Effect of Bortezomib on Global Gene Expression in PC12-Derived Nerve Cells
Abstract
1. Introduction
2. Results
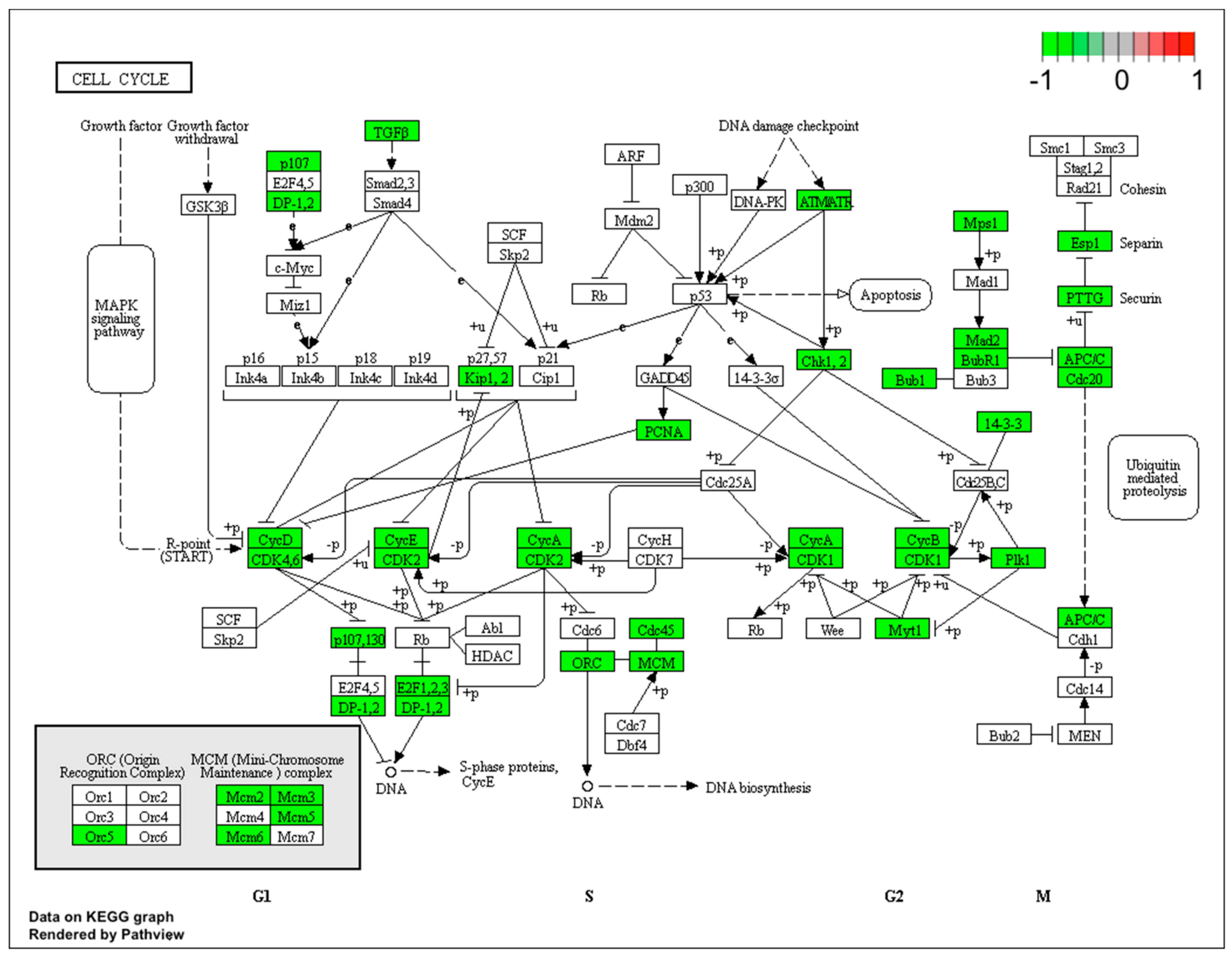
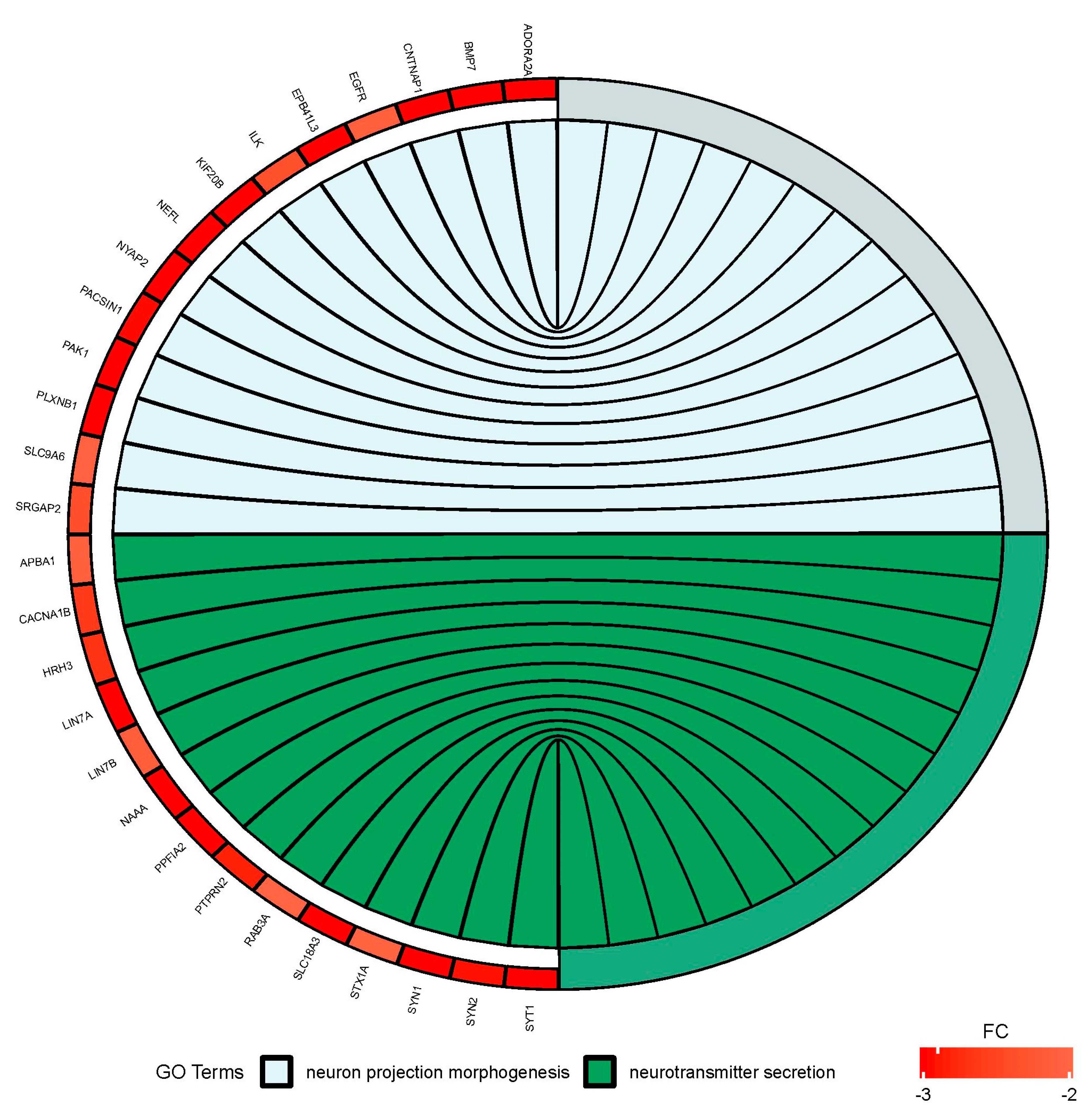
2.1. Gene Expression Profile in PC12-derived Nerve Cells
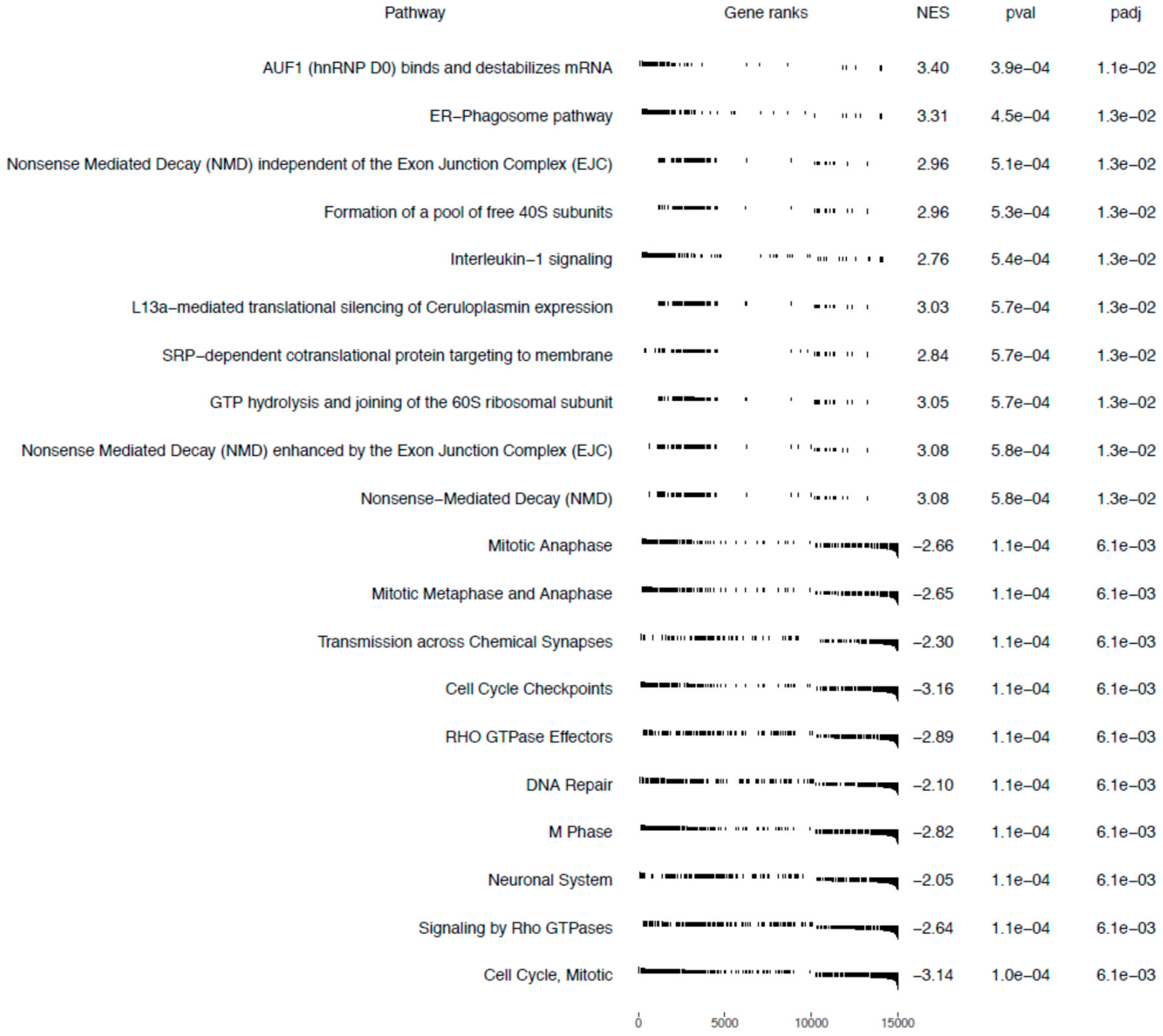
Gene Set Enrichment Analysis
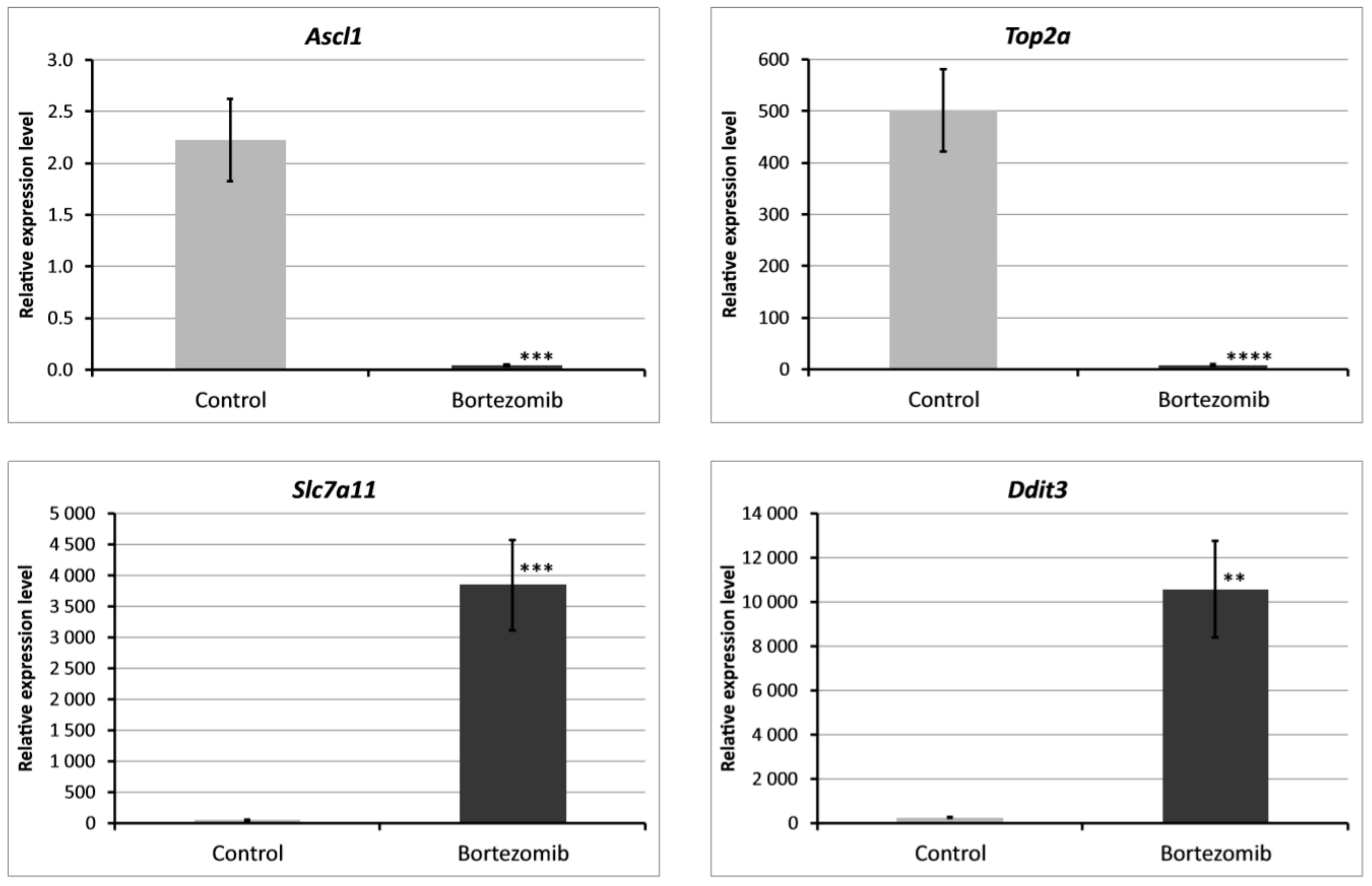
2.2. Validation of Dysregulated mRNAs
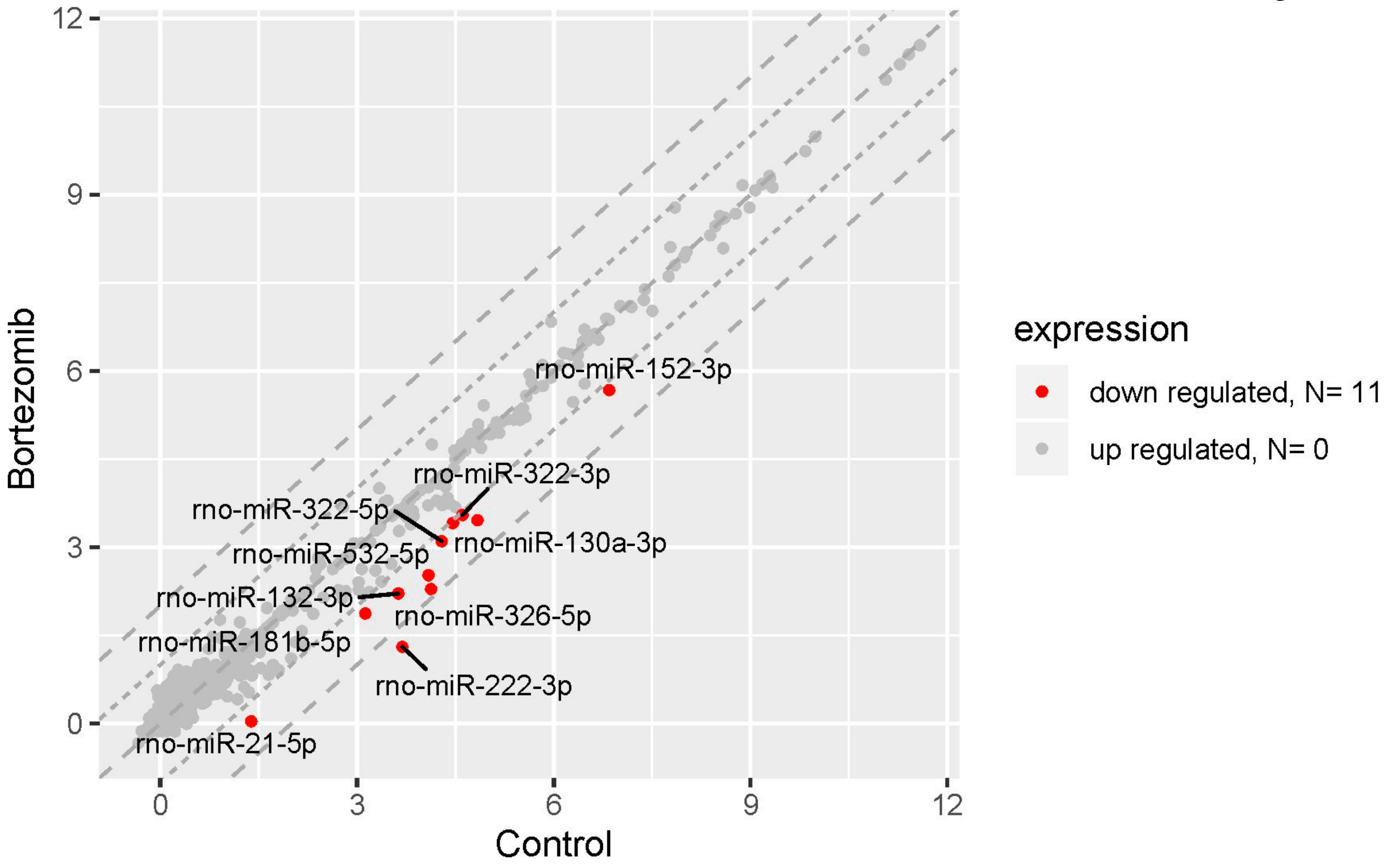
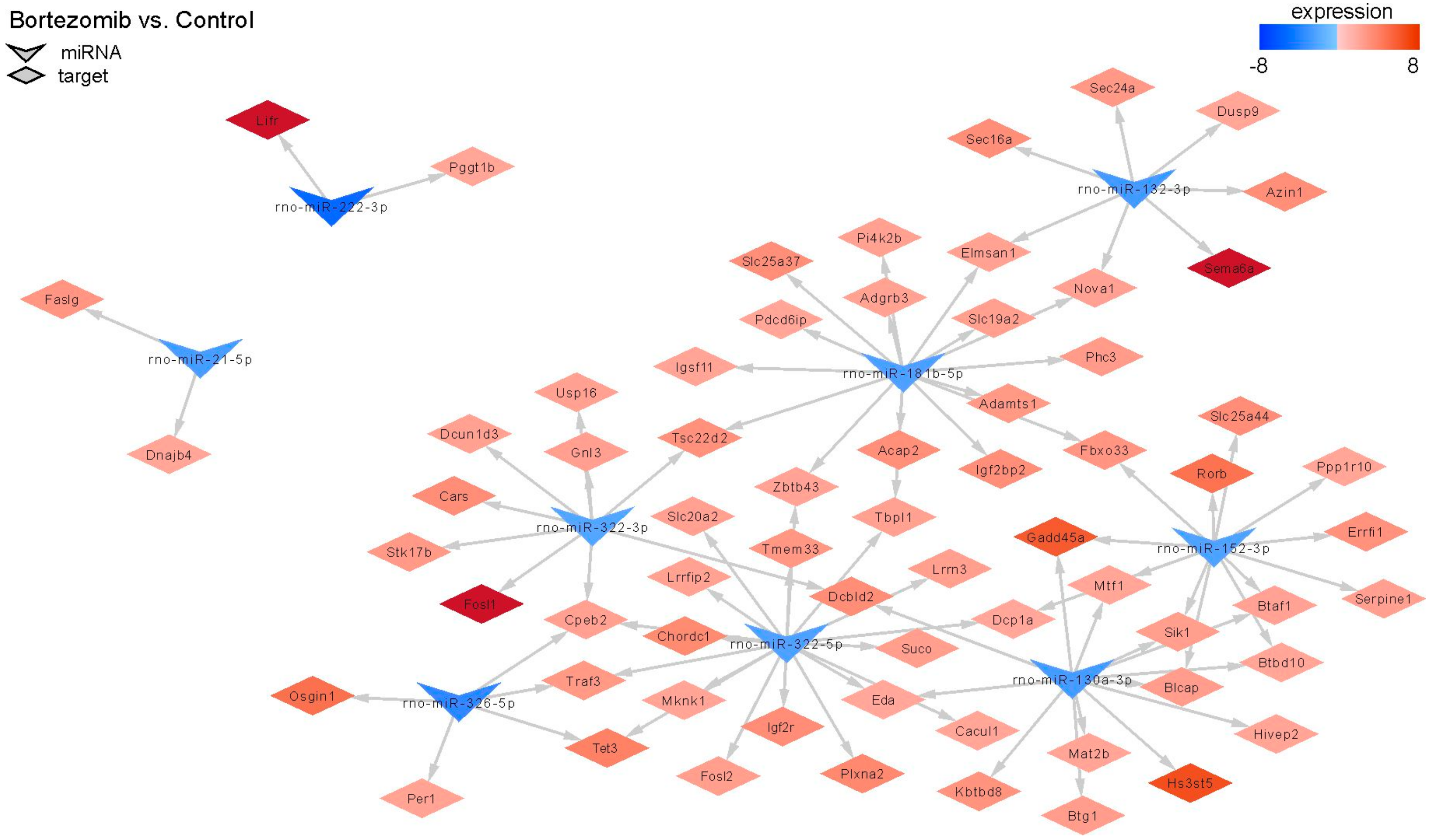
2.3. miRNAs Expression Profile in Neural Cells
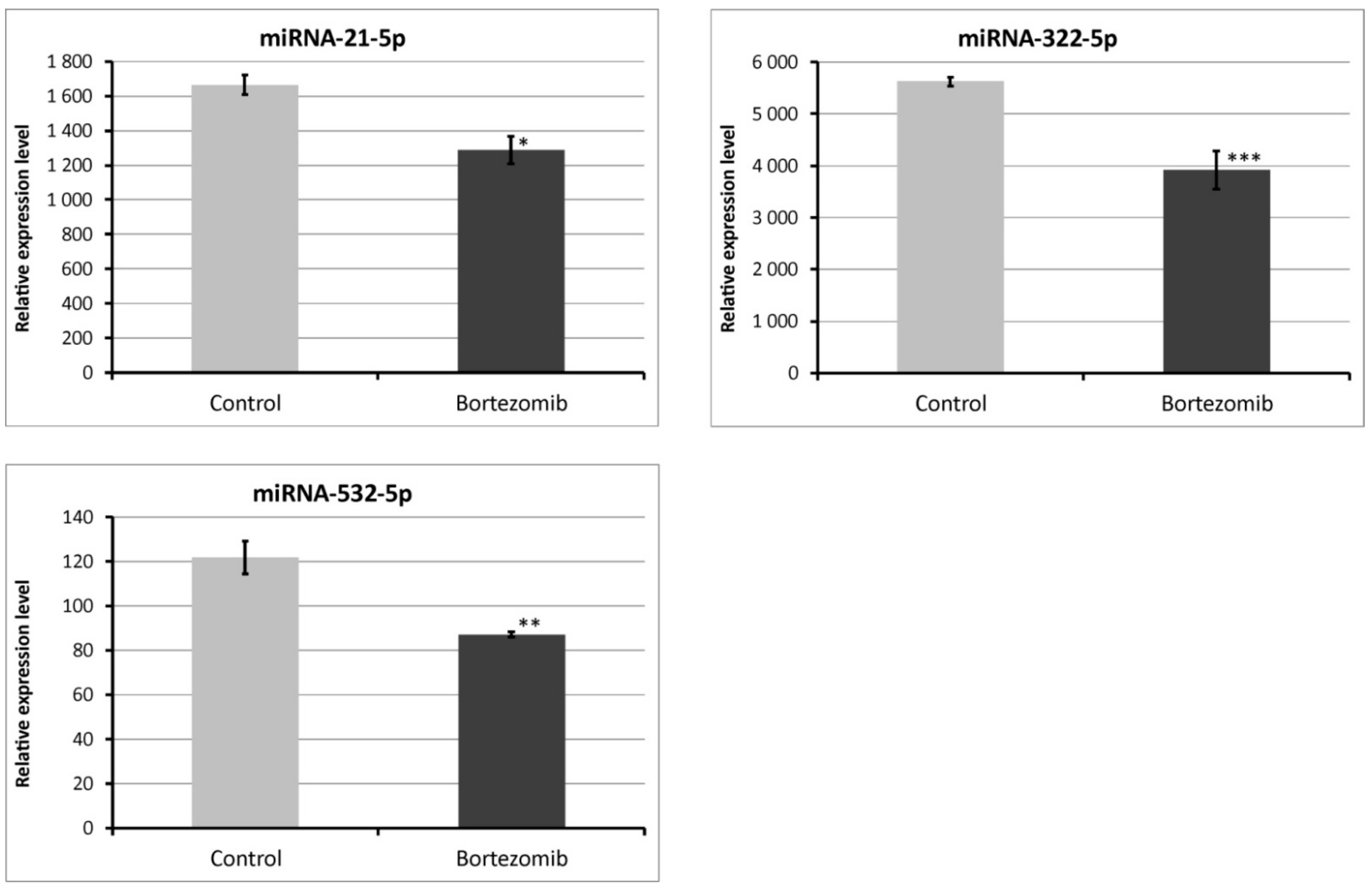
2.4. Validation of Dysregulated miRNAs
3. Discussion
Potential Study Limitations
4. Materials and Methods
4.1. Cell Culture and Incubation
4.2. RNA
4.3. Affymetrix GeneChip Microarray and Data Analysis
4.4. Affymetrix GeneChip miRNA Microarray and Data Analysis
4.5. DAVID
4.6. Validation of Data Obtained from Microarrays
4.6.1. Validation of Gene Expression
4.6.2. Validation of Selected miRNA Expression
4.7. Gene Set Enrichment Analysis (GSEA)
4.8. Statistical Methods
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ALS | amyotrophic lateral sclerosis |
| A-T | ataxia telangiectasia |
| BiPN | bortezomib-induced peripheral neuropathy |
| BZT | bortezomib |
| CNS | central nervous system |
| CS | Cockayne’s syndrome |
| DAVID | Database for Annotation, Visualization, and Integrated Discovery |
| DSB | DNA double-strand break |
| ES | enrichment score |
| FBS | fetal bovine serum |
| GO | Gene Ontology |
| GSEA | Gene Set Enrichment Analysis |
| HS | heat-inactivated horse serum |
| HSP | hereditary spastic paraplegia |
| IGF-1 | insulin-like growth factor 1 |
| IMID | including immunomodulatory drug |
| IL-6 | interleukin-6 |
| MAPK/ERK | mitogen-activated protein kinase/extracellular signal-regulated kinase |
| miRNAs | microRNAs |
| MM | multiple myeloma |
| MsigDB | Molecular Signatures Database |
| NER | nucleotide excision repair |
| NFκB | nuclear factor kappa B |
| NGF | nerve growth factor |
| NHEJ | non-homologous end joining |
| PN | peripheral neuropathy |
| PNS | peripheral nervous system |
| qRT-PCR | quantitative polymerase chain reaction |
| SCAN-1 | spinocerebellar ataxia with axonal neuropathy |
