Validation of a New Multicistronic Plasmid for the Efficient and Stable Expression of Transgenes in Microalgae
Abstract
:1. Introduction
2. Results and Discussion
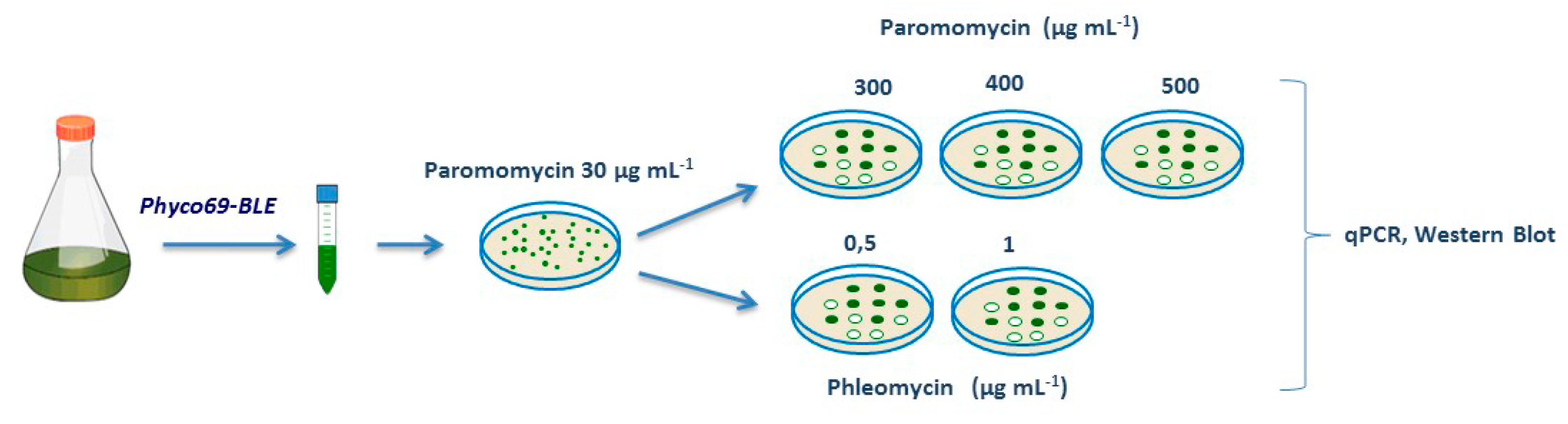
2.1. Construction and Transformation Efficiency of Several Multicistronic APHVIII-based Fusion Plasmids
2.2. Phyco69 Enables the Selection of Transformants with High Expression Levels of the Gene of Interest
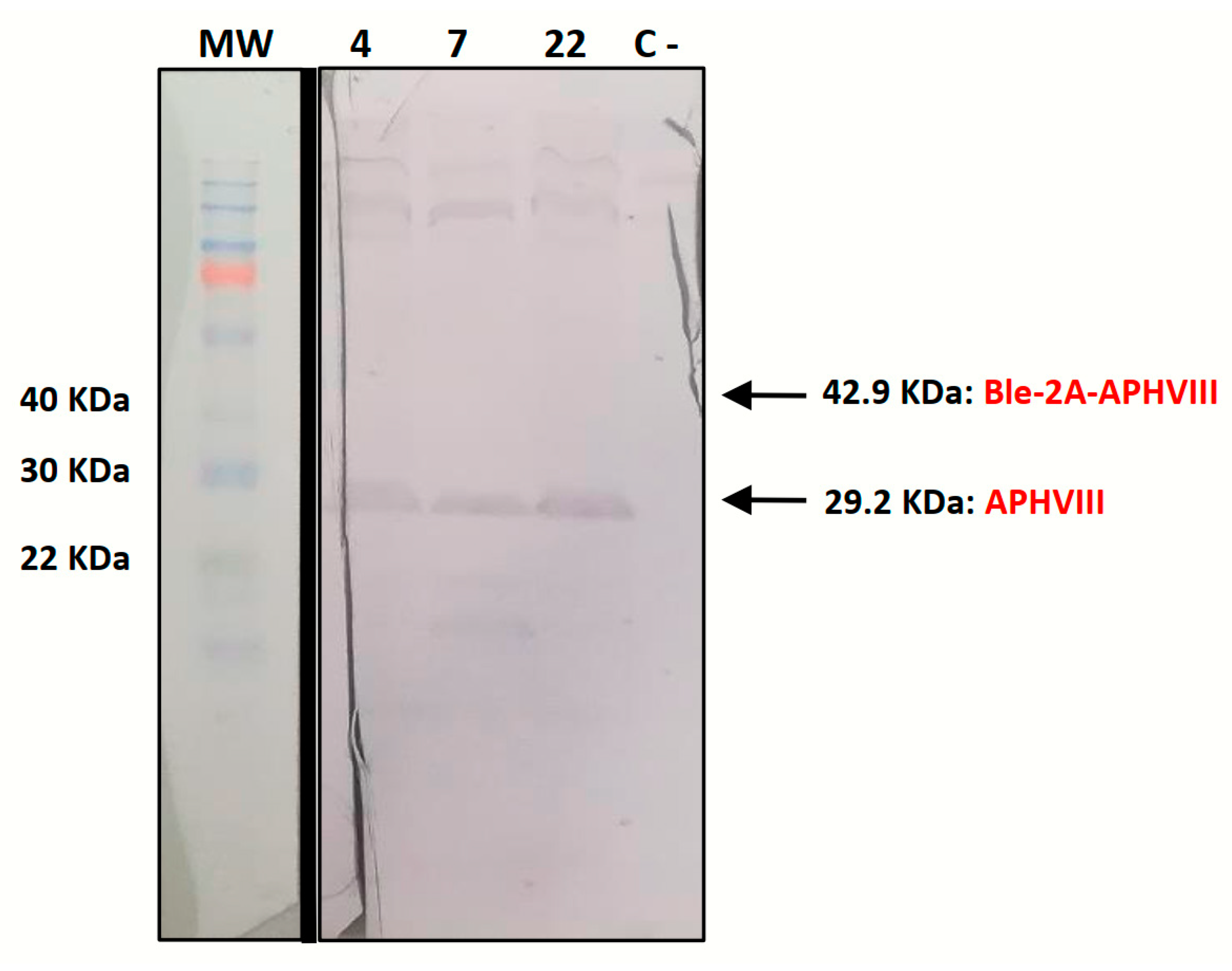
2.3. Phyco69 Enables the Simultaneous Expression of Two Genes, Which are Efficiently Processed Generating Independent Proteins
3. Materials and Methods
3.1. Strains and Culture Conditions
3.2. Plasmids Constructions
3.3. Chlamydomonas Nuclear Transformation
3.4. Real Time PCR
3.5. Western Blot
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A


| Target Gene | Primer Name | Sequence |
|---|---|---|
| BLE | qBLE-F | CGACTTCGCCGGTGTGGTC |
| qBLE-R | CACGAAGTGCACGCAGTTGC | |
| APHVIII | qAPHVIIIF | GAGGATCTGGACGAGGAGCGGAA |
| qAPHVIIIR | CCCTCAGAAGAACTCGTCCAACAGC | |
| UBC8 | qUBC8 | GTACAGCGGCGGCTAGAGGCAC |
| qUBC8 | AGCGTCAGCGGCGGTTGCAGGTATCT |
References
- Leu, S.; Boussiba, S. Advances in the Production of High-Value Products by Microalgae. Ind. Biotechnol. 2014, 10, 169–183. [Google Scholar] [CrossRef]
- Benemann, J. Microalgae for Biofuels and Animal Feeds. Energies 2013, 6, 5869–5886. [Google Scholar] [CrossRef] [Green Version]
- Varela, J.C.; Pereira, H.; Vila, M.; León, R. Production of carotenoids by microalgae: Achievements and challenges. Photosynth. Res. 2015, 125, 423–436. [Google Scholar] [CrossRef] [PubMed]
- Gangl, D.; Zedler, J.A.Z.; Rajakumar, P.D.; Martinez, E.M.R.; Riseley, A.; Włodarczyk, A.; Purton, S.; Sakuragi, Y.; Howe, C.J.; Jensen, P.E.; et al. Biotechnological exploitation of microalgae. J. Exp. Bot. 2015, 66, 6975–6990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valverde, F.; Romero-Campero, F.J.; León, R.; Guerrero, M.G.; Serrano, A. New challenges in microalgae biotechnology. Eur. J. Protistol. 2016, 55, 95–101. [Google Scholar] [CrossRef]
- Gifuni, I.; Pollio, A.; Safi, C.; Marzocchella, A.; Olivieri, G. Current Bottlenecks and Challenges of the Microalgal Biorefinery. Trends Biotechnol. 2019, 37, 242–252. [Google Scholar] [CrossRef]
- Sharon-Gojman, R.; Maimon, E.; Leu, S.; Zarka, A.; Boussiba, S. Advanced methods for genetic engineering of Haematococcus pluvialis (Chlorophyceae, Volvocales). Algal Res. 2015, 10, 8–15. [Google Scholar] [CrossRef]
- Doron, L.; Segal, N.; Shapira, M. Transgene Expression in Microalgae-From Tools to Applications. Front. Plant Sci. 2016, 7, 505. [Google Scholar] [CrossRef]
