Abstract
Marine biofilms are composed of many species of bacteria, unicellular algae, and protozoa. Biofilms can induce, inhibit, or have no effect on settlement of larvae and spores of algae. In this review, we focus on induction of larval settlement by marine bacteria and unicellular eukaryotes and review publications from 2010 to September 2019. This review provides insights from meta-analysis on what is known about the effect of marine biofilms on larval settlement. Of great interest is the impact of different components of marine biofilms, such as bacteria and diatoms, extracellular polymeric substances, quorum sensing signals, unique inductive compounds, exoenzymes, and structural protein degradation products on larval settlement and metamorphosis. Molecular aspects of larval settlement and impact of climate change are reviewed and, finally, potential areas of future investigations are provided.
1. Introduction
Any clean substratum submerged in seawater is quickly colonized by molecules, micro-organisms [1], and propagules [2]. On the surface of the substratum, microbial colonizers form complex biofilms composed of bacteria, archaea, fungi, protozoa, and unicellular microalgae [3]. Marine biofilms are mixed complex microbial communities dominated by bacteria and diatoms (Bacillariophyceae) [4]. Biofilms are three dimensional in structure. Micro-organisms in biofilms are surrounded by a self-produced matrix of extracellular polymeric substances (EPS), composed of polysaccharides, proteins, nucleic acids, and lipids [5]. EPS keep microbial cells together and provide suitable micro-environmental conditions [6]. Marine biofilms are highly dynamic. Changes in temperature, salinity, solar radiation, etc., result in corresponding changes in microbial communities inhabiting biofilms [3]. Additionally, in the presence and absence of macro-organisms micro-organisms interact with each other within a biofilm, which changes microbial densities, biofilm’s species composition, and chemistries in space and time [7]. Products of exoenzymes, part of degradative microbial processes on structural materials, including proteins and proteinaceous glues, are integral to the organization of marine communities [8].
Investigations of marine biofilms, their composition, and dynamics started less than 100 years ago with the pioneering work of Claude Zobell and Ester Allen [9] who observed microbial biofilms and developed culture-dependent methods to isolate and cultivate marine bacteria. This pioneering work on isolation and culturing of bacteria began a fruitful area of research [10]. Culture-dependent methods continue to be intensively used and have been supplemented by development of molecular and metagenomic methods (see reviews [3,4,7,11]). Polymerase chain reaction combined with high throughput next generation sequencing methods allow characterization of non-cultivable and rare microorganisms present in biofilms [12]. Additionally, these methods allow identification of genes and prediction of their functions [3]. Recently developed metabolomic methods are employed to identify metabolites produced by microbes in biofilms in response to environmental stress [13,14]. Combinations of metagenomics, transcriptomics, proteomics, and metabolomics methods enables study of changes in composition of biofilms and chemical compounds produced as well as their functions. Though still in their infancy with respect to analysis and interpretation, these new methods have already demonstrated remarkable potential.
Cell-to-cell communication called quorum sensing (QS) allows bacteria in biofilms to coordinate their adhesion, swarming, luminescence, and production of bioactive compounds [15,16]. Bacteria produce and release small inductive molecules (autoinducers) and respond to them once they reach to a threshold concentration [17,18]. QS autoinducers depend on the density of producing bacteria. Different Gram-positive and Gram-negative bacteria use different QS signals for communication (see reviews [15,16,19]). The best studied QS autoinducers are acyl homoserine lactones (AHLs). Specificity of this autoinducers is related to the length of R-group side chain. Bacteria not only produce QS signals but also inhibit or disrupt QS of other bacteria (see reviews [20,21,22]). Additionally, some marine eukaryotes respond to bacterial QS signals and disrupt them [16].
Marine organisms that are sessile as adults routinely have free living planktonic dispersal stages called larvae and sporelings and other kinds of propagules [11]. These are referred to generally as propagules. During the planktonic stage, propagules may or may not, feed, grow, and develop. All propagules disperse some distance in the water column before they settle [23,24]. While some species of marine invertebrates are highly specialized and settle in the particular habitats, other species are generalists and settle and attach to any available substratum [25]. For most propagules, the selected substratum is particularly important for subsequent survival and reproduction. Once larvae or spores attach to the substratum, they metamorphose into juveniles and gather energy growing quickly or slowly into adult organisms. Many macro-fouling organisms involved in fouling of man-made surfaces grow very quickly and are reproductive within weeks of settlement.
The relationship of propagules to the substrate is complex. Microbial biofilms can either induce, inhibit or have no effect on the settlement of marine organisms (see reviews [7,25,26]). Depending on the species, settlement can be rapid, even faster than initial attachment by bacteria and can be triggered by surface energy and organic films [27,28,29,30]. In many cases, specialist larvae require general or specific bacterial films for settlement [25,26,31].
All larvae of specialist species and most larvae of generalist species respond to cues and pheromones produced by symbionts and conspecifics. Differences in a biofilm’s species composition, age, and the density of different species influence the attractiveness of the substratum to larvae through modification of its physical and chemical properties [26,31,32,33].
Biofouling is undesirable growth of micro- and macro-organisms on man-made structures. Biofouling causes huge economic losses to maritime industries, thus development of antifouling solutions that prevent settlement of larvae is of particular importance [34,35,36]. Microbes are so adaptive that usually even antifouling coatings are colonized by bacteria and diatoms [13,37].
The process of larval settlement is particularly important for biofouling and aquaculture research [11]. Sessile species, like the barnacle Amphibalanus (=Balanus) amphitrite, the polychaete Hydroides elegans, and the bryozoan Bugula neritina are global major biofouling species [25]. Larval settlement is important for aquaculture of mussels, oysters, abalone, scallops, and algae. Important to restoration ecology is the rehabilitation of habitat-forming species, like hard corals and oysters, and restoration of the stocks of commercially important shellfish [11].
Over the past several decades few reviews about marine biofilms have been published and there are even fewer reviews of larval-biofilm interactions [3,4,7,11,38]. Previous reports show that biofilms can induce or inhibit larval settlement and metamorphosis [7,26,28,39].
Since the discovery of natural products inhibitors of biofouling [29,40,41], natural products from marine organisms, including bacteria and diatoms, that inhibit biofouling have been reviewed [7,42,43,44,45,46]. Several reviews analyzed chemical ecology and natural products of microbes, invertebrates, vertebrates, and plants [47,48,49,50].
Hadfield and Paul [51] provided a comprehensive review of induction of larval settlement of invertebrates from 10 phyla by biofilms. Subsequent reviews focused either on biofouling and commercially important species [11,25] or particular processes, like quorum sensing and its inhibition [16,22]. Molecular mechanisms of larval settlement and inhibition were recently reviewed as well [52,53,54,55]. The development of novel omic-methods (metagenomics, metabolomics, transcriptomics, proteomics) resulted in many new findings in larval settlement and attachment. Thus, it is timely to summarize findings on induction of larval settlement by marine biofilms.
The main aim of this publication is to review recent publications on the induction of propagule settlement of macro-organisms by microbial biofilms published from January 2010 to September 2019. For earlier scientific publications readers should refer to above cited reviews and papers on this subject. Of great interest of this review is the impact of different components of marine biofilms, such as exoenzymes, EPS, bacteria and diatoms, QS autoinducers on larval settlement and metamorphosis, and unique inductive compounds from marine microorganisms. Molecular aspects of larval settlement are reviewed and, finally, future perspectives, and conclusions are provided. Additionally, the effect of factors associated with climate change on biofilms and larval settlement is discussed. Our ultimate goal is to begin to provide a coherent picture of the range of critical roles that microbes play in the development of marine communities.
2. Meta-Analysis of Recent Publications
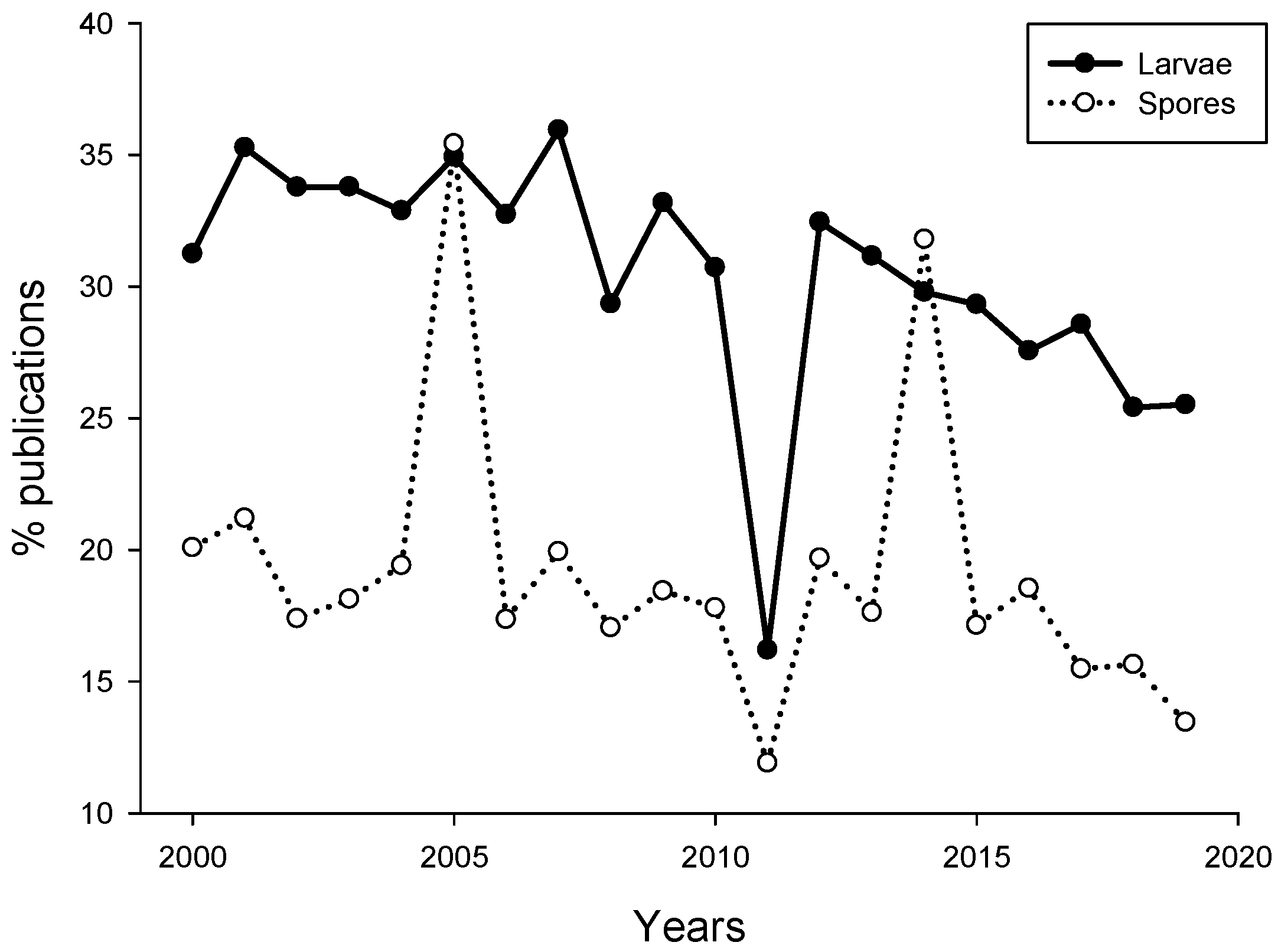
Analysis of publications suggested that the number of papers dealing with induction or inhibition of propagules settlement published for the last 19 years remains quite low (469). The percentage of publications dealing with larval settlement in response to biofilms did not exceed 35%, while the percentage of algal spore publications even lower (~21%, Figure 1). There was a sudden drop in the percentage of larval settlement and algal spore publications in 2011.
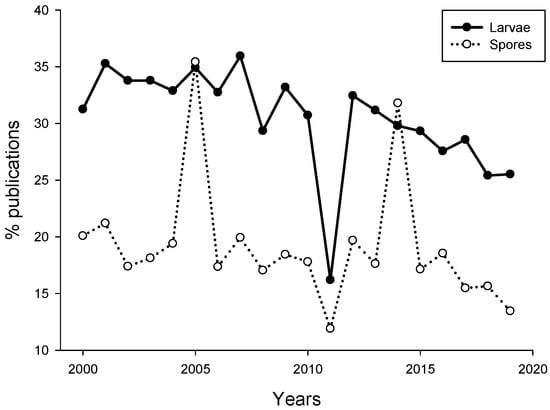
Figure 1.
Percent of publications dealing with settlement of invertebrate larvae (black circle) and algal spores (open circle) and microbes. The search was performed using Google Scholar for the period of 2000–2019. The search keywords were “marine microbe” and “larva” or “marine microbe” and “spore”.
