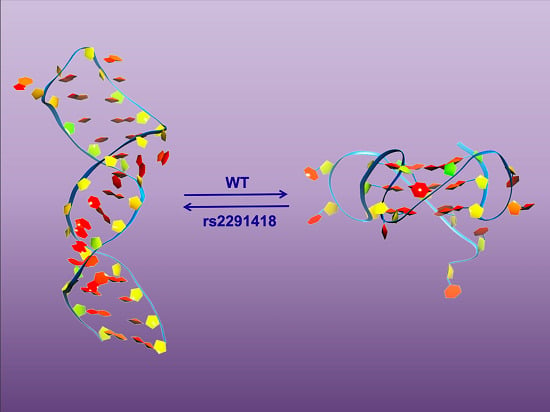
Characterization of a G-Quadruplex Structure in Pre-miRNA-1229 and in Its Alzheimer’s Disease-Associated Variant rs2291418: Implications for miRNA-1229 Maturation
Abstract
:1. Introduction
2. Results
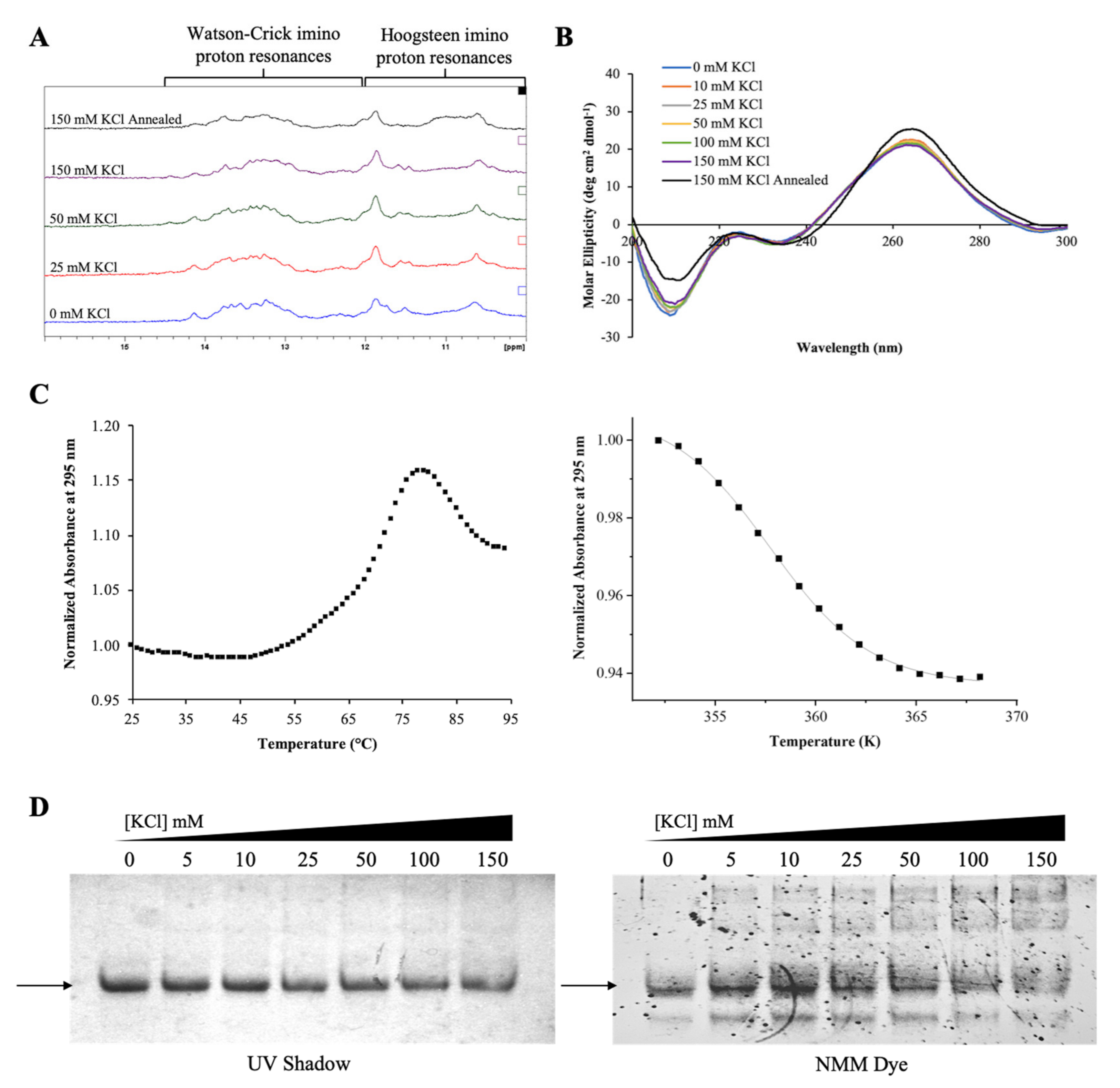
2.1. The G-Rich Region of Pre-miRNA-1229 Folds into a GQ Structure
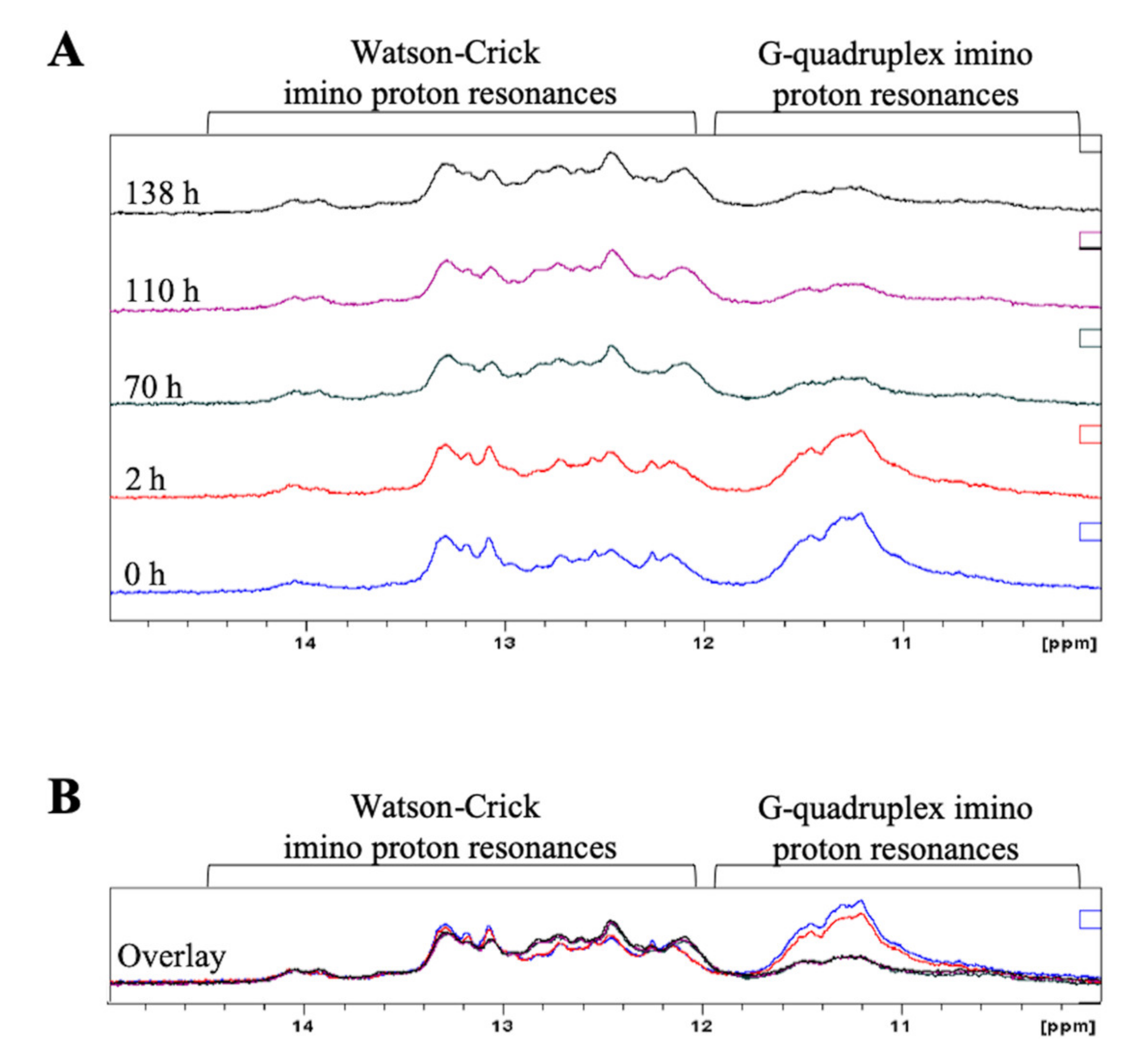
2.2. Full-Length Pre-miRNA-1229 Forms a GQ Structure That Coexists in Equilibrium with an Extended Hairpin Structure
2.3. The GQ Structure Formed by the G-Rich Region of Pre-miRNA-1229 Is Retained in the AD-Associated rs2291418 Variant
2.4. The AD-Associated rs2291418 Mutation within the Full-Length Pre-miRNA-1229 Shifts the Equilibrium from the GQ Structure to the Extended Hairpin Structure
3. Discussion
4. Materials and Methods
4.1. In Vitro RNA Synthesis
4.2. 1H NMR Spectroscopy
4.3. Circular Dichroism Spectroscopy
4.4. UV Spectroscopy Thermal Denaturation
4.5. Nondenaturing Polyacrylamide Gel Electrophoresis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AD | Alzheimer’s disease |
| Aβ | β-amyloid |
| SORL1 | Sortilin-related receptor |
| miRNA | microRNA |
| nt | nucleotide |
| RISC | RNA-induced silencing complex |
| GQ | G-quadruplex |
| SNP | Single nucleotide polymorphism |
| NMR | Nuclear magnetic resonance |
| CD | Circular dichroism |
| UV | Ultraviolet |
| Tm | Melting temperature |
| NMM | N-methyl mesoporphyrin IX |
References