| TNF-α | tumor necrosis factor-α |
| VEGF | endothelial growth factor |
References
- Rajkumar, S.V.; Kumar, S. Multiple Myeloma: Diagnosis and Treatment. Mayo Clin. Proc. 2016, 91, 101–119. [Google Scholar] [CrossRef] [PubMed]
- Quach, H.; Ritchie, D.; Stewart, A.K.; Neeson, P.; Harrison, S.; Smyth, M.J.; Prince, H.M. Mechanism of action of immunomodulatory drugs (IMiDS) in multiple myeloma. Leukemia 2010, 24, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Adams, J.; Kauffman, M. Development of the proteasome inhibitor Velcade (Bortezomib). Cancer Investig. 2004, 22, 304–311. [Google Scholar] [CrossRef] [PubMed]
- Hideshima, T.; Mitsiades, C.; Akiyama, M.; Hayashi, T.; Chauhan, D.; Richardson, P.; Schlossman, R.; Podar, K.; Munshi, N.C.; Mitsiades, N.; et al. Molecular mechanisms mediating antimyeloma activity of proteasome inhibitor PS-341. Blood 2003, 101, 1530–1534. [Google Scholar] [CrossRef]
- Rajkumar, S.V.; Richardson, P.G.; Hideshima, T.; Anderson, K.C. Proteasome inhibition as a novel therapeutic target in human cancer. J. Clin. Oncol. 2005, 23, 630–639. [Google Scholar] [CrossRef]
- Landowski, T.H.; Megli, C.J.; Dorr, R.T.; Dalton, W.S. Calcium-mediated mitochondrial perturbation is a critical determinant of Velcade (PS-341/bortezomib) cytotoxicity in myeloma cell lines. Proc. Am. Assoc. Cancer Res. 2004, 67, 520. [Google Scholar]
- Richardson, P.G.; Delforge, M.; Beksac, M.; Wen, P.; Jongen, J.L.; Sezer, O.; Terpos, E.; Munshi, N.; Palumbo, A.; Rajkumar, S.V.; et al. Management of treatment-emergent peripheral neuropathy in multiple myeloma. Leukemia 2012, 26, 595–608. [Google Scholar] [CrossRef]
- Argyriou, A.A.; Iconomou, G.; Kalofonos, H.P. Bortezomib-induced peripheral neuropathy in multiple myeloma: A comprehensive review of the literature. Blood 2008, 112, 1593–1599. [Google Scholar] [CrossRef]
- Chen, D.; Frezza, M.; Schmitt, S.; Kanwar, J.; Dou, Q.P. Bortezomib as the first proteasome inhibitor anticancer drug: Current status and future perspectives. Curr. Cancer Drug Targets 2011, 11, 239–253. [Google Scholar] [CrossRef]