The reason for that drop in the percentage of publications is not clear. Overall, the percentage of studies investigating propagule settlement decreased from 2000 to 2019. This may be related to the decrease of biofouling-related publications in recent years due to decreased funding opportunities in basic research on marine larvae and increased funding for basic and applied studies related to biofouling management [56].
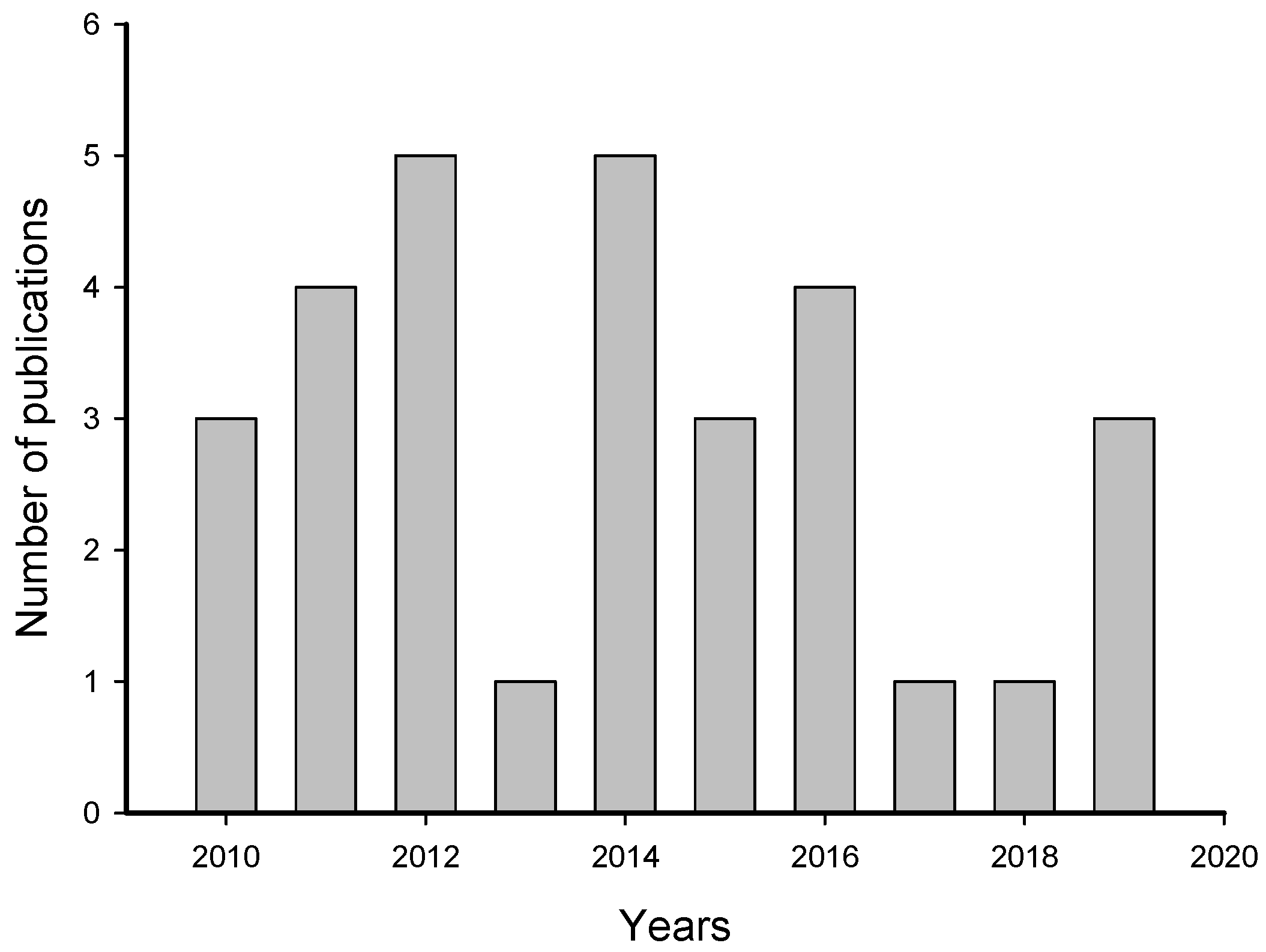
The systematic review of 469 publications by the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) method resulted, after citation screening, in 31 articles about induction of larval settlement by biofilms (Table S1). During the screening, duplicates and articles not related to the topic as well as reviews were eliminated. The number of publications that met our criteria increased from 2010 to 2012 and decreased from 2014 to 2019 (Figure 2). The highest numbers of publications (5) about induction of larval settlement by biofilms were published in 2012 and 2014 and the lowest number (1) was published in 2013, 2017, and 2018. Overall, the average rate of publication was 3.1 papers per year.
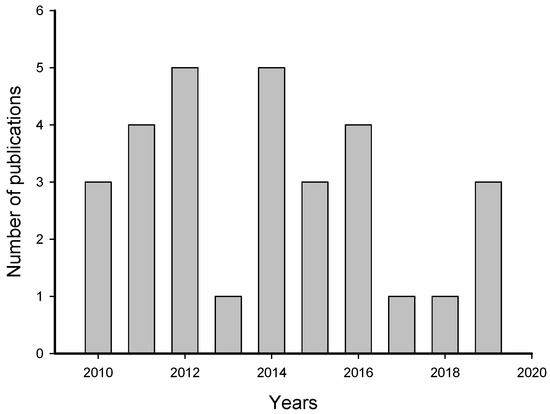
Figure 2.
Number of publications that examined the induction of larval settlement by marine biofilms. The publications include from January 2010 to September 2019. This figure only includes the publications that met all inclusion criteria.
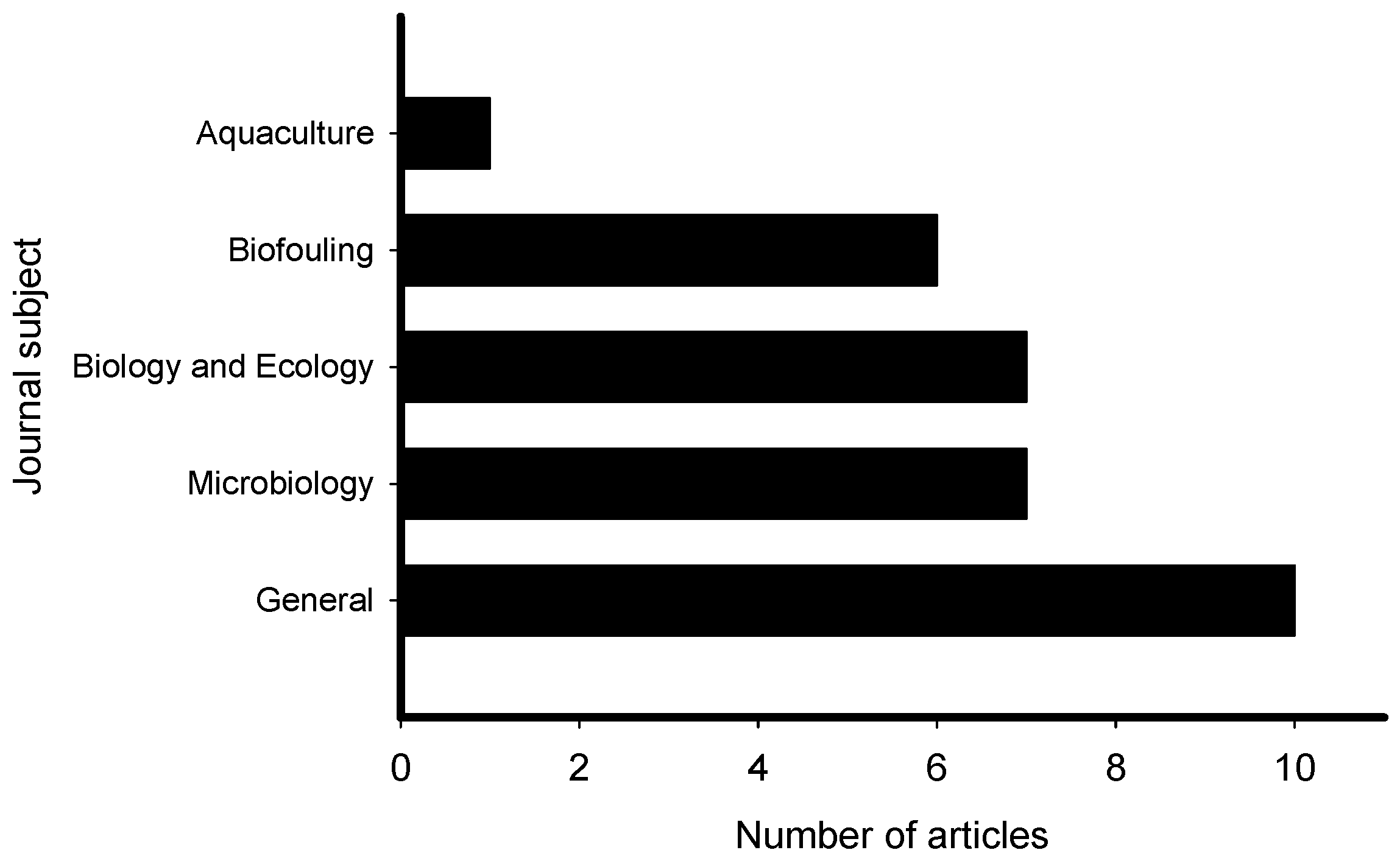
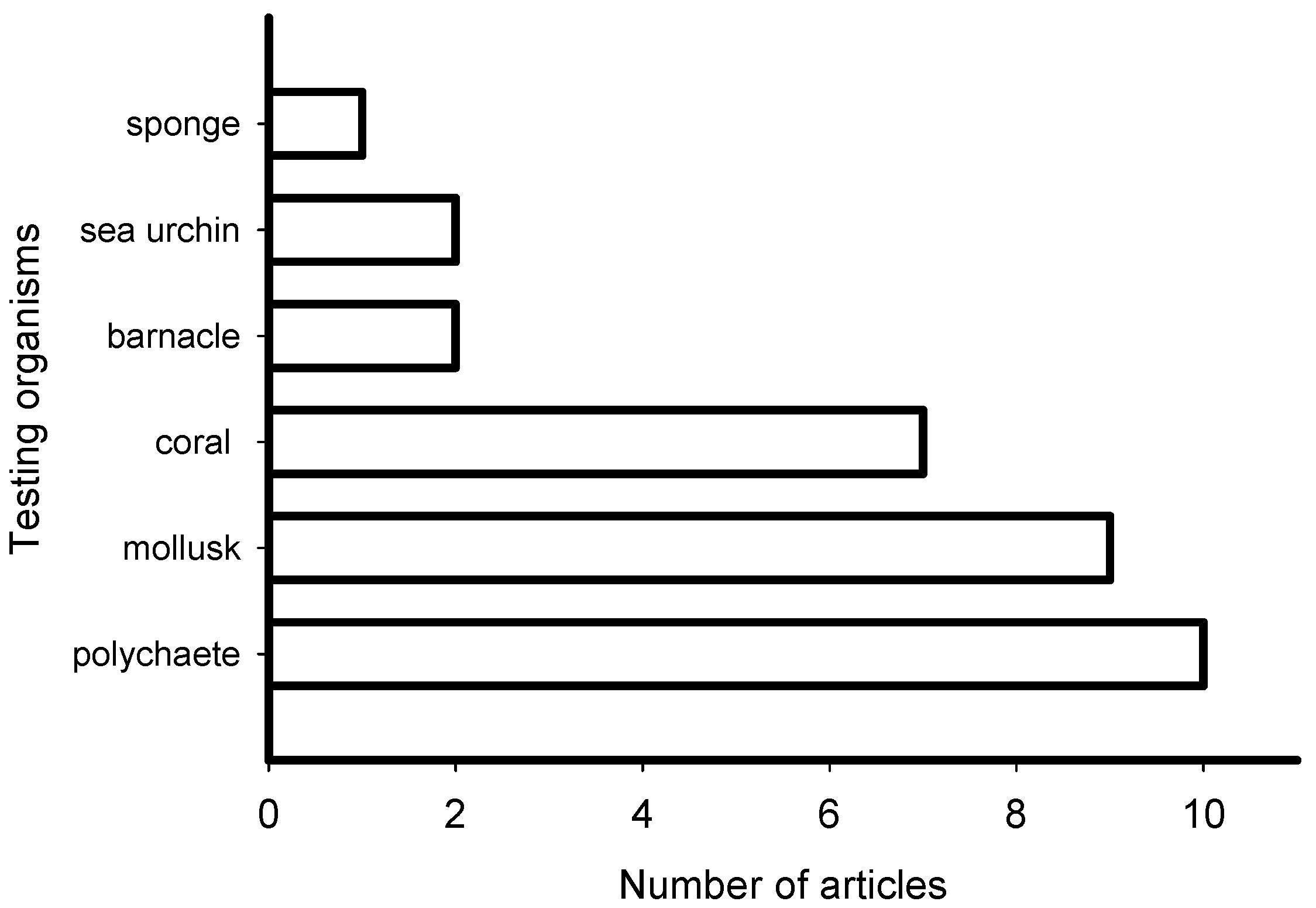
The reviewed articles on induction of larval settlement by marine biofilms were published in journals that represented general interest (e.g., PLoS ONE, Science, Scientific Reports), microbiology (ISME Journal, Microbial Ecology), marine ecology and biology (Marine Ecology Progress Series), biofouling, and aquaculture fields (Figure 3). The highest number of articles (10) was published in general field journals with open access and the lowest number (1) was published in an aquaculture journal. However, though low in number the collective group of papers was represented in very high impact factor journals. All 31 articles reported induction of larval settlement by bacteria or biofilms (Table S1). Not any diatom-related study reporting induction of larval settlement was published in the last 10 years. Fifteen of 31 articles reported effects of monospecies films of bacteria, while 16 of 31 reported effects of multispecies biofilms. The highest number of articles reported induction of polychaete settlement (mostly Hydroides elegans) followed by mussel (mostly Mytilus spp.) larval settlement (Figure 4). Induction of coral planulae settlement by bacteria and biofilms is represented by seven articles. One article was published on induction of sponge larvae settlement by bacterial biofilms.