- Hebert, L.E.; Weuve, J.; Scherr, P.A.; Evans, D.A. Alzheimer disease in the United States (2010-2050) estimated using the 2010 census. Neurology 2013, 80, 1778–1783. [Google Scholar] [CrossRef] [Green Version]
- Förstl, H.; Kurz, A. Clinical features of Alzheimer’s disease. Eur. Arch. Psychiatry Clin. Neurosci. 1999, 249, 288–290. [Google Scholar] [CrossRef] [PubMed]
- Barker, W.W.; Luis, C.A.; Kashuba, A.; Luis, M.; Harwood, D.G.; Loewenstein, D.; Waters, C.; Jimison, P.; Shepherd, E.; Sevush, S.; et al. Relative frequencies of Alzheimer’s disease, Lewy body, vascular and frontotemporal dementia, and hippocampal sclerosis in the state of Florida brain bank. Alzheimer Dis. Assoc. Disord. 2002, 16, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.S.; Segawa, E.; Boyle, P.A.; Anagnos, S.E.; Hizel, L.P.; Bennett, D.A. The natural history of cognitive decline in Alzheimer’s disease. Psychol. Aging 2012, 27, 1008–1017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selkoe, D.J. Alzheimer’s disease: Genes, proteins, and therapy. Physiol. Rev. 2001, 81, 741–766. [Google Scholar] [CrossRef]
- Glenner, G.G.; Wong, C.W. Alzheimer’s disease: Initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem. Biophys. Res. 1984, 120, 885–890. [Google Scholar] [CrossRef]
- Iwatsubo, T.; Odaka, A.; Suzuki, N.; Mizusawa, H.; Nukina, N.; Ihara, Y. Visualization of Aβ42 and Aβ40 in senile plaques with end-specific Aβ monoclonals: evidence that an initially deposited species is Aβ42. Neuron 1994, 13, 45–53. [Google Scholar] [CrossRef]