- Chu, Y.-Y.; Ko, C.-Y.; Wang, S.-M.; Lin, P.-I.; Wang, H.-Y.; Lin, W.-C.; Wu, D.-Y.; Wang, L.-H.; Wang, J.-M. Bortezomib-induced miRNAs direct epigenetic silencing of locus genes and trigger apoptosis in leukemia. Cell Death Dis. 2017, 8, e3167. [Google Scholar] [CrossRef]
- Łuczkowska, K.; Rogińska, D.; Ulańczyk, Z.; Paczkowska, E.; Schmidt, A.C.; Machaliński, B. Molecular Mechanisms of Bortezomib Action: Novel Evidence for the miRNA–mRNA Interaction Involvement. Int. J. Mol. Sci. 2020, 21, 350. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chen, J.; Sen, S. MicroRNA as Biomarkers and Diagnostics. J. Cell. Physiol. 2016, 231, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Schraml, E.; Hackl, M.; Grillari, J. MicroRNAs and toxicology: A love marriage. Toxicol. Rep. 2017, 4, 634–636. [Google Scholar] [CrossRef] [PubMed]
- Łuczkowska, K.; Litwińska, Z.; Paczkowska, E.; Machaliński, B. Pathophysiology of drug-induced peripheral neuropathy in multiple myeloma patients. J. Physiol. Pharmacol. 2018, 69, 165–172. [Google Scholar] [CrossRef]
- Field-Smith, A.; Morgan, G.J.; Davies, F.E. Bortezomib (Velcade™) in the Treatment of Multiple Myeloma. Ther. Clin. Risk Manag. 2006, 2, 271–279. [Google Scholar] [CrossRef]
- Landowski, T.H.; Megli, C.J.; Nullmeyer, K.D.; Lynch, R.M.; Dorr, R.T. Mitochondrial-mediated disregulation of Ca2+ is a critical determinant of Velcade (PS-341/bortezomib) cytotoxicity in myeloma cell lines. Cancer Res. 2005, 65, 3828–3836. [Google Scholar] [CrossRef]
- Karademir, B.; Sari, G.; Jannuzzi, A.T.; Musunuri, S.; Wicher, G.; Grune, T.; Mi, J.; Hacioglu-Bay, H.; Forsberg-Nilsson, K.; Bergquist, J.; et al. Proteomic approach for understanding milder neurotoxicity of Carfilzomib against Bortezomib. Sci. Rep. 2018, 8, 16318. [Google Scholar] [CrossRef]
- Su, Y.; Ming, G.L.; Song, H. DNA damage and repair regulate neuronal gene expression. Cell Res. 2015, 25, 993–994. [Google Scholar] [CrossRef]
- Englander, E.W. DNA Damage Response in Peripheral Nervous System: Coping with Cancer Therapy-Induced DNA Lesions. DNA Repair (Amst.) 2013, 12, 685–690. [Google Scholar] [CrossRef]
- McKinnon, P.J. Topoisomerases and the regulation of neural function. Nat. Rev. Neurosci. 2016, 17, 673–679. [Google Scholar] [CrossRef]