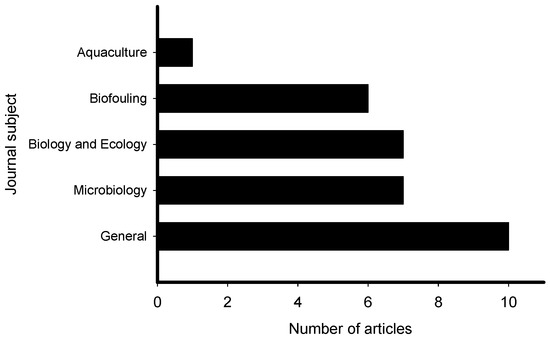
Figure 3.
Number of articles that examined the induction of larval settlement by marine biofilms from January 2010 to September 2019 according to the subject area. This figure only includes the publications that met all inclusion criteria.
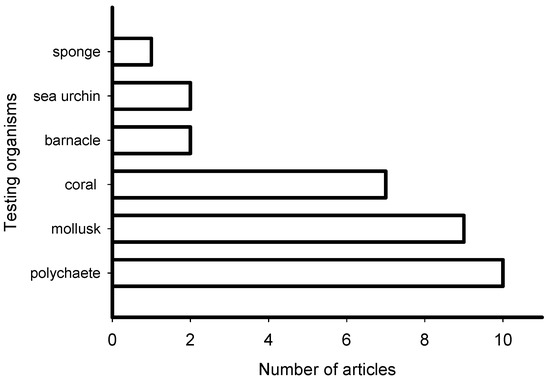
Figure 4.
Number of articles that examined the induction of larval settlement by marine biofilms from January 2010 to September 2019 according to the group of larvae. This figure only includes the publications that met all inclusion criteria.
Most publications focused on species with high economic significance, either because they can cause a significant biofouling problem (e.g., barnacles and polychaetes) or because they are commercially exploited (e.g., mussels and corals). By in large, our conclusion on the topic of publications is due to the decrease in availability of funding and need to target focused bacteria-larvae interactions of specialist settlers, like Hydroides elegans in order to be competitive for that limited funding. Support for this view is limited to observations by Holm [56] on productivity of publications on the best-studied model organisms for fouling management, barnacles, as compared to other model organisms with global distribution such as calcareous tube worms and bushy bryozoans. Holm showed reports of studies using barnacles are outstripping other biofouling models due to a focus on barnacles as globally important biofoulers. Because biofouling is a major driver of a major source of funding, when several funding agencies switched from biological solutions for biofouling management materials science solutions both micro- and macro-fouling research were less supported. These changes contributed to the decrease of reports of bacterial induction of settlement of generalist species, especially of less mature models such as bryozoans and ascidians that have highly variable responses to microbial films, are under-represented in the last decade. In this field, part of the changes represents a shift in focus from interaction with bacteria to studies of biological glues, novel materials, and less environmentally damaging forms of fouling management.
3. Induction of Larval Settlement by Bacteria and Biofilms
3.1. Multispecies Biofilms and Macrofouler Settlement
From 2010 to 2019, the majority of researchers studied the effect of multispecies biofilms on larval settlement of polychaetes, mollusks, and corals (Figure 4; Table S1). Researchers observed highly diverse bacterial biofilms developed on different substrata in intertidal and subtidal zones [57]. The composition of these biofilms varied with season and tidal level [57], as well as with biofilm age [58]. Additionally, biofilm age and the type of substratum alter metabolites produced by microorganisms (biofilm’s chemical profile) [59].
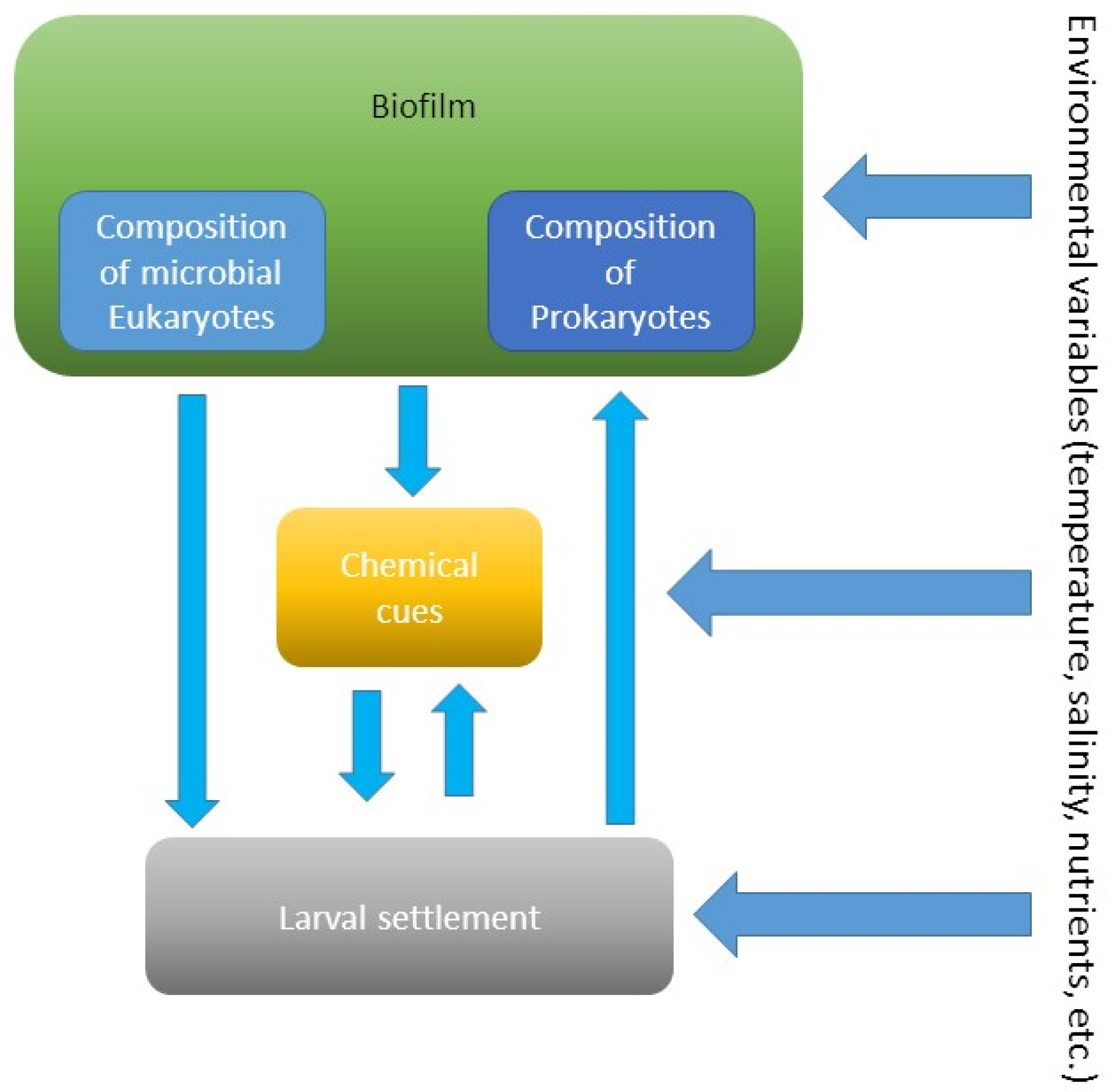
The studies demonstrated that age-dependent composition of biofilms determines settlement of invertebrate larvae (Figure 5). For example, older biofilms induced higher larval settlement of Mytilus coruscus compared to young biofilms [58]. The authors concluded that the change in bacterial community composition could explain the change in biofilm’s attractiveness. Similar findings were observed for larval settlement of the bivalve Mytilus edulis [60]. Moreover, the inductiveness of biofilms for the filter-feeding bivalve was higher in the absence of food (phytoplankton). This suggests that the influence of composition of biofilms could be more critical larval settlement when nutrient resources are absent or limited (Figure 5).
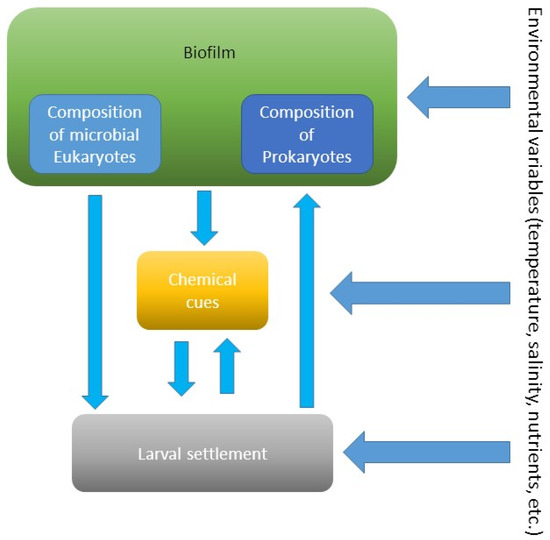
Figure 5.
Interactions between biofilms, their chemical compounds, and larvae of invertebrates at variable environmental conditions.
Larvae of the sponge Rhopaloeides odorabile settle more on biofilms developed over longer periods and at higher temperatures [61]. This suggests that environmental variables, such as temperature, affect the composition and probably metabolites of biofilms, which, in turn, affect settlement of larvae (Figure 5). During settlement, larvae of the sea urchin Heliocidaris erythrogramma respond to the specific biofilm composition on the surface of macroalgae [62]. In contrast, another sea urchin Holopneustes purpurascens does not respond to epibiotic biofilms [62].
Settlement of the barnacle Amphibalanus (=Balanus) amphitrite on biofilms developed in the intertidal zone was higher than on biofilms developed in the subtidal zone, which correlated with the presence of Proteobacteria and Cyanobacteria species [57]. While composition of biofilms developed at inshore and offshore sites was different, larvae of the specialist species H. elegans indiscriminately settled on all biofilms [63]. Settlement of H. elegans was strongly correlated with bacterial abundances in biofilms, which confirmed previous findings [64,65]. Since the density of bacteria and their exopolymers usually increases over time, it is not surprising that extracts of older biofilms induced higher settlement of H. elegans than the younger ones [59].
The presence of pollutants, like heavy metals, can change microbial composition of biofilms and, in turn, can affect larval settlement (Figure 5). The settlement of the polychaete H. elegans on biofilms developed in the presence of copper was lower compared to pristine biofilms [66]. Moreover, the inductiveness of young biofilms was more easily altered by the presence of copper than that of older biofilms. Bao et al. [66] suggest that these changes were due to changes in biofilm composition and production of microbial chemical compounds.
3.2. Monospecies Bacterial Films and Macrofouler Settlement
Most natural biofilms are complex communities which include Prokaryotes and unicellular Eukaryotes. Because this complexity cannot be duplicated in controlled conditions most researchers work with cultivable bacteria in monospecies biofilms in the laboratory (Table S1). While such biofilms never exist in the marine environment, they are useful tools for investigation of mechanisms of larval settlement. Cell cultures of α-Proteobacterial strain, Roseivivax sp. 46E8, significantly increased larval settlement in Porites astreoides only during active growth phase [67]. Treatment of monospecies biofilms of Shewanella sp. 1 with antibiotics, formalin, and ultra-violate radiation significantly reduce settlement of Mytilus coruscus [68]. This indicates that live bacteria produce inductive cues. While the red colored bacterium Pseudoalteromonas sp. SF57 inhibits larval settlement of the polychaete H. elegans, the white mutant WM1 of this bacterium is inductive [69]. Analysis of the disrupted genes suggests that the type II secretion pathway, the LysR transcriptional regulator, NAD(P)-binding proteins, exonuclease, pyruvate metabolism, flagella assembly, and cell membrane processes play a role in the regulation of pigmentation and inductiveness of Pseudoalteromonas sp. [69].