- Jeon, S.; Lim, J.-M.; Lee, H.-G.; Shin, S.-E.; Kang, N.K.; Park, Y.-I.; Oh, H.-M.; Jeong, W.-J.; Jeong, B.; Chang, Y.K. Current status and perspectives of genome editing technology for microalgae. Biotechnol. Biofuels 2017, 10, 267. [Google Scholar] [CrossRef] [Green Version]
- León-Bañares, R.; González-Ballester, D.; Galván, A.; Fernández, E. Transgenic microalgae as green cell-factories. Trends Biotechnol. 2004, 22, 45–52. [Google Scholar] [CrossRef]
- Vazquez-Villegas, P.; Torres-Acosta, M.A.; Garcia-Echauri, S.A.; Aguilar-Yanez, J.M.; Rito-Palomares, M.; Ruiz-Ruiz, F. Genetic manipulation of microalgae for the production of bioproducts. Front. Biosci. 2018, 10, 254–275. [Google Scholar]
- Charoonnart, P.; Purton, S.; Saksmerprome, V. Applications of Microalgal Biotechnology for Disease Control in Aquaculture. Biology 2018, 7, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Potvin, G.; Zhang, Z. Strategies for high-level recombinant protein expression in transgenic microalgae: A review. Biotechnol. Adv. 2010, 28, 910–918. [Google Scholar] [CrossRef] [PubMed]
- Purton, S.; Szaub, J.B.; Wannathong, T.; Young, R.; Economou, C.K. Genetic engineering of algal chloroplasts: Progress and prospects. Russ. J. Plant Physiol. 2013, 60, 491–499. [Google Scholar] [CrossRef]
- Surzycki, R.; Greenham, K.; Kitayama, K.; Dibal, F.; Wagner, R.; Rochaix, J.-D.; Ajam, T.; Surzycki, S. Factors effecting expression of vaccines in microalgae. Biologicals 2009, 37, 133–138. [Google Scholar] [CrossRef]
- Rasala, B.A.; Barrera, D.J.; Ng, J.; Plucinak, T.M.; Rosenberg, J.N.; Weeks, D.P.; Oyler, G.A.; Peterson, T.C.; Haerizadeh, F.; Mayfield, S.P. Expanding the spectral palette of fluorescent proteins for the green microalga Chlamydomonas reinhardtii. Plant J. 2013, 74, 545–556. [Google Scholar] [CrossRef]
- Hallmann, A. Algal Transgenics and Biotechnology. Transgenic Plant J. 2007, 1, 81–98. [Google Scholar]
- Lauersen, K.J.; Kruse, O.; Mussgnug, J.H. Targeted expression of nuclear transgenes in Chlamydomonas reinhardtii with a versatile, modular vector toolkit. Appl. Microbiol. Biotechnol. 2015, 99, 3491–3503. [Google Scholar] [CrossRef]
- Schroda, M.; Beck, C.F.; Vallon, O. Sequence elements within an HSP70 promoter counteract transcriptional transgene silencing in Chlamydomonas. Plant J. 2002, 31, 445–455. [Google Scholar] [CrossRef] [Green Version]
- Fischer, N.; Rochaix, J.D. The flanking regions of PsaD drive efficient gene expression in the nucleus of the green alga Chlamydomonas reinhardtii. Mol. Genet. Genom. 2001, 265, 888–894. [Google Scholar] [CrossRef]