- Takashima, H.; Boerkoel, C.F.; John, J.; Saifi, G.M.; Salih, M.A.; Armstrong, D.; Mao, Y.; Quiocho, F.A.; Roa, B.B.; Nakagawa, M.; et al. Mutation of TDP1, encoding a topoisomerase I-dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nat. Genet. 2002, 32, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Katyal, S.; Downing, S.M.; Zhao, J.; Russell, H.R.; McKinnon, P.J. Neurogenesis requires TopBP1 to prevent catastrophic replicative DNA damage in early progenitors. Nat. Neurosci. 2012, 15, 819–826. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Duan, J.; Shu, S.; Wang, X.; Gao, L.; Guo, J.; Zhang, Y. Ligase I and ligase III mediate the DNA double-strand break ligation in alternative end-joining. Proc. Natl. Acad. Sci. USA 2016, 113, 1256–1260. [Google Scholar] [CrossRef] [PubMed]
- Ellenberger, T.; Tomkinson, A.E. Eukaryotic DNA Ligases: Structural and Functional Insights. Annu. Rev. Biochem. 2008, 77, 313–338. [Google Scholar] [CrossRef]
- Troelstra, C.; van Gool, A.; de Wit, J.; Vermeulen, W.; Bootsma, D.; Hoeijmakers, J.H. ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne’s syndrome and preferential repair of active genes. Cell 1992, 71, 939–953. [Google Scholar] [CrossRef]
- Dzagnidze, A.; Katsarava, Z.; Makhalova, J.; Liedert, B.; Yoon, M.S.; Kaube, H.; Limmroth, V.; Thomale, J. Repair capacity for platinum-DNA adducts determines the severity of cisplatin-induced peripheral neuropathy. J. Neurosci. 2007, 27, 9451–9457. [Google Scholar] [CrossRef]
- Bermudez-Guzman, L.; Leal, A. DNA repair deficiency in neuropathogenesis: When all roads lead to mitochondria. Transl. Neurodegener. 2019, 8, 14. [Google Scholar] [CrossRef]
- Nam, D.E.; Yoo, D.H.; Choi, S.S.; Choi, B.O.; Chung, K.W. Wide phenotypic spectrum in axonal Charcot-Marie-Tooth neuropathy type 2 patients with KIF5A mutations. Genes Genom. 2018, 40, 77–84. [Google Scholar] [CrossRef]
- Rachana, K.S.; Manu, M.S.; Advirao, G.M. Insulin-induced upregulation of lipoprotein lipase in Schwann cells during diabetic peripheral neuropathy. Diabetes Metab. Syndr. 2018, 12, 525–530. [Google Scholar] [CrossRef]
- Hetman, M.; Vashishta, A.; Rempala, G. Neurotoxic mechanisms of DNA damage: Focus on transcriptional inhibition. J. Neurochem. 2010, 114, 1537–1549. [Google Scholar] [CrossRef]