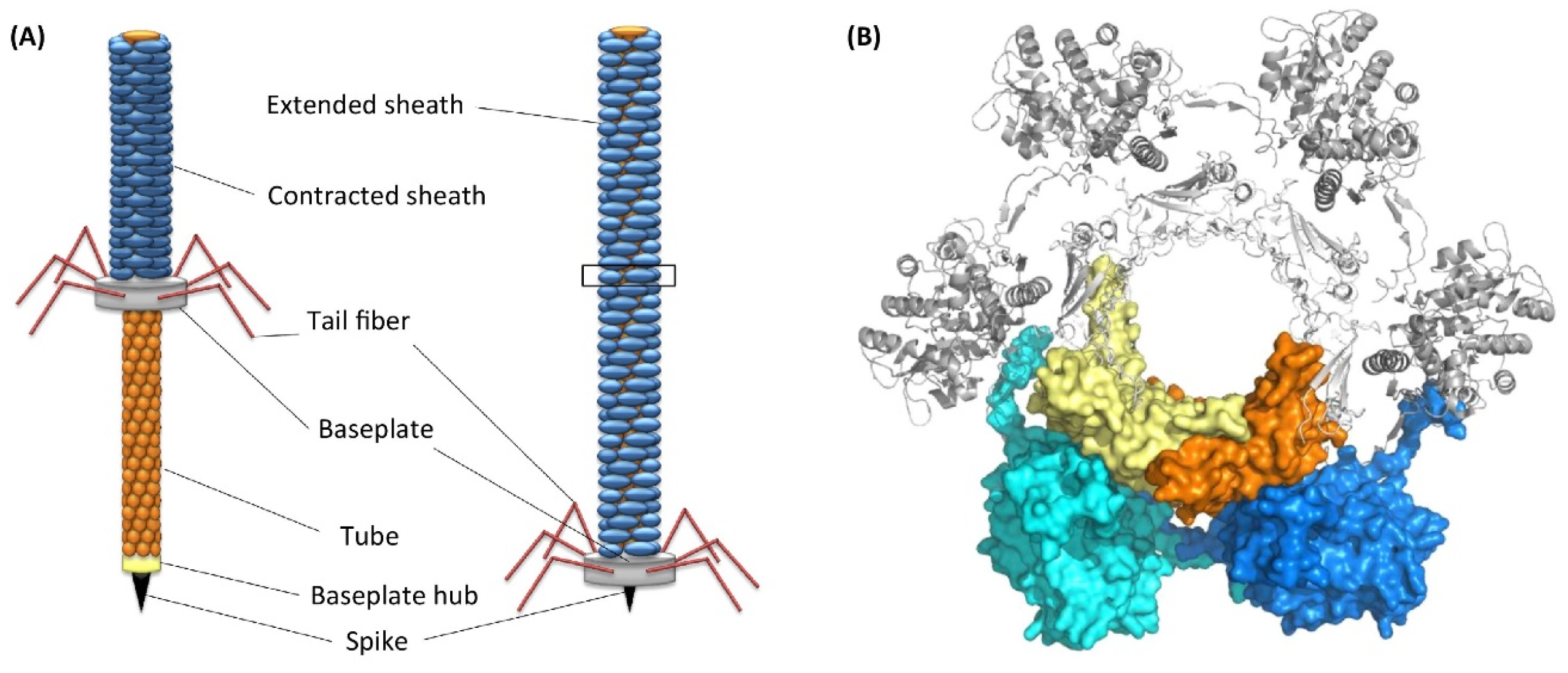
The marine bacterium Pseudoalteromonas luteoviolacea produces phage tail-like structures (tailocins) that comprise of contractile structures with outward-facing baseplates linked by tail fibers and a hexagonal sheath [70] (Figure 6). These structures have bacteriocin activities. It has been demonstrated using bacterial mutants that tailocins or metamorphosis-associated contractile structures (MACs) of P. luteoviolacea trigger metamorphosis of H. elegans [71,72]. Sequencing of the genome and transcripts of H. elegans before and after larval settlement revealed that MACs induce the regulation of groups of genes important for tissue remodeling, innate immunity, and mitogen-activated protein kinase signaling [71].
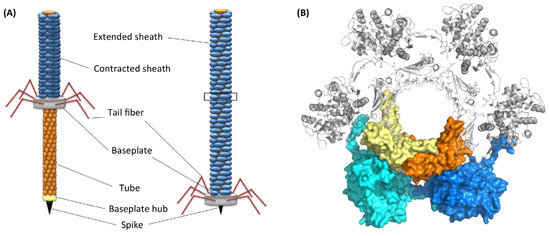
Figure 6.
(A) Structure of tailocins of bacteria. (B) Top view of a transverse section of tail tube. Reproduced with permission from Chequire and De Mot [70].
The following sequence of bacteria-induced metamorphic events was proposed. First, MACs induce larval settlement. Second, MACs encoded by a specific locus in P. luteoviolacea initiate larval cilia loss and activate metamorphosis-associated transcription. Finally, signaling through mitogen-activated protein kinase pathways alters gene expression and leads to initiation of metamorphosis [71]. A subsequent study investigated the presence of tailocins in other inductive species of bacteria, such as Cellulophaga lytica, Bacillus aquimaris, and Staphylococcus warneri [73]. Using electron microscopy and genomic analysis the authors found only P. luteoviolacea has tailocins. This suggests that tailocins are not universal inducers across inductive bacterial species.
Another study investigated genes of the marine bacterium P. luteoviolacea important for larval settlement of H. elegans [74]. Genes ORF1, ORF2, and ORF4 absent in non-inductive strain of P. luteoviolacea were necessary for induction of larval settlement. This is one of the first studies that identified bacterial genes required for induction of settlement and metamorphosis of a marine invertebrate. Future studies are needed in order to determine if these genes are present and active in other inductive bacteria and to identify gene functions.
4. Microbial Eukaryotes and Larval Settlement
Information on effects of microbial eukaryotes on larval settlement is limited (Table S1). Diatoms (Bacilariophytae) induce larval settlement of the polychaete H. elegans in laboratory experiments [75]. Lam et.al. [76] reported that carbohydrates not proteins from EPS of the diatoms Achnanthes sp. and Nitzschia constricta induced settlement of H. elegans. Metamorphosis of the abalone Haliotis asinina was significantly higher on biofilms of Navicula compare to unfilmed surfaces [77]. Similarly, settlement of the mussel Argopecten purpuratus was higher on natural diatom biofilms [78]. Cyprids of Amphibalanus (=Balanus) amphitrite predominantly settled on biofilms of Cocconeis sp. and Navicula ramosissima [79]. Higher densities of diatoms triggered higher settlement rate of the barnacle. Larval attachment of the bryozoan Bugula neritina correlated with the density of diatoms in biofilms [80]. Dahms et al. [81] demonstrated that different diatom species affect B. neritina settlement in different ways. The highest percentage of settlement of larvae of B. nertina was mediated by biofilms of Achnanthes sp., Amphora cofeaeformis, Amphora tenerrima, and Nitzschia constricta, while the lowest settlement was observed on Nitzschia frustulum biofilms. When low inductive and highly inductive diatom strains were combined together in a multispecies biofilm, their resulting activity was always highly inductive for B. neritina [81]. Thus, responses to multispecies and monospecies biofilms are different. There have been no publications reporting induction of larval settlement by diatoms from 2010 to 2019 (Table S1). While flagellates are the third largest group in marine biofilms [82], there is no evidence that this group can induce or inhibit larval settlement of invertebrate larvae.
Ciliates and other Protista are a ubiquitous component of biofilms [82]. However, only one study reported impacts of ciliates on settlement of larvae of invertebrates (Table S1). Watson et al. [83] investigated the effect of four different species of planktonic and vagile ciliates on the settlement of the polychaete Galeolaria caespotisa. Compared to control, settlement was significantly reduced in the presence of three species of ciliates: Amphisiella sp., Euplotes minuta, and Uronema marinum. Larval settlement was inhibited in the physical presence of ciliates but not with just their metabolites. Additionally, ciliates changed the distribution of bacteria in biofilms indicating that ciliates can control settlement indirectly [83].
5. Bioactive Compounds from Biofilms Inducing Settlement and Metamorphosis
5.1. Bacterial Chemical Signals
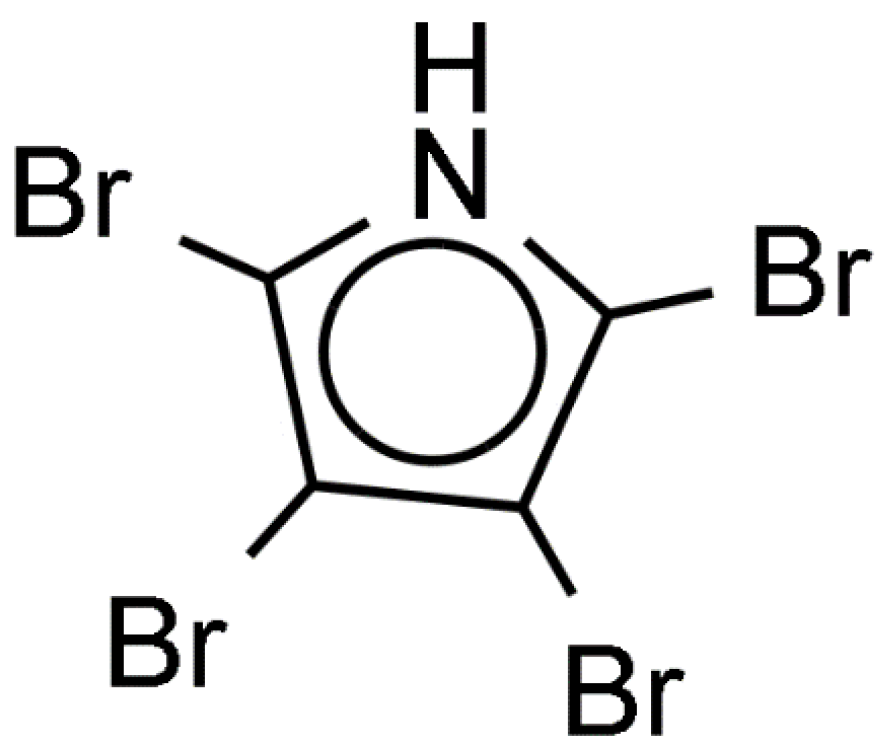
Review of publications dealing with induction of larval settlement and metamorphosis by biofilms showed a limited number of studies that identified chemical inducers produced by microorganisms (Table S1). The larvae of the acropoid coral Acropora millepora metamorphose in the presence of coralline algae Neogoniolithon fosliei and Hydrolithon onkodes [84]. Pseudoalteromonas bacteria isolated from coralline algae induce metamorphosis of larvae of the acropoid coral Acropora millepora. The bacterial metamorphosis cue was identified as tetrabromopyrrole (TBP, Figure 7). While other Pseudomonas species produce brominated compounds, only TBP induced almost 100% of larval metamorphosis without attachment [84]. Another study conducted with Pseudoalteromonas sp. PS5 associated with coralline algae showed that this strain also produces TBP [85]. TBP induced settlement of several species of corals, such as Porites astreoides, Orbicella (Montastraea) franksi, and Acropora palmata. The effect of TBP binding protein on gene expression of larvae A. millepora was studied using reverse-transcriptase quantitative PCR (RT-qPCR) [86]. TBP-containing bacterial extract changed expression of 24 out of 42 genes of interest. Expression of genes was concentration dependent. At the threshold TBP concentration only 14 of these genes were significantly regulated [86]. Amgalaxin-like-1 genes were upregulated and the SCRiP genes were down regulated during larval metamorphosis. Additionally, lectin encoding genes were either not affected or downregulated, suggesting that lectins are not involved in A. millepora larval metamorphosis but probably used in larval substrate exploration. This in accordance with previous findings that suggested that lectins, like concanavalin A of larvae Neodexiospira braziliensis are involved in larval recognition of biofilmed substrata through specific ‘lock-and-key’ binding with polysaccharides of the bacterial film [87].
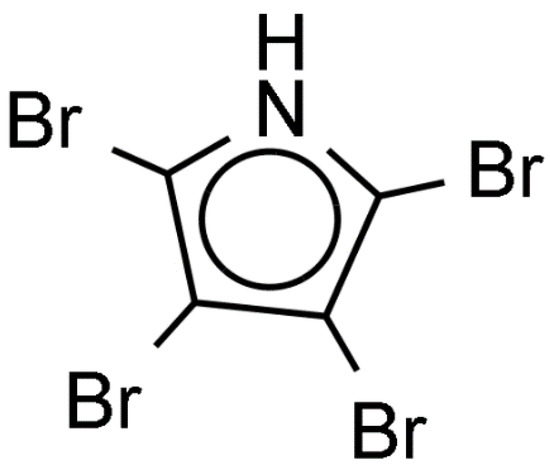
Figure 7.