- Wu, J.; Hu, Z.; Wang, C.; Li, S.; Lei, A. Efficient expression of green fluorescent protein (GFP) mediated by a chimeric promoter in Chlamydomonas reinhardtii. Chin. J. Oceanol. Limnol. 2008, 26, 242–247. [Google Scholar] [CrossRef]
- Scranton, M.A.; Ostrand, J.T.; Georgianna, D.R.; Lofgren, S.M.; Li, D.; Ellis, R.C.; Carruthers, D.N.; Dräger, A.; Masica, D.L.; Mayfield, S.P. Synthetic promoters capable of driving robust nuclear gene expression in the green alga Chlamydomonas reinhardtii. Algal Res. 2016, 15, 135–142. [Google Scholar] [CrossRef] [Green Version]
- Crozet, P.; Navarro, F.J.; Willmund, F.; Mehrshahi, P.; Bakowski, K.; Lauersen, K.J.; Pérez-Pérez, M.-E.; Auroy, P.; Gorchs Rovira, A.; Sauret-Gueto, S.; et al. Birth of a Photosynthetic Chassis: A MoClo Toolkit Enabling Synthetic Biology in the Microalga Chlamydomonas reinhardtii. ACS Synth. Biol. 2018, 7, 2074–2086. [Google Scholar] [CrossRef] [Green Version]
- Baier, T.; Kros, D.; Feiner, R.C.; Lauersen, K.J.; Müller, K.M.; Kruse, O. Engineered Fusion Proteins for Efficient Protein Secretion and Purification of a Human Growth Factor from the Green Microalga Chlamydomonas reinhardtii. ACS Synth. Biol. 2018, 7, 2547–2557. [Google Scholar] [CrossRef] [PubMed]
- Fuhrmann, M.; Oertel, W.; Hegemann, P. A synthetic gene coding for the green fluorescent protein (GFP) is a versatile reporter in Chlamydomonas reinhardtii. Plant J. 1999, 19, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Fuhrmann, M. Production of antigens in Chlamydomonas reinhardtii: Green microalgae as a novel source of recombinant proteins. Methods Mol. Med. 2004, 94, 191–195. [Google Scholar]
- Shao, N.; Bock, R. A codon-optimized luciferase from Gaussia princeps facilitates the in vivo monitoring of gene expression in the model alga Chlamydomonas reinhardtii. Curr. Genet. 2008, 53, 381–388. [Google Scholar] [CrossRef] [Green Version]
- Díaz-Santos, E.; Vila, M.; Vigara, J.; León, R. A new approach to express transgenes in microalgae and its use to increase the flocculation ability of Chlamydomonas reinhardtii. J. Appl. Phycol. 2016, 28, 1611–1621. [Google Scholar] [CrossRef]
- Neupert, J.; Karcher, D.; Bock, R. Generation of Chlamydomonas strains that efficiently express nuclear transgenes. Plant J. 2009, 57, 1140–1150. [Google Scholar] [CrossRef]
- Kong, F.; Yamasaki, T.; Kurniasih, S.D.; Hou, L.; Li, X.; Ivanova, N.; Okada, S.; Ohama, T. Robust expression of heterologous genes by selection marker fusion system in improved Chlamydomonas strains. J. Biosci. Bioeng. 2015, 120, 239–245. [Google Scholar] [CrossRef] [Green Version]