- Blüthgen, N.; van Bentum, M.; Merz, B.; Kuhl, D.; Hermey, G. Profiling the MAPK/ERK dependent and independent activity regulated transcriptional programs in the murine hippocampus in vivo. Sci. Rep. 2017, 7, 45101. [Google Scholar] [CrossRef] [PubMed]
- Davis, S.; Vanhoutte, P.; Pages, C.; Caboche, J.; Laroche, S. The MAPK/ERK cascade targets both Elk-1 and cAMP response element-binding protein to control long-term potentiation-dependent gene expression in the dentate gyrus in vivo. J. Neurosci. 2000, 20, 4563–4572. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, L.A.; Murphy, P.R. MicroRNA: Biogenesis, Function and Role in Cancer. Curr. Genom. 2010, 11, 537–561. [Google Scholar] [CrossRef] [PubMed]
- Schafer, A. Gadd45 proteins: Key players of repair-mediated DNA demethylation. Adv. Exp. Med. Biol. 2013, 793, 35–50. [Google Scholar] [PubMed]
- Keilbaugh, S.A.; Prusoff, W.H.; Simpson, M.V. The PC12 cell as a model for studies of the mechanism of induction of peripheral neuropathy by anti-HIV-1 dideoxynucleoside analogs. Biochem. Pharmacol. 1991, 42, R5–R8. [Google Scholar] [CrossRef]
- Zhao, S.; Zhang, L.; Xu, Z.; Chen, W. Neurotoxic effects of iron overload under high glucose concentration. Neural Regen. Res. 2013, 8, 3423–3433. [Google Scholar]
- Slotkin, T.A.; MacKillop, E.A.; Ryde, I.T.; Tate, C.A.; Seidler, F.J. Screening for Developmental Neurotoxicity Using PC12 Cells: Comparisons of Organophosphates with a Carbamate, an Organochlorine, and Divalent Nickel. Environ. Health Perspect. 2007, 115, 93–101. [Google Scholar] [CrossRef]
- Christen, V.; Rusconi, M.; Crettaz, P.; Fent, K. Developmental neurotoxicity of different pesticides in PC-12 cells in vitro. Toxicol. Appl. Pharmacol. 2017, 325, 25–36. [Google Scholar] [CrossRef]
- Popova, D.; Karlsson, J.; Jacobsson, S.O.P. Comparison of neurons derived from mouse P19, rat PC12 and human SH-SY5Y cells in the assessment of chemical-and toxin-induced neurotoxicity. BMC Pharmacol. Toxicol. 2017, 18, 42. [Google Scholar] [CrossRef]