Tetrabromopyrrole produced by Pseudomonas bacteria inducing metamorphosis of coral larvae.
5.2. Quorum Sensing and Settlement
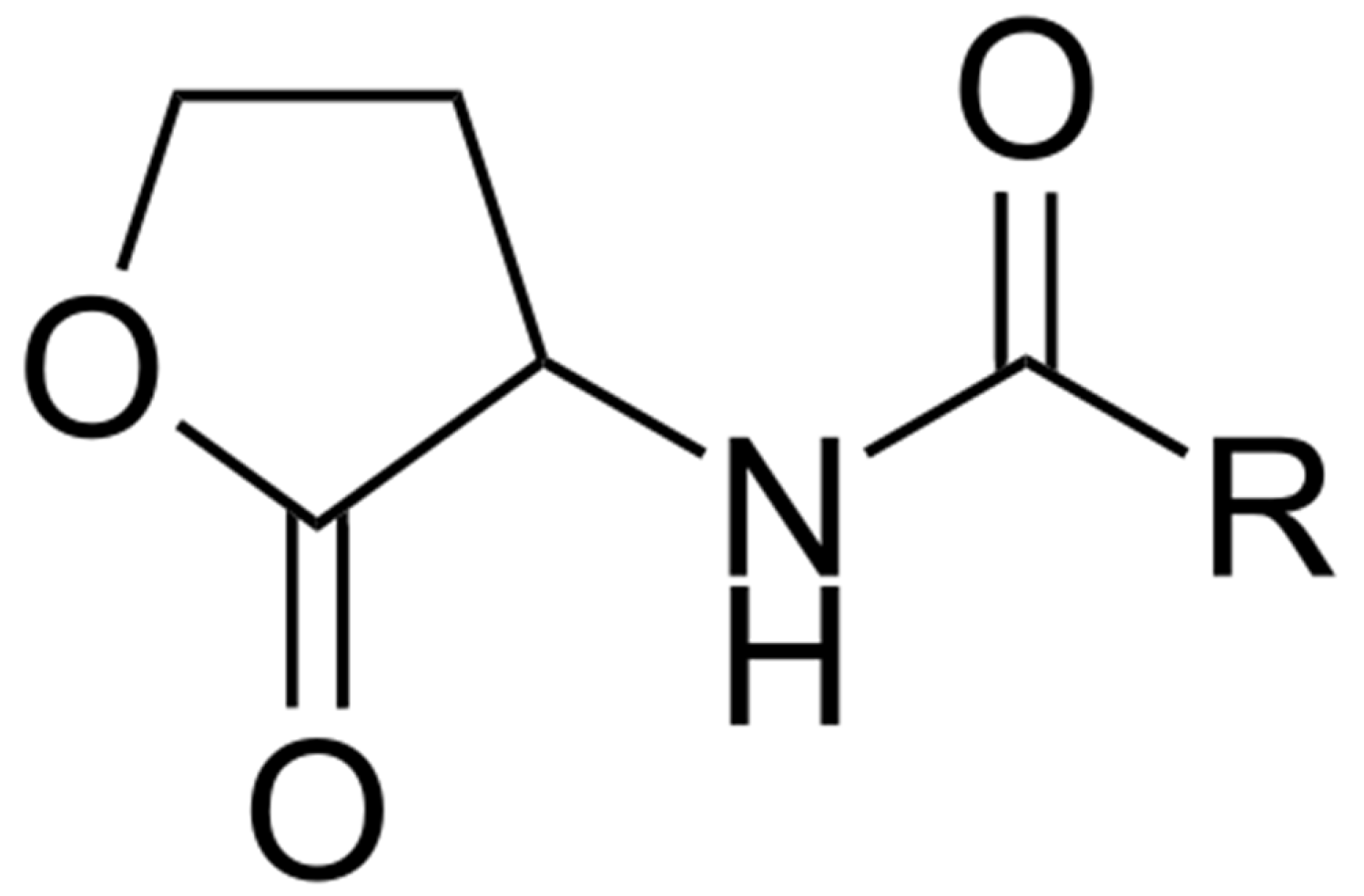
Although the first QS compound named acyl homoserine lactone (AHL, Figure 8) was isolated from the marine bacterium Vibrio fisheri associated with the light organ of the squid Euprymna scolopes [88], information concerning the presence of QS molecules in the marine environment is scarce [15]. AHL QS signals were found predominantly in Vibrio species and in the Roseobacter clade. The production of AHLs by bacteria colonizing different marine habitats—such as marine snow [89], marine sponges [90,91], corals [92], and dinoflagellates [93]—have been reported. N-dodecanoyl-L-homoserine lactone (C12-HSL) was found in marine tropical subtidal biofilms [94]. These findings suggest that QS signals are present in biofilms and produced by bacteria in vivo.
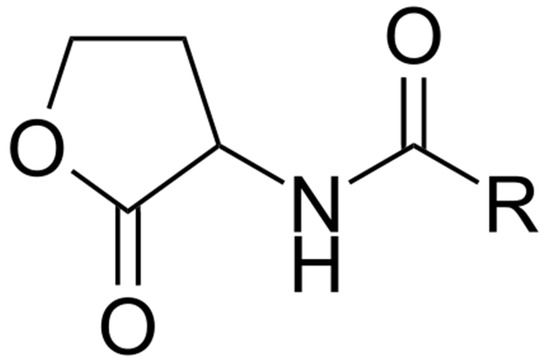
Figure 8.
Generalized structure of acyl homoserine lactones (AHLs). R—fatty acid acyl chain with number of carbons from 4 to 18.
QS autoinducer signals produced by bacteria can work as inter-kingdom signals (Table 1). For example, zoospores of the green alga Ulva (=Enteroporpha) sp. use AHLs produced by Vibrio anguillarum to attach [95,96]. Similarly, the epiphytic bacteria Sulfitobacter spp. and Shewanella spp. associated with the alga Ulva linza produce AHLs and these QS signals increased zoospore germination and growth [97]. AHLs produced by epibiotic bacteria associated with Gracilaria dura and Acrochaetium sp. induce algal spore liberation [98] (Table 1). Interestingly, unsaturated long chain hydrocarbons and dibutylphthalates also induce settlement in U. linza [99].

Table 1.
Quorum sensing (QS) compounds production and their effect on eukaryotes
Not only macro-algae respond to bacterial QS signals. AHL producing bacterial strains Vibrio anguillarum, Aeromonas hydrophila, and Sulfitobacter sp. induce larval settlement of barnacle larvae Balanus improvises [100] (Table 1). Mutants of bacterial strains that do not produce AHLs do not induce barnacle larval settlement. Additionally, synthetic AHLs at concentrations similar to those found within natural biofilms resulted in increased barnacle larval settlement. Similarly, larvae of the polychaete Hydroides elegans stop swimming and start crawling on the bottom when exposed to synthetic AHLs or biofilms producing AHLs [94]. These publications suggested that QS signals produced by bacteria could control larval and spore settlement as well as spore germination. There is no clear evidence that eukaryotes evolved with bacteria to sense QS signals and there is no known direct benefit from such communication to the bacteria.
6. Molecular Aspects of Induction of Settlement
Expanding perspective from consortia communication and tapping into that communication as described above, one can move to the ecosystem level. Ecosystems can be considered as complex symbiotic assemblages. Within estuarine ecosystems, three examples of a scale that provides a tractable experimental unit are clam beds, mussel patches, and oyster reefs. Clam beds, mussels, and oyster reefs are examples of what was classically described in the 1950s and 1960s as gregarious settlement [101,102,103,104,105]. Gregarious settlement is a complex process that involves flows, textures, and species-specific settlement cues. However, unlike QS cues, these settlement cues are much more primitive and less well understood. At least some cues are based upon degradation of structural protein substrates. The best understood of which are proteins degraded by exoenzymes [106].
Oyster reefs are interesting because the biological habitat generated extends off the substrate, undergoes succession and many ecological details are well studied. Oyster reef communities are organized and informed by the actions of symbiotic bacteria that are known mainly through the products of their exo-enzymatic activity on host structural proteins. All this biochemistry provides communication between the nodes of this complex community [8,101,105,107,108,109].
Rather than being based on production and secretion of signal molecules, larvae of gregarious settlers—like oysters—recognize species-specific degradative products that result from actions of exoenzymes on structural proteins and biological glues. The best studied of these settlement pheromones are those associated with gregarious settlement of barnacles and oysters. Though in separate phyla, settlement pheromones can be mimicked by the same pure synthetic and natural neutral amino acids, basic carboxyl terminal amino acid, tripeptides glycil-glycil-arginine that can be generated by the action of trypsin-like serine proteases on proteins found in periostraca, in the organic matrix of shells and by degradation of natural glues [8].
The basic structure of an oyster reef is organized by bacterial exoenzymes that produce oyster and barnacle settlement pheromones from barnacle and oyster structural proteins. That not all exoenzymes originate from microbes reflects the complexity and interdependence of complex symbiotic assemblages [8]. The settlement pheromones, in some cases in concert with specific bacteria [114,115], result in gregarious settlement of planktonic larvae. Growth of gregariously settled individuals results in the structure of the reef [108]. Gregarious settlement is important and often essential for reproduction [116].
Bacteria-barnacle interactions can at one level be negative [117,118,119]. Settling barnacles kill the bacteria on the surface they are gluing to by releasing reactive oxygen species, antibacterial peptides and activating enzyme cascades associated with the innate immune response [117,120,121]. Similarly, large mobile predators, like blue crabs, use enzymes to remove biofouling from places they cannot scratch, their gills, and their brooded eggs [117]. However, at another level, these processes produce species specific settlement cues and pheromones. Bacteria thrive on cured glues [122] by digesting them with proteolytic enzymes including serine proteinases. Enzyme generated peptides directly impact oysters and barnacles as well as other members of the community.
In the crabs, both fouling and cleanliness come a cost because they both generate body odors. Prey detect the odors of stone crabs and blue crabs, and avoid contact by fleeing or ‘hiding’ by stopping ventilation (clamming up) while the crab is in the immediate vicinity. In the reefs, body odors enable individual recognition. Snapping shrimp that live in pairs recognize their mates [123] and shrimp that have lost an aggressive encounter with another shrimp recognize the winner by its odor [124].
It is fascinating that barnacle settlement pheromones also attract predatory snails that eat barnacles and oysters and other sessile organisms [125,126]. Predatory snails have tapped into an essential sexual communication system for oysters and barnacles [125]. It is also likely that a parasitic symbiotic crab that lives in oysters uses these cues as do the intermediate hosts of trematode parasites that cycle through oysters fish and birds.
Over the last 15 years, understanding of the roles of exoenzyme actions is expanding [106,117,118,119,120,121,122,125,126,127,128]. However, this is just the tip of the iceberg. New shotgun proteomics technologies and microbiome technologies provide huge potential for study of this most basic of positive interactions between microbes and gregarious macro-organisms.
7. Impact of Climate Change on Biofilms and Larval Settlement
Release of anthropogenic CO2 in the atmosphere increases temperatures of our planet [129]. This increases sea surface temperatures of the oceans and leads to ocean acidification [130]. Such changes will have a direct and indirect impact on biofilms, chemical cues, and larval settlement [131]. In several studies, the impact of increased sea temperature and acidification on different species of biofouling organisms has been studied. It is predicted that acidification will impact aragonite and magnesium calcite producers—such as coralline algae, corals, mussels, barnacles, and some bryozoans [132,133,134,135]—and will increase abundances of soft-bodied organisms [136]. At the same time, information about the impact of climate change on biofilms and larvae of invertebrates is limited [131].
Climate change can alter the structure of microbial communities, which, in turn, can have impact on the structure of macro-fouling communities [60]. Microbial communities developed at 23 °C and 30 °C had different structure from those developed at 16 °C [137]. Settlement of larvae of barnacles of Amphibalanus (=Balanus) amphitrite and Balanus trigonus on biofilms developed at high temperature was different from settlement on biofilms developed at low temperatures. Similarly, only biofilms developed at elevated temperatures stimulated sponge larval settlement [60].
Acidification also affects microbial communities. For example, seawater acidification (a decrease of 0.2 pH units) over 100 days decreased the density of diatoms, which in turn, increased densities of sponges, tunicates, and decreased the density of spirorbids [136]. These examples show that factors associated with climate change directly affect the composition and densities of microorganisms in biofilms and indirectly reduce or enhance larval settlement of macro-fouling species.