- Szymczak, A.L.; Workman, C.J.; Wang, Y.; Vignali, K.M.; Dilioglou, S.; Vanin, E.F.; Vignali, D.A.A. Correction of multi-gene deficiency in vivo using a single “self-cleaving” 2A peptide-based retroviral vector. Nat. Biotechnol. 2004, 22, 589–594. [Google Scholar] [CrossRef] [PubMed]
- Minskaia, E.; Nicholson, J.; Ryan, M.D. Optimisation of the foot-and-mouth disease virus 2A co-expression system for biomedical applications. BMC Biotechnol. 2013, 13, 67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subramanian, V.; Schuster, L.A.; Moore, K.T.; Taylor, L.E.; Baker, J.O.; Vander Wall, T.A.; Linger, J.G.; Himmel, M.E.; Decker, S.R. A versatile 2A peptide-based bicistronic protein expressing platform for the industrial cellulase producing fungus, Trichoderma reesei. Biotechnol. Biofuels 2017, 10, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luke, G.A.; De Felipe, P.; Cowton, V.M.; Hughes, L.E.; Halpin, C.; Ryan, M.D. Self-processing polyproteins: A strategy for co-expression of multiple proteins in plants. Biotechnol. Genet. Eng. Rev. 2006, 23, 239–252. [Google Scholar] [CrossRef] [PubMed]
- Rasala, B.A.; Lee, P.A.; Shen, Z.; Briggs, S.P.; Mendez, M.; Mayfield, S.P. Robust expression and secretion of Xylanase1 in Chlamydomonas reinhardtii by fusion to a selection gene and processing with the FMDV 2A peptide. PLoS ONE 2012, 7, e43349. [Google Scholar] [CrossRef] [Green Version]
- Rasala, B.A.; Chao, S.-S.; Pier, M.; Barrera, D.J.; Mayfield, S.P. Enhanced Genetic Tools for Engineering Multigene Traits into Green Algae. PLoS ONE 2014, 9, e94028. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez-Ballester, D.; de Montaigu, A.; Higuera, J.J.; Galvan, A.; Fernandez, E. Functional genomics of the regulation of the nitrate assimilation pathway in Chlamydomonas. Plant Physiol. 2005, 137, 522–533. [Google Scholar] [CrossRef] [Green Version]
- Kong, F.; Yamaoka, Y.; Ohama, T.; Lee, Y.; Li-Beisson, Y. Molecular Genetic Tools and Emerging Synthetic Biology Strategies to Increase Cellular Oil Content in Chlamydomonas reinhardtii. Plant Cell Physiol. 2019, 60, 1184–1196. [Google Scholar] [CrossRef]
- Sizova, I.A.; Lapina, T.V.; Frolova, O.N.; Alexandrova, N.N.; Akopiants, K.E.; Danilenko, V.N. Stable nuclear transformation of Chlamydomonas reinhardtii with a Streptomyces rimosus gene as the selective marker. Gene 1996, 181, 13–18. [Google Scholar] [CrossRef]
- Sizova, I.; Fuhrmann, M.; Hegemann, P. A Streptomyces rimosus aphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii. Gene 2001, 277, 221–229. [Google Scholar] [CrossRef]