- Gautier, L.; Cope, L.; Bolstad, B.M.; Irizarry, R.A. Affy—Analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004, 20, 307–315. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Dennis, G., Jr.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003, 4, R60. [Google Scholar] [CrossRef]
- Fresno, C.; Fernandez, E.A. RDAVIDWebService: A versatile R interface to DAVID. Bioinformatics 2013, 29, 2810–2811. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Birger, C.; Thorvaldsdottir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
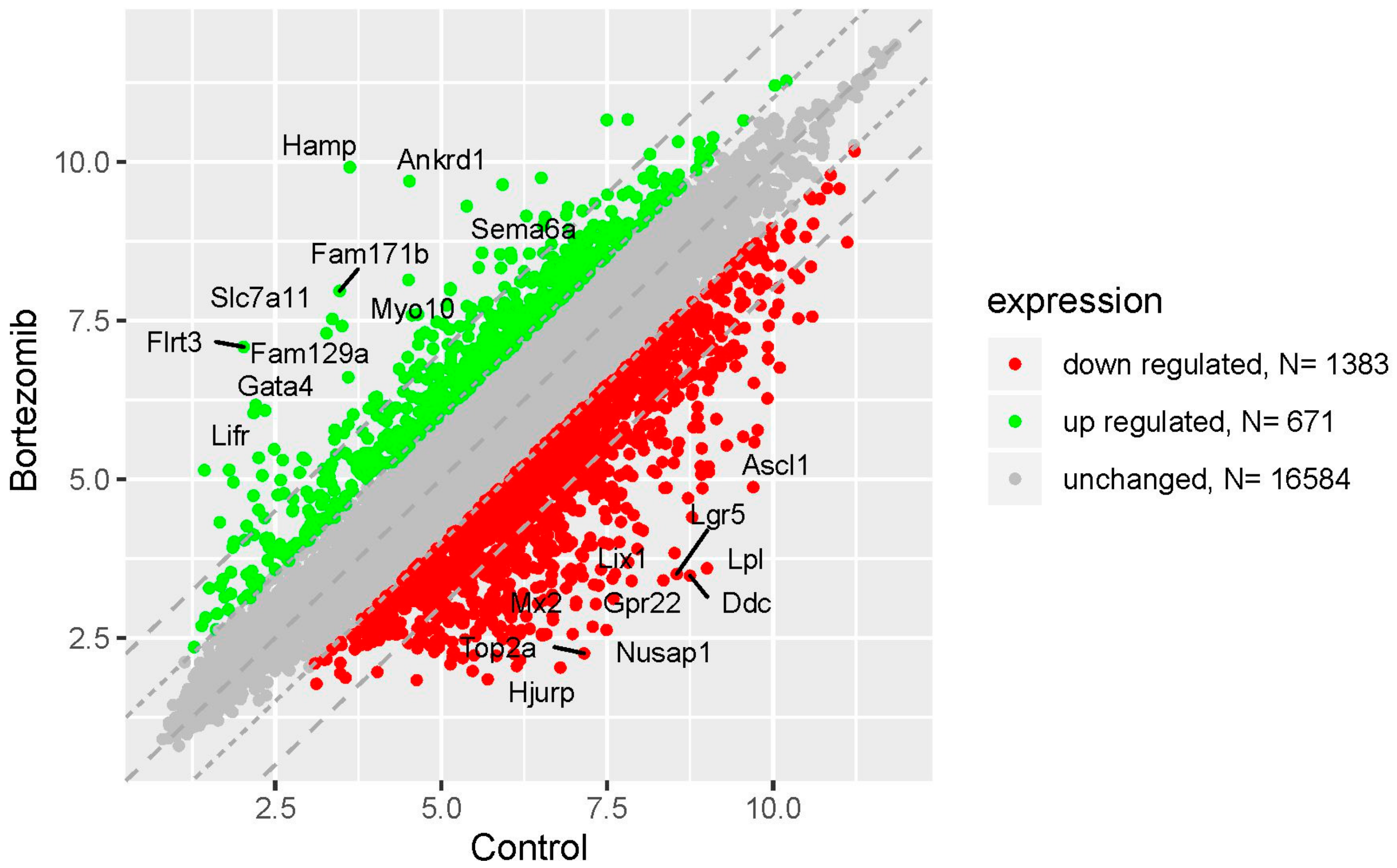
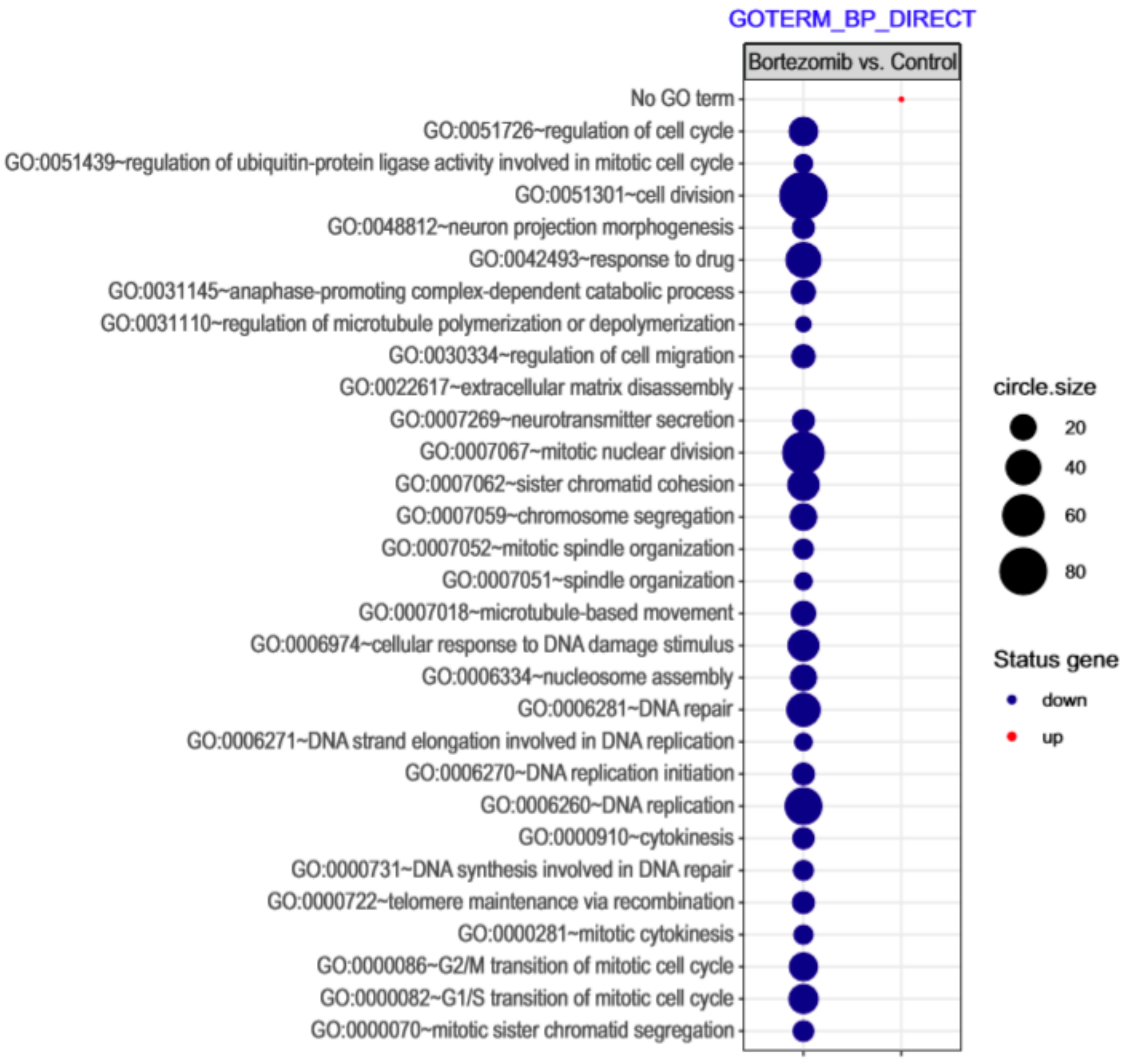
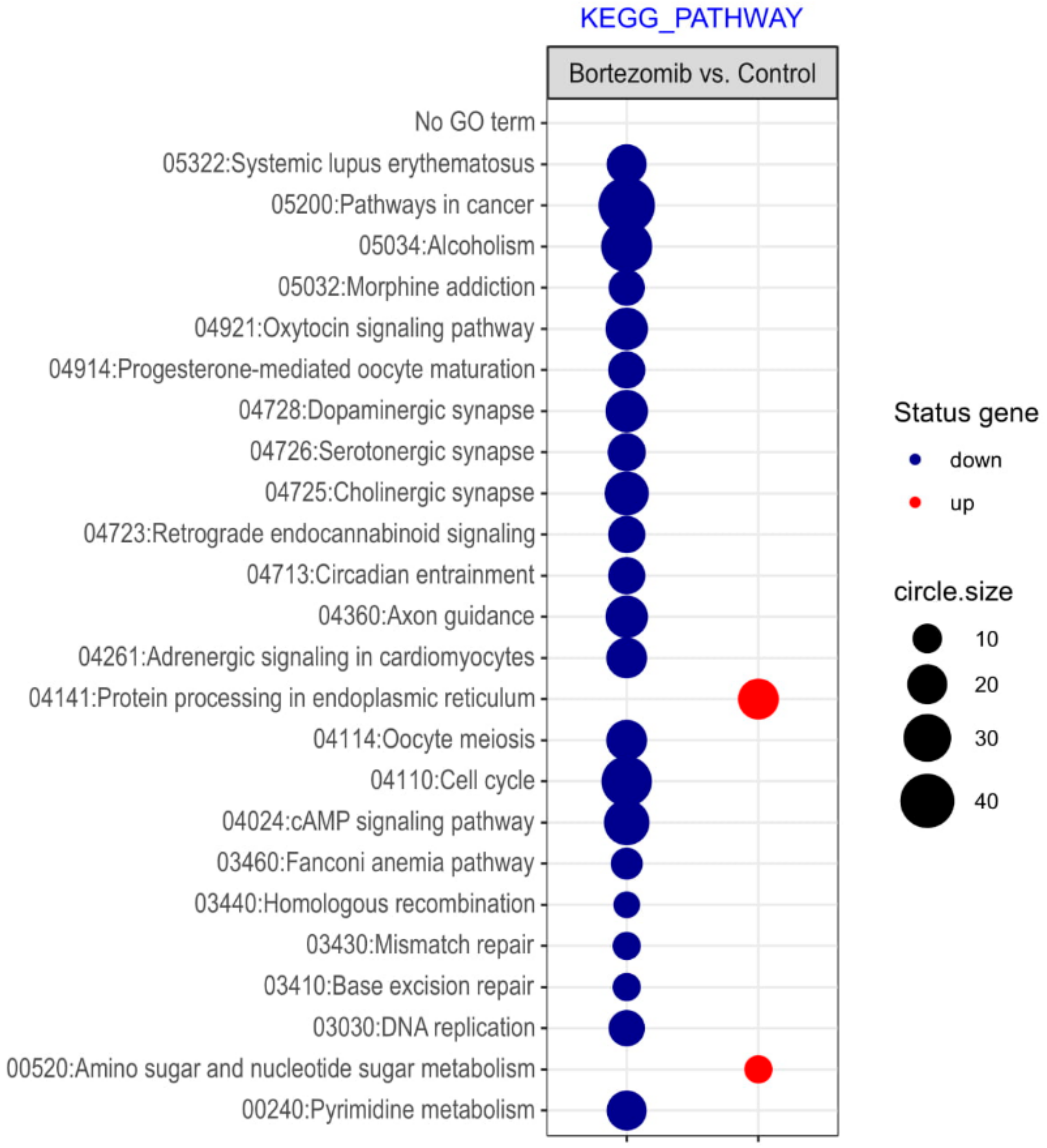
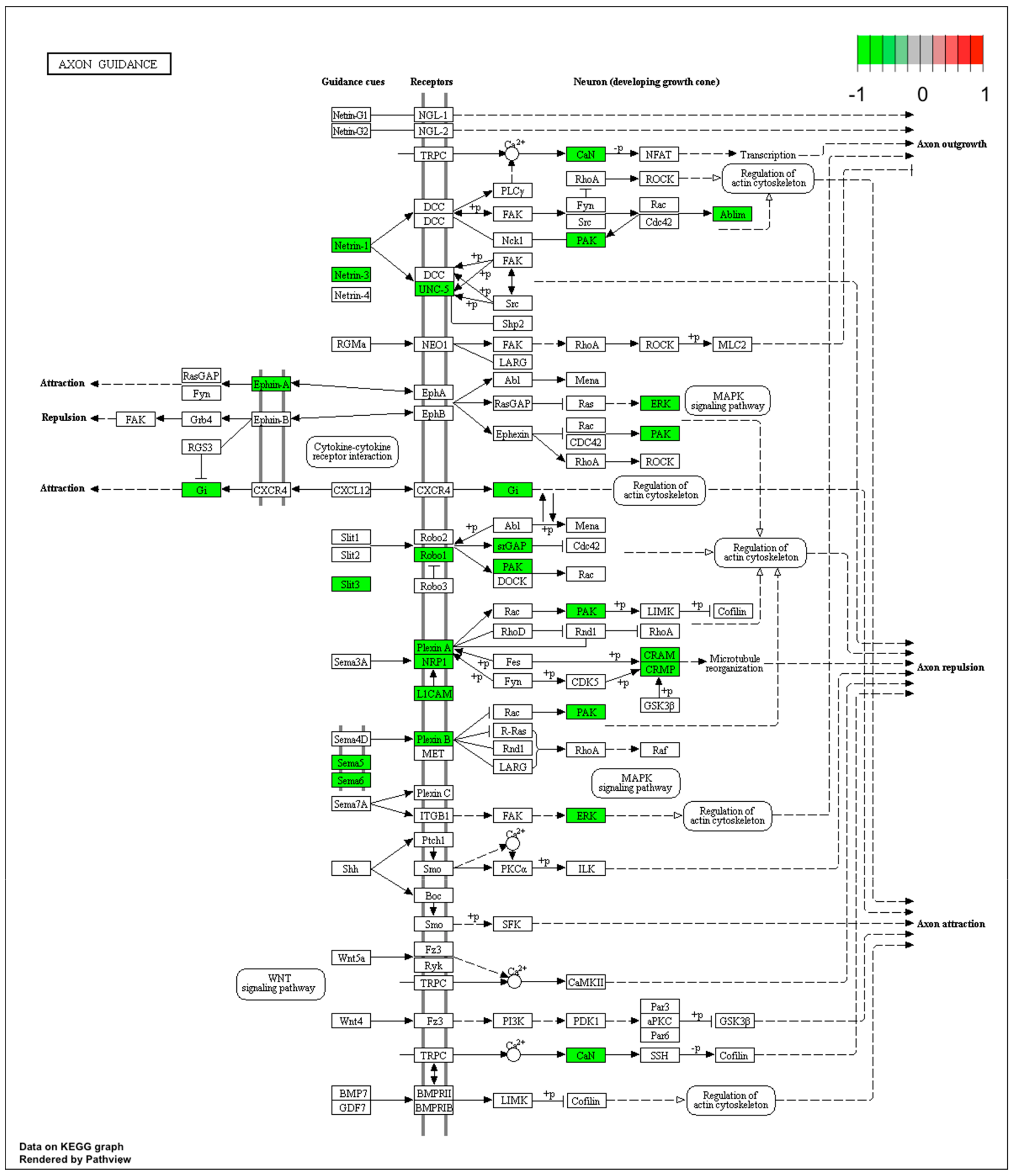

| Gene Symbol | Gene Name | Gene Function | Fold Change |
|---|---|---|---|
| Lpl | lipoprotein lipase | Recruited to its site of action on the luminal surface of vascular endothelium by binding to GPIHBP1 and cell surface heparan sulfate proteoglycans | −42.493 |
| Ddc | dopa decarboxylase (aromatic L-amino acid decarboxylase) | important in the brain and nervous system. This enzyme takes part in the pathway that produces dopamine and serotonin, which are chemical messengers that transmit signals between nerve cells (neurotransmitters). | −38.562 |
| Lgr5 | leucine rich repeat containing G protein coupled receptor 5 | expressed across a diverse range of tissue such as in the muscle, placenta, spinal cord and brain and particularly as a biomarker of adult stem cells in certain tissues | −32.995 |
| Gpr22 | G protein-coupled receptor 22 | play an essential role in the regulation of cardiovascular function | −30.754 |
| Top2a | topoisomerase (DNA) II alpha | controls and alters the topologic states of DNA during transcription; involved in processes such as chromosome condensation, chromatid separation, and the relief of torsional stress that occurs during DNA transcription and replication | −29.831 |
| Nusap1 | nucleolar and spindle associated protein 1 | binds and stabilizes microtubules (by similarity); can promote the organization of mitotic spindle microtubules around them | −29.200 |
| Ascl1 | achaete-scute family bHLH transcription factor 1 | neuronal commitment and differentiation and in the generation of olfactory and autonomic neurons | −28.366 |