8. Future Studies
In general, there was a substantial amount of work on the roles of microbes and larval settlement from the 1980s until the first decade of the 2000s and this work is comprehensively reviewed by Hadfield and Paul [51]. Most, if not all, of this work focused on microbial films on inert surfaces and features of the bacteria, like age of film, and components of the film, like EPS. The majority of studies are about induction of larval settlement by bacteria. The new omic techniques—such as metagenomics, metabolomics, proteomics, and transcriptomics—allow to characterize microbes in biofilms, identifying metabolites produced and genes responsible for their production [11,12,13,14,86]. In the future, the development of such techniques and the lower cost of such methods will result in an increase of molecular studies of microbe–larva interactions. This will enhance our understanding of receptors, genes, and pathways responsible for larval settlement and metamorphosis.
The future of ‘omics’ technologies is extension of these technologies to interactions between larvae and bacteria living on complex biological substrates like macroalgae, shells, biological glues, and wood. This aspect is especially important for understanding gregarious settlement, which often depends on interactions of exoenzymes from bacteria and symbionts on structural components during feeding and cleaning. Although there is ecological evidence that there is induction of especially gregarious larval settlement by less investigated groups of microorganisms—like diatoms, ciliates, and flagellates—the technology is now available to study these phenomena.
Specific bacterial–larval interactions are productive and worthy of continued investigation. Another area understudied by modern techniques is settlement due to constitutive and induced exoenzymes and their products should be a fruitful area of research especially with microbiome technology, proteomic and metabolomic and shotgun sequencing techniques [52,138]. The majority of bacteria secrete enzymes when they feed on biological substrates. We postulate products that escape consumption may be central to the organization of marine communities [8]. Degradation products of structural proteins generated by serine proteases and hydrolysis of complex amino sugars are the only examples to date [106] and more future research is required.
9. Conclusions
Analysis of published literature suggested that induction of larval settlement by marine microbes is by three major routes: (1) release of inductive molecules including those involved in quorum sensing from established biofilms and consortia; (2) release of exoenzymes during microbial feeding generating degradation products that act as settlement cues and pheromones; (3) inductive molecules and even physical viral-like structures, like tailocins found within bacteria. In the last decade, a relatively small number of very high impact reports significantly advanced the field of microbe macro-fouler interactions to include genes involved in settlement and metamorphosis, specific groups of inductive microbes, and physically unique induction mechanisms. Most recent advances employed sophisticated and new molecular biology techniques. These reports are exciting and at the same time humbling in that they support our contention that understanding of the roles of bacterial induction of settlement are in their research infancy. The future in this area is in communities, large symbiotic assemblages, and the impact of environmental change due to human impacts on complex ecosystems.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/21/3/731/s1.
Author Contributions
Both authors are equally contributed to design, writing, review, and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by TRC grant RC/AGR/FISH/16/01 (ORG/EBR/15/003) and SQU grants CL/SQU-SA/18/01 and IG/AGR/FISH/18/01 to S.D.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Wahl, M. Marine epibiosis. I. Fouling and antifouling: Some basic aspects. Mar. Ecol. Prog. Ser. 1989, 58, 175–189. [Google Scholar] [CrossRef]
- Clare, A.S.; Rittschof, D.; Gerhart, D.J.; Maki, J.S. Molecular approaches to nontoxic antifouling. Invertebr. Reprod. Dev. 1992, 22, 67–76. [Google Scholar] [CrossRef]
- Salta, M.; Wharton, J.A.; Blache, Y.; Stokes, K.R.; Briand, J.-F. Marine biofilms on artificial surfaces: Structure and dynamics. Environ. Microbiol. 2013, 15, 2879–2893. [Google Scholar] [CrossRef]
- Antunes, J.; Leão, P.; Vasconcelos, V. Marine biofilms: Diversity of communities and of chemical cues. Environ. Microbiol. Rep. 2019, 11, 287–305. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.-C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.-C.; Neu, T.R.; Wozniak, D.J. The EPS matrix: The “house of biofilm cells ”. J. Bacteriol. 2007, 189, 7945–7947. [Google Scholar] [CrossRef]
- Dobretsov, S.; Dahms, H.-U.; Qian, P.-Y. Inhibition of biofouling by marine microorganisms and their metabolites. Biofouling 2006, 22, 43–54. [Google Scholar] [CrossRef]
- Rittschof, D. Trypsins: Keystone enzymes in marine communities. JSM Enzymol. Protein Sci. 2017, 2, 1009. [Google Scholar]
- Zobell, C.E.; Allen, E.C. The significance of marine bacteria in the fouling of submerged surfaces. J. Bacteriol. 1935, 29, 239–251. [Google Scholar] [CrossRef]
- O’Toole, G.A. Classic spotlight: Before they were biofilms. J. Bacteriol. 2015, 198, 5. [Google Scholar] [CrossRef]
- Qian, P.-Y.; Lau, S.C.K.; Dahms, H.-U.; Dobretsov, S.; Harder, T. Marine biofilms as mediators of colonization by marine macroorganisms: Implications for antifouling and aquaculture. Mar. Biotechnol. 2007, 9, 399–410. [Google Scholar] [CrossRef] [PubMed]
- Franklin, M.J.; Chang, C.; Akiyama, T.; Bothner, B. New technologies for studying biofilms. Microbiol. Spectr. 2015, 3. [Google Scholar] [CrossRef] [PubMed]
- Briand, J.-F.; Pochon, X.; Wood, S.A.; Bressy, C.; Garnier, C.; Réhel, K.; Urvois, F.; Culioli, G.; Zaiko, A. Metabarcoding and metabolomics offer complementarity in deciphering marine eukaryotic biofouling community shifts. Biofouling 2018, 34, 657–672. [Google Scholar] [CrossRef] [PubMed]
- Brauer, J.I.; Makama, Z.; Bonifay, V.; Aydin, E.; Kaufman, E.D.; Beech, I.B.; Sunner, J. Mass spectrometric metabolomic imaging of biofilms on corroding steel surfaces using laser ablation and solvent capture by aspiration. Biointerphases 2015, 10, 019003. [Google Scholar] [CrossRef]
- Hmelo, L.R. Quorum sensing in marine microbial environments. Annu. Rev. Mar. Sci. 2017, 9, 257–281. [Google Scholar] [CrossRef]
- Dobretsov, S.; Teplitski, M.; Paul, V. Mini-review: Quorum sensing in the marine environment and its relationship to biofouling. Biofouling 2009, 25, 413–427. [Google Scholar] [CrossRef]
- Kjelleberg, S.; Molin, S. Is there a role for quorum sensing signals in bacterial biofilms? Curr. Opin. Microbiol. 2002, 5, 254–258. [Google Scholar] [CrossRef]
- Miller, M.B.; Bassler, B.L. Quorum sensing in bacteria. Annu. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar] [CrossRef]
- Mukherjee, S.; Bassler, B.L. Bacterial quorum sensing in complex and dynamically changing environments. Nat. Rev. Microbiol. 2019, 17, 371–382. [Google Scholar] [CrossRef]
- Dobretsov, S.; Teplitski, M.; Bayer, M.; Gunasekera, S.; Proksch, P.; Paul, V.J. Inhibition of marine biofouling by bacterial quorum sensing inhibitors. Biofouling 2011, 27, 893–905. [Google Scholar] [CrossRef]
- Grandclément, C.; Tannières, M.; Moréra, S.; Dessaux, Y.; Faure, D. Quorum quenching: Role in nature and applied developments. FEMS Microbiol. Rev. 2016, 40, 86–116. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wang, B.; Lu, Y.; Guo, Y.; Sun, J.; Wei, B.; Zhang, H.; Wang, H. Quorum sensing inhibitors from marine microorganisms and their synthetic derivatives. Mar. Drugs 2019, 17, 80. [Google Scholar] [CrossRef] [PubMed]
- Crisp, D.J. The larval stages of balanus hameri (ascanius, 1767). Crustaceana 1962, 4, 123–130. [Google Scholar] [CrossRef]
- Thorson, G. Reproductive and larval ecology of marine bottom invertebrates. Biol. Rev. 1950, 25, 1–45. [Google Scholar] [CrossRef] [PubMed]
- Hadfield, M.G. Biofilms and marine invertebrate larvae: What bacteria produce that larvae use to choose settlement sites. Annu. Rev. Mar. Sci. 2011, 3, 453–470. [Google Scholar] [CrossRef] [PubMed]
- Wieczorek, S.K.; Todd, C.D. Inhibition and facilitation of settlement of epifaunal marine invertebrate larvae by microbial biofilm cues. Biofouling 1998, 12, 81–118. [Google Scholar] [CrossRef]
- Mullineaux, L.S. The role of settlement in structuring a hard-substratum community in the deep sea. J. Exp. Mar. Biol. Ecol. 1988, 120, 247–261. [Google Scholar] [CrossRef]
- Rittschof, D.; Forward, R.B., Jr.; Cannon, G.; Welch, J.M.; McClary, M., Jr.; Holm, E.R.; Clare, A.S.; Conova, S.; McKelvey, L.M.; Bryan, P.; et al. Cues and context: Larval responses to physical and chemical cues. Biofouling 1998, 12, 31–44. [Google Scholar] [CrossRef]
- Rittschof, D.; Hooper, I.R.; Costlow, J.D. Barnacle settlement inhibitors from sea pansies, Renilla reniformis. Bull. Mar. Sci. 1986, 39, 376–382. [Google Scholar]
- Roberts, D.; Rittschof, D.; Holm, E.; Schmidt, A.R. Factors influencing initial larval settlement: Temporal, spatial and surface molecular components. J. Exp. Mar. Biol. Ecol. 1991, 150, 203–221. [Google Scholar] [CrossRef]
- Tamburri, M.N.; Luckenbach, M.W.; Breitburg, D.L.; Bonniwell, S.M. Settlement of Crassostrea ariakensis larvae: Effects of substrate, biofilms, sediment and adult chemical cues. J. Shellfish Res. 2008, 27, 601–608. [Google Scholar] [CrossRef]
- Dobretsov, S.V. Effects of macroalgae and biofilm on settlement of blue mussel (Mytilus edulis L.) larvae. Biofouling 1999, 14, 153–165. [Google Scholar] [CrossRef]
- Unabia, C.R.C.; Hadfield, M.G. Role of bacteria in larval settlement and metamorphosis of the polychaete Hydroides elegans. Mar. Biol. 1999, 133, 55–64. [Google Scholar] [CrossRef]
- Alberte, R.S.; Snyder, S.; Zahuranec, B.J.; Whetstone, M. Biofouling research needs for the United States Navy: Program history and goals. Biofouling 1992, 6, 91–95. [Google Scholar] [CrossRef]
- Maréchal, J.-P.; Hellio, C. Challenges for the development of new non-toxic antifouling solutions. Int. J. Mol. Sci. 2009, 10, 4623–4637. [Google Scholar] [CrossRef] [PubMed]
- Yebra, D.M.; Kiil, S.; Dam-Johansen, K. Antifouling technology—Past, present and future steps towards efficient and environmentally friendly antifouling coatings. Prog. Org. Coat. 2004, 50, 75–104. [Google Scholar] [CrossRef]
- Muthukrishnan, T.; Abed, R.M.M.; Dobretsov, S.; Kidd, B.; Finnie, A.A. Long-term microfouling on commercial biocidal fouling control coatings. Biofouling 2014, 30, 1155–1164. [Google Scholar] [CrossRef]
- Mieszkin, S.; Callow, M.E.; Callow, J.A. Interactions between microbial biofilms and marine fouling algae: A mini review. Biofouling 2013, 29, 1097–1113. [Google Scholar] [CrossRef]
