang, X.C.; Fu, T.; Gu, J.Z.; Wang, L.; Wang, Y.; Lai, Y.B.; Wang, Y.Q.; Guo, Y. Omega-3 polyunsaturated fatty acids inhibit the proliferation of the lung adenocarcinoma cell line A549 in vitro. Mol. Med. Rep. 2014, 9, 401–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, G.; Fu, C.; Wang, H.; Adoligbe, C.; Wei, S.; Li, S.; Jiang, B.; Wang, H.; Zan, L. Production of transgenic beef cattle rich in n-3 PUFAs by somatic cell nuclear transfer. Biotechnol. Lett. 2015, 37, 1565–1571. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Chen, H.; Wu, X.; Zhou, Y.; Lu, J.; Chen, H.; Deng, J. A modified n–3 fatty acid desaturase gene from Caenorhabditis briggsae produced high proportion of DHA and DPA in transgenic mice. Transgenic. Res. 2008, 17, 717–725. [Google Scholar] [CrossRef]
- Das, U.N. Essential fatty acids and their metabolites as modulators of stem cell biology with reference to inflammation, cancer, and metastasis. Cancer Metastasis Rev. 2011, 30, 311–324. [Google Scholar] [CrossRef]
- Nowak, J.; Weylandt, K.H.; Habbel, P.; Wang, J.; Dignass, A.; Glickman, J.N.; Kang, J.X. Colitis-associated colon tumorigenesis is suppressed in transgenic mice rich in endogenous n-3 fatty acids. Carcinogenesis 2007, 28, 1991–1995. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Xu, X.; Liu, Y.; Zhang, L.; Odle, J.; Lin, X.; Zhu, H.; Wang, X.; Liu, Y. EPA and DHA Inhibit Myogenesis and Downregulate the Expression of Muscle-related Genes in C2C12 Myoblasts. Genes 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Halade, G.V.; Rahman, M.M.; Bhattacharya, A.; Barnes, J.L.; Chandrasekar, B.; Fernandes, G. Docosahexaenoic acid-enriched fish oil attenuates kidney disease and prolongs median and maximal life span of autoimmune lupus-prone mice. J. Immunol. 2010, 184, 5280–5286. [Google Scholar] [CrossRef] [Green Version]
- Sam, M.R.; Tavakoli-Mehr, M.; Safaralizadeh, R. Omega-3 fatty acid DHA modulates p53, survivin, and microRNA-16-1 expression in KRAS-mutant colorectal cancer stem-like cells. Genes. Nutr. 2018, 13, 8. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Li, M.; Geng, F. Omega-3 Polyunsaturated Fatty Acids Eicosapentaenoic Acid and Docosahexaenoic Acid Enhance Dexamethasone Sensitivity in Multiple Myeloma Cells by the p53/miR-34a/Bcl-2 Axis. Biochemistry (Mosc) 2017, 82, 826–833. [Google Scholar] [CrossRef] [PubMed]
- Borghetti, G.; Yamaguchi, A.A.; Aikawa, J.; Yamazaki, R.K.; de Brito, G.A.; Fernandes, L.C. Fish oil administration mediates apoptosis of Walker 256 tumor cells by modulation of p53, Bcl-2, caspase-7 and caspase-3 protein expression. Lipids. Health Dis. 2015, 14, 94. [Google Scholar] [CrossRef] [Green Version]
- Zand, H.; Rhimipour, A.; Bakhshayesh, M.; Shafiee, M.; Nour, M.I.; Salimi, S. Involvement of PPAR-gamma and p53 in DHA-induced apoptosis in Reh cells. Mol. Cell. Biohem. 2007, 304, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wu, Y.; Zhang, Z.; Zheng, X.; Wang, Y.; Yu, M.; Liu, G. Effects of the rs3834458 Single Nucleotide Polymorphism in FADS2 on Levels of n-3 Long-chain Polyunsaturated Fatty Acids: A Meta-analysis. Prostaglandins Leukot. Essent. Fatty Acids 2019, 150, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Lee, H.; Kang, S.; Park, W. Fatty Acid Desaturases, Polyunsaturated Fatty Acid Regulation, and Biotechnological Advances. Nutrients 2016, 8, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome. Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, R.; Zheng, Z.; Yang, C.; Zhang, X.; Cheng, L.; Su, G.; Bai, C.; Li, G. Comparative Transcriptome Analysis Provides Insights into the Polyunsaturated Fatty Acid Synthesis Regulation of Fat-1 Transgenic Sheep. Int. J. Mol. Sci. 2020, 21, 1121. https://doi.org/10.3390/ijms21031121
Luo R, Zheng Z, Yang C, Zhang X, Cheng L, Su G, Bai C, Li G. Comparative Transcriptome Analysis Provides Insights into the Polyunsaturated Fatty Acid Synthesis Regulation of Fat-1 Transgenic Sheep. International Journal of Molecular Sciences. 2020; 21(3):1121. https://doi.org/10.3390/ijms21031121
Chicago/Turabian StyleLuo, Rongsong, Zhong Zheng, Chunrong Yang, Xiaoran Zhang, Lei Cheng, Guanghua Su, Chunling Bai, and Guangpeng Li. 2020. "Comparative Transcriptome Analysis Provides Insights into the Polyunsaturated Fatty Acid Synthesis Regulation of Fat-1 Transgenic Sheep" International Journal of Molecular Sciences 21, no. 3: 1121. https://doi.org/10.3390/ijms21031121