| Hjurp | Holliday junction recognition protein | plays a central role in the incorporation and maintenance of histone H3-like variant CENPA at centromeres | −27.183 |
| Mx2 | myxovirus (influenza virus) resistance 2 | exhibits antiviral activity; play a role in regulating nucleocytoplasmic transport and cell-cycle progression | −24.350 |
| RGD1564463 | similar to Mdes protein | able to upregulation of proliferation-related gene expression, such as that of CDX2 and PCNA | −22.386 |
| S100a5 | S100 calcium binding protein A5 | involved in the regulation of a number of cellular processes such as cell cycle progression and differentiation. | −22.222 |
| Bub1 | BUB1 mitotic checkpoint serine/threonine kinase | establishment of the mitotic spindle checkpoint and chromosome congression | −21.434 |
| Ndufa4l2 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2 | codes for a subunit of Complex I of the respiratory chain, which transfers electrons from NADH to ubiquinone | −20.830 |
| Nuf2 | NUF2, NDC80 kinetochore complex component | chromosome segregation | −19.673 |
| Faxdc2 | fatty acid hydroxylase domain containing 2 | catalyzes the synthesis of 2-hydroxysphingolipids | −17.710 |
| Gene Symbol | Gene Name | Gene Function | Fold Change |
|---|---|---|---|
| Hamp | hepcidin antimicrobial peptide | maintaining iron balance in the body | 78.308 |
| Ankrd1 | ankyrin repeat domain 1 | transcription factor involved in development and under conditions of stress | 36.103 |
| Flrt3 | fibronectin leucine rich transmembrane protein 3 | cell adhesion and receptor signalling | 33.410 |
| Fam171b | family with sequence similarity 171, member B | associated with neurodevelopmental disorders | 22.564 |
| Slc7a11 | solute carrier family 7 | regulates synaptic activity by stimulating extrasynaptic receptors and performs nonvesicular glutamate release | 17.940 |
| Fam129a | family with sequence similarity 129, member A | marker for certain cancers; regulates p53-mediated apoptosis | 16.321 |
| Gata4 | GATA binding protein 4 | transcriptional regulator for many cardiac genes; regulates hypertrophic growth of the heart; promotes cardiac morphogenesis, cardiomyocytes survival, and maintains cardiac function in the adult heart. | 15.599 |