- Maki, J.S.; Mitchell, R. Biofouling in the marine environment. In Encyclopedia of Environmental Microbiology; American Cancer Society: New York, NY, USA, 2003; ISBN 978-0-471-26339-5. [Google Scholar]
- Bakus, G.J.; Targett, N.M.; Schulte, B. Chemical ecology of marine organisms: An overview. J. Chem. Ecol. 1986, 12, 951–987. [Google Scholar] [CrossRef]
- Standing, J.D.; Hooper, I.R.; Costlow, J.D. Inhibition and induction of barnacle settlement by natural products present in octocorals. J. Chem. Ecol. 1984, 10, 823–834. [Google Scholar] [CrossRef]
- Dobretsov, S.; Abed, R.M.M.; Teplitski, M. Mini-review: Inhibition of biofouling by marine microorganisms. Biofouling 2013, 29, 423–441. [Google Scholar] [CrossRef] [PubMed]
- Qian, P.-Y.; Li, Z.; Xu, Y.; Li, Y.; Fusetani, N. Mini-review: Marine natural products and their synthetic analogs as antifouling compounds: 2009–2014. Biofouling 2015, 31, 101–122. [Google Scholar] [CrossRef] [PubMed]
- Saha, M.; Goecke, F.; Bhadury, P. Minireview: Algal natural compounds and extracts as antifoulants. J. Appl. Phycol. 2018, 30, 1859–1874. [Google Scholar] [CrossRef] [PubMed]
- Satheesh, S.; Ba-akdah, M.A.; Al-Sofyani, A.A. Natural antifouling compound production by microbes associated with marine macroorganisms—A review. Electron. J. Biotechnol. 2016, 21, 26–35. [Google Scholar] [CrossRef]
- Wang, K.-L.; Wu, Z.-H.; Wang, Y.; Wang, C.-Y.; Xu, Y. Mini-review: Antifouling natural products from marine microorganisms and their synthetic analogs. Mar. Drugs 2017, 15, 266. [Google Scholar] [CrossRef]
- Hay, M.E. Marine chemical ecology: Chemical signals and cues structure marine populations, communities, and ecosystems. Annu. Rev. Mar. Sci. 2009, 1, 193–212. [Google Scholar] [CrossRef]
- Paul, V.J.; Ritson-Williams, R.; Sharp, K. Marine chemical ecology in benthic environments. Nat. Prod. Rep. 2011, 28, 345–387. [Google Scholar] [CrossRef]
- Paul, V.J.; Puglisi, M.P.; Ritson-Williams, R. Marine chemical ecology. Nat. Prod. Rep. 2006, 23, 153–180. [Google Scholar] [CrossRef]
- Paul, V.J.; Ritson-Williams, R. Marine chemical ecology. Nat. Prod. Rep. 2008, 25, 662–695. [Google Scholar] [CrossRef]
- Hadfield, M.; Paul, V. Natural chemical cues for settlement and metamorphosis of marine-invertebrate larvae. In Marine Chemical Ecology; McClintock, J., Baker, B., Eds.; CRC Press: Boca Raton, FL, USA, 2001; Volume 20015660, pp. 431–461. ISBN 978-0-8493-9064-7. [Google Scholar]
- Chandramouli, K.H.; Qian, P.-Y.; Ravasi, T. Proteomics insights: Proteins related to larval attachment and metamorphosis of marine invertebrates. Front. Mar. Sci. 2014, 1. [Google Scholar] [CrossRef]
- Chen, L.; Qian, P.-Y. Review on molecular mechanisms of antifouling compounds: An update since 2012. Mar. Drugs 2017, 15, 264. [Google Scholar] [CrossRef] [PubMed]
- Qian, P.-Y.; Chen, L.; Xu, Y. Mini-review: Molecular mechanisms of antifouling compounds. Biofouling 2013, 29, 381–400. [Google Scholar] [CrossRef] [PubMed]
- Thiyagarajan, V. A review on the role of chemical cues in habitat selection by barnacles: New insights from larval proteomics. J. Exp. Mar. Biol. Ecol. 2010, 392, 22–36. [Google Scholar] [CrossRef]
- Holm, E. Barnacles and biofouling. Integr. Comp. Biol. 2012, 52, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Lee, O.O.; Chung, H.C.; Yang, J.; Wang, Y.; Dash, S.; Wang, H.; Qian, P.-Y. Molecular techniques revealed highly diverse microbial communities in natural marine biofilms on polystyrene dishes for invertebrate larval settlement. Microb. Ecol. 2014, 68, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Bao, W.-Y.; Gu, Z.-Q.; Li, Y.-F.; Liang, X.; Ling, Y.; Cai, S.-L.; Shen, H.-D.; Yang, J.-L. Larval settlement and metamorphosis of the mussel Mytilus coruscus in response to natural biofilms. Biofouling 2012, 28, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.C.; Lee, O.O.; Huang, Y.-L.; Mok, S.Y.; Kolter, R.; Qian, P.-Y. Bacterial community succession and chemical profiles of subtidal biofilms in relation to larval settlement of the polychaete Hydroides elegans. ISME J. 2010, 4, 817–828. [Google Scholar] [CrossRef]
- Toupoint, N.; Mohit, V.; Linossier, I.; Bourgougnon, N.; Myrand, B.; Olivier, F.; Lovejoy, C.; Tremblay, R. Effect of biofilm age on settlement of Mytilus edulis. Biofouling 2012, 28, 985–1001. [Google Scholar] [CrossRef]
- Whalan, S.; Webster, N.S. Sponge larval settlement cues: The role of microbial biofilms in a warming ocean. Sci. Rep. 2014, 4, 4072. [Google Scholar] [CrossRef]
- Nielsen, S.J.; Harder, T.; Steinberg, P.D. Sea urchin larvae decipher the epiphytic bacterial community composition when selecting sites for attachment and metamorphosis. FEMS Microbiol. Ecol. 2015, 91, 1–9. [Google Scholar] [CrossRef][Green Version]
- Lema, K.A.; Constancias, F.; Rice, S.A.; Hadfield, M.G. High bacterial diversity in nearshore and oceanic biofilms and their influence on larval settlement by Hydroides elegans (Polychaeta). Environ. Microbiol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Hadfield, M.G. Composition and density of bacterial biofilms determine larval settlement of the polychaete Hydroides elegans. Mar. Ecol. Prog. Ser. 2003, 260, 161–172. [Google Scholar] [CrossRef]
- Lau, S.C.K.; Mak, K.K.W.; Chen, F.; Qian, P.-Y. Bioactivity of bacterial strains isolated from marine biofilms in Hong Kong waters for the induction of larval settlement in the marine polychaete Hydroides elegans. Mar. Ecol. Prog. Ser. 2002, 226, 301–310. [Google Scholar] [CrossRef][Green Version]
- Bao, W.-Y.; Lee, O.-O.; Chung, H.-C.; Li, M.; Qian, P.-Y. Copper affects biofilm inductiveness to larval settlement of the serpulid polychaete Hydroides elegans (Haswell). Biofouling 2010, 26, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Sharp, K.H.; Sneed, J.M.; Ritchie, K.B.; Mcdaniel, L.; Paul, V.J. Induction of larval settlement in the reef coral Porites astreoides by a cultivated marine roseobacter strain. Biol. Bull. 2015, 228, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.-L.; Shen, P.-J.; Liang, X.; Li, Y.-F.; Bao, W.-Y.; Li, J.-L. Larval settlement and metamorphosis of the mussel Mytilus coruscus in response to monospecific bacterial biofilms. Biofouling 2013, 29, 247–259. [Google Scholar] [CrossRef]
- Huang, Y.-L.; Li, M.; Yu, Z.; Qian, P.-Y. Correlation between pigmentation and larval settlement deterrence by Pseudoalteromonas sp. sf57. Biofouling 2011, 27, 287–293. [Google Scholar] [CrossRef]
- Ghequire, M.G.K.; De Mot, R. The tailocin tale: Peeling off phage tails. Trends Microbiol. 2015, 23, 587–590. [Google Scholar] [CrossRef]
- Shikuma, N.J.; Antoshechkin, I.; Medeiros, J.M.; Pilhofer, M.; Newman, D.K. Stepwise metamorphosis of the tubeworm Hydroides elegans is mediated by a bacterial inducer and MAPK signaling. Proc. Natl. Acad. Sci. USA 2016, 113, 10097–10102. [Google Scholar] [CrossRef]
- Shikuma, N.J.; Pilhofer, M.; Weiss, G.L.; Hadfield, M.G.; Jensen, G.J.; Newman, D.K. Marine tubeworm metamorphosis induced by arrays of bacterial phage tail-like structures. Science 2014, 343, 529–533. [Google Scholar] [CrossRef]
- Freckelton, M.L.; Nedved, B.T.; Hadfield, M.G. Induction of invertebrate larval settlement; Different bacteria, different mechanisms? Sci. Rep. 2017, 7, 42557. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Callahan, S.; Hadfield, M.G. Recruitment in the sea: Bacterial genes required for inducing larval settlement in a polychaete worm. Sci. Rep. 2012, 2, 228. [Google Scholar] [CrossRef] [PubMed]
- Harder, T.; Lam, C.; Qian, P.-Y. Induction of larval settlement in the polychaete Hydroides elegans by marine biofilms: An investigation of monospecific diatom films as settlement cues. Mar. Ecol. Prog. Ser. 2002, 229, 105–112. [Google Scholar] [CrossRef]
- Lam, C.; Harder, T.; Qian, P.-Y. Induction of larval settlement in the polychaete Hydroides elegans by extracellular polymers of benthic diatoms. Mar. Ecol. Prog. Ser. 2005, 286, 145–154. [Google Scholar] [CrossRef]
- Gallardo, W.G.; Buen, S.M.A. Evaluation of mucus, Navicula, and mixed diatoms as larval settlement inducers for the tropical abalone Haliotis asinina. Aquaculture 2003, 221, 357–364. [Google Scholar] [CrossRef]
- Avendaño-Herrera, R.; Riquelmes, C.; Silva, F.; Avendañod, M.; Irgang, R. Optimization of settlement of larval Argopecten purpuratus using natural diatom biofilms. J. Shellfish Res. 2003, 22, 393–399. [Google Scholar]
- Jouuchi, T.; Satuito, C.G.; Kitamura, H. Sugar compound products of the periphytic diatom Navicula ramosissima induce larval settlement in the barnacle, Amphibalanus amphitrite. Mar. Biol. 2007, 152, 1065–1076. [Google Scholar] [CrossRef]
- Kitamura, H.; Hirayama, K. Effect of primary films on the settlement of larvae of a bryozoan Bugula neritina. Nippon Suisan Gakkaishi 1987, 53, 1377–1381. [Google Scholar] [CrossRef]
- Dahms, H.-U.; Dobretsov, S.; Qian, P.-Y. The effect of bacterial and diatom biofilms on the settlement of the bryozoan Bugula neritina. J. Exp. Mar. Biol. Ecol. 2004, 313, 191–209. [Google Scholar] [CrossRef]
- Railkin, A.I. Marine Biofouling: Colonization Processes and Defenses; CRC Press: Boca Raton, FL, USA, 2003; ISBN 978-0-203-50323-2. [Google Scholar]
- Watson, M.G.; Scardino, A.J.; Zalizniak, L.; Shimeta, J. Inhibition of invertebrate larval settlement by biofilm ciliates. Mar. Ecol. Prog. Ser. 2016, 557, 77–90. [Google Scholar] [CrossRef]
- Tebben, J.; Tapiolas, D.M.; Motti, C.A.; Abrego, D.; Negri, A.P.; Blackall, L.L.; Steinberg, P.D.; Harder, T. Induction of larval metamorphosis of the coral Acropora millepora by tetrabromopyrrole isolated from a Pseudoalteromonas Bacterium. PLoS ONE 2011, 6, e19082. [Google Scholar] [CrossRef] [PubMed]
- Sneed Jennifer, M.; Sharp Koty, H.; Ritchie Kimberly, B.; Paul Valerie, J. The chemical cue tetrabromopyrrole from a biofilm bacterium induces settlement of multiple Caribbean corals. Proc. R. Soc. B Biol. Sci. 2014, 281, 20133086. [Google Scholar] [CrossRef] [PubMed]
- Siboni, N.; Abrego, D.; Seneca, F.; Motti, C.A.; Andreakis, N.; Tebben, J.; Blackall, L.L.; Harder, T. Using bacterial extract along with differential gene expression in Acropora millepora larvae to decouple the processes of attachment and metamorphosis. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Kirchman, D.; Graham, S.; Reish, D.; Mitchell, R. Bacteria induce settlement and metamorphosis of Janua (Dexiospira) brasiliensis Grube (Polychaeta:Spirprbidae). J. Exp. Mar. Biol. Ecol. 1981, 56, 153–163. [Google Scholar] [CrossRef]
- Eberhard, A.; Burlingame, A.L.; Eberhard, C.; Kenyon, G.L.; Nealson, K.H.; Oppenheimer, N.J. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry 1981, 20, 2444–2449. [Google Scholar] [CrossRef] [PubMed]
- Gram, L.; Grossart, H.-P.; Schlingloff, A.; Kiørboe, T. Possible quorum sensing in marine snow bacteria: Production of acylated homoserine lactones by Roseobacter strains isolated from marine snow. Appl. Environ. Microbiol. 2002, 68, 4111–4116. [Google Scholar] [CrossRef]