- Jinkerson, R.E.; Jonikas, M.C. Molecular techniques to interrogate and edit the Chlamydomonas nuclear genome. Plant J. 2015, 82, 393–412. [Google Scholar] [CrossRef] [PubMed]
- Boyko, K.M.; Gorbacheva, M.A.; Korzhenevskiy, D.A.; Alekseeva, M.G.; Mavletova, D.A.; Zakharevich, N.V.; Elizarov, S.M.; Rudakova, N.N.; Danilenko, V.N.; Popov, V.O. Structural characterization of the novel aminoglycoside phosphotransferase AphVIII from Streptomyces rimosus with enzymatic activity modulated by phosphorylation. Biochem. Biophys. Res. Commun. 2016, 477, 595–601. [Google Scholar] [CrossRef]
- Fordham-Skelton, A.P.; Chilley, P.; Lumbreras, V.; Reignoux, S.; Fenton, T.R.; Dahm, C.C.; Pages, M.; Gatehouse, J.A. A novel higher plant protein tyrosine phosphatase interacts with SNF1-related protein kinases via a KIS (kinase interaction sequence) domain. Plant J. 2002, 29, 705–715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffmann, G.R.; Ronan, M.V.; Sylvia, K.E.; Tartaglione, J.P. Enhancement of the recombinagenic and mutagenic activities of bleomycin in yeast by intercalation of acridine compounds into DNA. Mutagenesis 2009, 24, 317–329. [Google Scholar] [CrossRef] [Green Version]
- Jiang, W.; Brueggeman, A.J.; Horken, K.M.; Plucinak, T.M.; Weeks, D.P. Successful transient expression of Cas9 and single guide RNA genes in Chlamydomonas reinhardtii. Eukaryot. Cell 2014, 13, 1465–1469. [Google Scholar] [CrossRef] [Green Version]
- Xu, T.; Ripp, S.; Sayler, G.S.; Close, D.M. Expression of a Humanized Viral 2A-Mediated lux Operon Efficiently Generates Autonomous Bioluminescence in Human Cells. PLoS ONE 2014, 9, e96347. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.; Zhou, N.; Wang, H.; Huang, D.; Lang, Z. Processing and targeting of proteins derived from polyprotein with 2A and LP4/2A as peptide linkers in a maize expression system. PLoS ONE 2017, 12, e0174804. [Google Scholar] [CrossRef] [Green Version]
- Onishi, M.; Pringle, J.R. Robust transgene expression from bicistronic mRNA in the green alga Chlamydomonas reinhardtii. G3 2016, 6, 4115–4125. [Google Scholar] [CrossRef] [Green Version]
- Loppes, R.; Radoux, M.; Ohresser, M.C.; Matagne, R.F. Transcriptional regulation of the Nia1 gene encoding nitrate reductase in Chlamydomonas reinhardtii: Effects of various environmental factors on the expression of a reporter gene under the control of the Nia1 promoter. Plant Mol. Biol. 1999, 41, 701–711. [Google Scholar] [CrossRef]
- León, R.; Couso, I.; Fernández, E. Metabolic engineering of ketocarotenoids biosynthesis in the unicelullar microalga Chlamydomonas reinhardtii. J. Biotechnol. 2007, 130, 143–152. [Google Scholar] [CrossRef]