| Sema6a | sema domain, transmembrane domain ™ | promotes reorganization of the actin cytoskeleton;plays an important role in axon guidance in the developing central nervous system | 15.091 |
| Myo10 | myosin X | regulates cell shape, cell spreading and cell adhesion | 15.012 |
| Lifr | leukemia inhibitory factor receptor alpha | controls several cellular processes, including growth and division (proliferation), maturation (differentiation), and survival | 14.617 |
| Prss46 | protease, serine, 46 | serine-type endopeptidase activity; involved in proteolysis | 13.349 |
| Plaur | plasminogen activator, urokinase receptor | influences many normal and pathological processes related to cell-surface plasminogen activation and localized degradation of the extracellular matrix | 13.144 |
| Dcx | doublecortin | involved in the movement of nerve cells to their proper locations in the developing brain, a process called neuronal migration | 13.099 |
| Fosl1 | fos-like antigen 1 | regulates tumor cell proliferation and survival | 12.403 |
| Ddit3 | DNA-damage inducible transcript 3 | adipogenesis, erythropoiesis, growth arrest and endoplasmic reticulum stress response | 7.306 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Łuczkowska, K.; Rogińska, D.; Ulańczyk, Z.; Machaliński, B. Effect of Bortezomib on Global Gene Expression in PC12-Derived Nerve Cells. Int. J. Mol. Sci. 2020, 21, 751. https://doi.org/10.3390/ijms21030751
Łuczkowska K, Rogińska D, Ulańczyk Z, Machaliński B. Effect of Bortezomib on Global Gene Expression in PC12-Derived Nerve Cells. International Journal of Molecular Sciences. 2020; 21(3):751. https://doi.org/10.3390/ijms21030751
Chicago/Turabian StyleŁuczkowska, Karolina, Dorota Rogińska, Zofia Ulańczyk, and Bogusław Machaliński. 2020. "Effect of Bortezomib on Global Gene Expression in PC12-Derived Nerve Cells" International Journal of Molecular Sciences 21, no. 3: 751. https://doi.org/10.3390/ijms21030751
APA StyleŁuczkowska, K., Rogińska, D., Ulańczyk, Z., & Machaliński, B. (2020). Effect of Bortezomib on Global Gene Expression in PC12-Derived Nerve Cells. International Journal of Molecular Sciences, 21(3), 751. https://doi.org/10.3390/ijms21030751