- Mohamed, N.M.; Cicirelli, E.M.; Kan, J.; Chen, F.; Fuqua, C.; Hill, R.T. Diversity and quorum-sensing signal production of proteobacteria associated with marine sponges. Environ. Microbiol. 2008, 10, 75–86. [Google Scholar] [CrossRef]
- Taylor, M.W.; Schupp, P.J.; Baillie, H.J.; Charlton, T.S.; de Nys, R.; Kjelleberg, S.; Steinberg, P.D. Evidence for Acyl homoserine lactone signal production in bacteria associated with marine sponges. Appl. Environ. Microbiol. 2004, 70, 4387–4389. [Google Scholar] [CrossRef]
- Golberg, K.; Eltzov, E.; Shnit-Orland, M.; Marks, R.S.; Kushmaro, A. Characterization of quorum sensing signals in coral-associated bacteria. Microb. Ecol. 2011, 61, 783–792. [Google Scholar] [CrossRef]
- Wagner-Döbler, I.; Thiel, V.; Eberl, L.; Allgaier, M.; Bodor, A.; Meyer, S.; Ebner, S.; Hennig, A.; Pukall, R.; Schulz, S. Discovery of complex mixtures of novel long-chain quorum sensing signals in free-living and host-associated marine alphaproteobacteria. ChemBioChem 2005, 6, 2195–2206. [Google Scholar] [CrossRef]
- Huang, Y.-L.; Dobretsov, S.; Ki, J.-S.; Yang, L.-H.; Qian, P.-Y. Presence of Acyl-homoserine lactone in subtidal biofilm and the implication in larval behavioral response in the polychaete Hydroides elegans. Microb. Ecol. 2007, 54, 384–392. [Google Scholar] [CrossRef] [PubMed]
- Joint, I.; Tait, K.; Wheeler, G. Cross-kingdom signalling: Exploitation of bacterial quorum sensing molecules by the green seaweed Ulva. Philos. Trans. R. Soc. B Biol. Sci. 2007, 362, 1223–1233. [Google Scholar] [CrossRef] [PubMed]
- Tait, K.; Joint, I.; Daykin, M.; Milton, D.L.; Williams, P.; Cámara, M. Disruption of quorum sensing in seawater abolishes attraction of zoospores of the green alga Ulva to bacterial biofilms. Environ. Microbiol. 2005, 7, 229–240. [Google Scholar] [CrossRef] [PubMed]
- Twigg, M.S.; Tait, K.; Williams, P.; Atkinson, S.; Cámara, M. Interference with the germination and growth of Ulva zoospores by quorum-sensing molecules from Ulva-associated epiphytic bacteria. Environ. Microbiol. 2014, 16, 445–453. [Google Scholar] [CrossRef] [PubMed]
- Weinberger, F.; Beltran, J.; Correa, J.A.; Lion, U.; Pohnert, G.; Kumar, N.; Steinberg, P.; Kloareg, B.; Potin, P. Spore release in Acrochaetium sp. (Rhodophyta) is bacterially controlled. J. Phycol. 2007, 43, 235–241. [Google Scholar] [CrossRef]
- Qi, Y.; Wu, Z.; He, J.; Rittschof, D.; Su, P.; Ke, C.; Feng, D. Conspecific cues that induce spore settlement in the biofouling and green tide-forming alga Ulva tepida provide a potential aggregation mechanism. Int. Biodeterior. Biodegrad. 2019, 145, 104807. [Google Scholar] [CrossRef]
- Tait, K.; Havenhand, J. Investigating a possible role for the bacterial signal molecules N-acylhomoserine lactones in Balanus improvisus cyprid settlement. Mol. Ecol. 2013, 22, 2588–2602. [Google Scholar] [CrossRef]
- Browne, K.A.; Tamburri, M.N.; Zimmer-Faust, R.K. Modelling quantitative structure-activity relationships between animal behaviour and environmental signal molecules. J. Exp. Biol. 1998, 201, 245–258. [Google Scholar]
- Crisp, D.J. Chemical factors inducing settlement in Crassostrea virginica (Gmelin). J. Anim. Ecol. 1967, 36, 329–335. [Google Scholar] [CrossRef]
- Crisp, D.J.; Meadows, P.S.; Brambell, F.W.R. Adsorbed layers: The stimulus to settlement in barnacles. Proc. R. Soc. Lond. Ser. B Biol. Sci. 1963, 158, 364–387. [Google Scholar]
- Crisp, D.J.; Meadows, P.S.; Brambell, F.W.R. The chemical basis of gregariousness in cirripedes. Proc. R. Soc. Lond. Ser. B Biol. Sci. 1962, 156, 500–520. [Google Scholar] [CrossRef]
- Tamburri, M.N.; Zimmer-Faust, R.K.; Tamplin, M.L. Natural sources and properties of chemical inducers mediating settlement of oyster larvae: A re-examination. Biol. Bull. 1992, 183, 327–338. [Google Scholar] [CrossRef] [PubMed]
- Rittschof, D.; Cohen, J.H. Crustacean peptide and peptide-like pheromones and kairomones. Peptides 2004, 25, 1503–1516. [Google Scholar] [CrossRef] [PubMed]
- Decho, A.W.; Browne, K.A.; Zimmer-Faust, R.K. Chemical cues: Why basic peptides are signal molecules in marine environments. Limnol. Oceanogr. 1998, 43, 1410–1417. [Google Scholar] [CrossRef]
- Hidu, H. Gregarious setting in the American oyster Crassostrea virginica Gmelin. Chesap. Sci. 1969, 10, 85–92. [Google Scholar] [CrossRef]
- Zimmer-Faust, R.K.; Tamburri, M.N. Chemical identity and ecological implications of a waterborne, larval settlement cue. Limnol. Oceanogr. 1994, 39, 1075–1087. [Google Scholar] [CrossRef]
- Wheeler, G.L.; Tait, K.; Taylor, A.; Brownlee, C.; Joint, I. Acyl-homoserine lactones modulate the settlement rate of zoospores of the marine alga Ulva intestinalis via a novel chemokinetic mechanism. Plant Cell Environ. 2006, 29, 608–618. [Google Scholar] [CrossRef]
- Tait, K.; Williamson, H.; Atkinson, S.; Williams, P.; Cámara, M.; Joint, I. Turnover of quorum sensing signal molecules modulates cross-kingdom signalling. Environ. Microbiol. 2009, 11, 1792–1802. [Google Scholar] [CrossRef]
- Singh, R.P.; Baghel, R.S.; Reddy, C.R.K.; Jha, B. Effect of quorum sensing signals produced by seaweed-associated bacteria on carpospore liberation from Gracilaria dura. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef]
- Zhou, J.; Lyu, Y.; Richlen, M.L.; Anderson, D.M.; Cai, Z. Quorum sensing is a language of chemical signals and plays an ecological role in algal-bacterial interactions. Crit. Rev. Plant Sci. 2016, 35, 81–105. [Google Scholar] [CrossRef]
- Bonar, D.B.; Coon, S.L.; Walch, M.; Weiner, R.M.; Fitt, W. Control of oyster settlement and metamorphosis by endogenous and exogenous chemical cues. Bull. Mar. Sci. 1990, 46, 484–498. [Google Scholar]
- Bonar, D.B.; Weiner, R.M.; Colwell, R.R. Microbial-invertebrate interactions and potential for biotechnology. Microb. Ecol. 1986, 12, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Knight-Jones, E.W.; Crisp, D.J. Gregariousness in barnacles in relation to the fouling of ships and to anti-fouling research. Nature 1953, 171, 1109–1110. [Google Scholar] [CrossRef] [PubMed]
- Essock-Burns, T.; Wepprich, A.; Thompson, A.; Rittschof, D. Enzymes manage biofilms on crab surfaces aiding in feeding and antifouling. J. Exp. Mar. Biol. Ecol. 2016, 479, 106–113. [Google Scholar] [CrossRef]
- Essock-Burns, T.; Gohad, N.V.; Orihuela, B.; Mount, A.S.; Spillmann, C.M.; Wahl, K.J.; Rittschof, D. Barnacle biology before, during and after settlement and metamorphosis: A study of the interface. J. Exp. Biol. 2017, 220, 194–207. [Google Scholar] [CrossRef]
- Fears, K.P.; Orihuela, B.; Rittschof, D.; Wahl, K.J. Acorn barnacles secrete phase-separating fluid to clear surfaces ahead of cement deposition. Adv. Sci. 2018, 5, 1700762. [Google Scholar] [CrossRef]
- Dickinson, G.H.; Vega, I.E.; Wahl, K.J.; Orihuela, B.; Beyley, V.; Rodriguez, E.N.; Everett, R.K.; Bonaventura, J.; Rittschof, D. Barnacle cement: A polymerization model based on evolutionary concepts. J. Exp. Biol. 2009, 212, 3499–3510. [Google Scholar] [CrossRef]
- So, C.R.; Scancella, J.M.; Fears, K.P.; Essock-Burns, T.; Haynes, S.E.; Leary, D.H.; Diana, Z.; Wang, C.; North, S.; Oh, C.S.; et al. Oxidase activity of the barnacle adhesive interface involves peroxide-dependent catechol oxidase and lysyl oxidase enzymes. ACS Appl. Mater. Interfaces 2017, 9, 11493–11505. [Google Scholar] [CrossRef]
- Essock-Burns, T. Exploring the Interface between Macroorganisms and Microorganisms: Biochemical, Ecological, and Evolutionary Contexts. Ph.D. Thesis, Duke University, Durham, North Carolina, 2015. [Google Scholar]
- Hughes, M. The function of concurrent signals: Visual and chemical communication in snapping shrimp. Anim. Behav. 1996, 52, 247–257. [Google Scholar] [CrossRef]
- Caldwell, R.L. Cavity occupation and defensive behaviour in the stomatopod Gonodactylus festai: Evidence for chemically mediated individual recognition. Anim. Behav. 1979, 27, 194–201. [Google Scholar] [CrossRef]
- Rittschof, D. Oyster drills and the frontiers of chemical ecology: Unsettling ideas. Am. Malacol. Bull. 1985, 1, 111–116. [Google Scholar]
- Rittschof, D. Body odors and neutral-basic peptide mimics: A review of responses by marine organisms. Integr. Comp. Biol. 1993, 33, 487–493. [Google Scholar] [CrossRef]
- Endrizzi, B.J.; Stewart, R.J. Glueomics: An expression survey of the adhesive gland of the sandcastle worm. J. Adhes. 2009, 85, 546–559. [Google Scholar] [CrossRef]
- Essock-Burns, T.; Soderblom, E.J.; Orihuela, B.; Moseley, M.A.; Rittschof, D. Hypothesis testing with proteomics: A case study using wound healing mechanisms in fluids associated with barnacle glue. Front. Mar. Sci. 2019, 6. [Google Scholar] [CrossRef]
- Reports—IPCC. Available online: https://www.ipcc.ch/reports/ (accessed on 17 November 2019).
- Doney, S.C.; Fabry, V.J.; Feely, R.A.; Kleypas, J.A. Ocean acidification: The other CO2 problem. Annu. Rev. Mar. Sci. 2009, 1, 169–192. [Google Scholar] [CrossRef] [PubMed]
- Dobretsov, S.; Coutinho, R.; Rittschof, D.; Salta, M.; Ragazzola, F.; Hellio, C. The oceans are changing: Impact of ocean warming and acidification on biofouling communities. Biofouling 2019, 35, 585–595. [Google Scholar] [CrossRef]
- Chan, V.B.S.; Li, C.; Lane, A.C.; Wang, Y.; Lu, X.; Shih, K.; Zhang, T.; Thiyagarajan, V. CO2-driven ocean acidification alters and weakens integrity of the calcareous tubes produced by the serpulid tubeworm, Hydroides elegans. PLoS ONE 2012, 7, e42718. [Google Scholar] [CrossRef]
- Doney, S.C.; Ruckelshaus, M.; Emmett Duffy, J.; Barry, J.P.; Chan, F.; English, C.A.; Galindo, H.M.; Grebmeier, J.M.; Hollowed, A.B.; Knowlton, N.; et al. Climate change impacts on marine ecosystems. Annu. Rev. Mar. Sci. 2012, 4, 11–37. [Google Scholar] [CrossRef]
- Lane, A.C.; Mukherjee, J.; Chan, V.B.S.; Thiyagarajan, V. Decreased pH does not alter metamorphosis but compromises juvenile calcification of the tube worm Hydroides elegans. Mar. Biol. 2013, 160, 1983–1993. [Google Scholar] [CrossRef][Green Version]
- Meng, Y.; Li, C.; Li, H.; Shih, K.; He, C.; Yao, H.; Thiyagarajan, V. Recoverable impacts of ocean acidification on the tubeworm, Hydroides elegans: Implication for biofouling in future coastal oceans. Biofouling 2019, 35, 945–957. [Google Scholar] [CrossRef]
- Peck, L.S.; Clark, M.S.; Power, D.; Reis, J.; Batista, F.M.; Harper, E.M. Acidification effects on biofouling communities: Winners and losers. Glob. Chang. Biol. 2015, 21, 1907–1913. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.C.K.; Thiyagarajan, V.; Cheung, S.C.K.; Qian, P.-Y. Roles of bacterial community composition in biofilms as a mediator for larval settlement of three marine invertebrates. Aquat. Microb. Ecol. 2005, 38, 41–51. [Google Scholar] [CrossRef]
- Zhang, Y.; Fonslow, B.R.; Shan, B.; Baek, M.-C.; Yates, J.R. Protein analysis by shotgun/bottom-up proteomics. Chem. Rev. 2013, 113, 2343–2394. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).