- Kindle, K.L. High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 1990, 87, 1228–1232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez-Ballester, D.; Pootakham, W.; Mus, F.; Yang, W.; Catalanotti, C.; Magneschi, L.; de Montaigu, A.; Higuera, J.J.; Prior, M.; Galvan, A.; et al. Reverse genetics in Chlamydomonas: A platform for isolating insertional mutants. Plant Methods 2011, 7, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vila, M.; Couso, I.; León, R. Carotenoid content in mutants of the chlorophyte Chlamydomonas reinhardtii with low expression levels of phytoene desaturase. Process. Biochem. 2008, 43, 1147–1152. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Santos, E.; de la Vega, M.; Vila, M.; Vigara, J.; León, R. Efficiency of different heterologous promoters in the unicellular microalga Chlamydomonas reinhardtii. Biotechnol. Prog. 2013, 29, 319–328. [Google Scholar] [CrossRef]



| Plasmid | 200 ng | 500 ng | 1 µg | 2 µg | Transformants µg−1 DNA |
|---|---|---|---|---|---|
| Phyco69 | 98 ± 22 | 215 ± 38 | 380 ± 58 | 820 ± 70 | 427 |
| pSI103 | 28 ± 12 | 60 ± 21 | 140 ± 32 | 250 ± 45 | 131 |
| PhycoC67 | 0 | 5 ± 2 | 7 ± 4 | 18 ± 6 | 6.5 |
| PhycoC67FL | 0 | 8 ± 4 | 12 ± 7 | 3.5 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Molina-Márquez, A.; Vila, M.; Rengel, R.; Fernández, E.; García-Maroto, F.; Vigara, J.; León, R. Validation of a New Multicistronic Plasmid for the Efficient and Stable Expression of Transgenes in Microalgae. Int. J. Mol. Sci. 2020, 21, 718. https://doi.org/10.3390/ijms21030718
Molina-Márquez A, Vila M, Rengel R, Fernández E, García-Maroto F, Vigara J, León R. Validation of a New Multicistronic Plasmid for the Efficient and Stable Expression of Transgenes in Microalgae. International Journal of Molecular Sciences. 2020; 21(3):718. https://doi.org/10.3390/ijms21030718
Chicago/Turabian StyleMolina-Márquez, Ana, Marta Vila, Rocío Rengel, Emilio Fernández, Federico García-Maroto, Javier Vigara, and Rosa León. 2020. "Validation of a New Multicistronic Plasmid for the Efficient and Stable Expression of Transgenes in Microalgae" International Journal of Molecular Sciences 21, no. 3: 718. https://doi.org/10.3390/ijms21030718
APA StyleMolina-Márquez, A., Vila, M., Rengel, R., Fernández, E., García-Maroto, F., Vigara, J., & León, R. (2020). Validation of a New Multicistronic Plasmid for the Efficient and Stable Expression of Transgenes in Microalgae. International Journal of Molecular Sciences, 21(3), 718. https://doi.org/10.3390/ijms21030718