- Hardy, J.A.; Higgins, G.A. Alzheimer’s disease: The amyloid cascade hypothesis. Science 1992, 256, 184–185. [Google Scholar] [CrossRef]
- Citron, M.; Oltersdorf, T.; Haass, C.; McConlogue, L.; Hung, A.Y.; Seubert, P.; Vigo-Pelfrey, C.; Lieberburg, I.; Selkoe, D.J. Mutation of the β-amyloid precursor protein in familial Alzheimer’s disease increases β-protein production. Nature 1992, 360, 672–674. [Google Scholar] [CrossRef]
- Suzuki, N.; Cheung, T.T.; Cai, X.; Odaka, A.; Otvos, L. Jr.; Eckman, C.; Golde, T.E.; Younkin, S.G. An increased percentage of long amyloid β protein secreted by familial amyloid β protein precursor (βAPP717) mutants. Science 1994, 264, 1336–1340. [Google Scholar] [CrossRef]
- Strittmatter, W.J.; Saunders, A.M.; Schmechel, D.; Pericak-Vance, M.; Enghild, J.; Salvesen, G.S.; Roses, A.D. Apolipoprotein E: high-avidity binding to β-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc. Natl. Acad. Sci. USA 1993, 90, 1977–1981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Grupe, A.; Rowland, C.; Nowotny, P.; Kauwe, J.S.K.; Smemo, S.; Hinrichs, A.; Tacey, K.; Toombs, T.A.; Kwok, S.; et al. DAPK1 variants are associated with Alzheimer’s disease and allele-specific expression. Human Mol. Genet. 2006, 15, 2560–2568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, K.; Liu, S.; Yao, S.; Wang, B.; Dai, D.; Yao, L. Interaction between interleukin-8 and methylenetetrahydrofolate reductase genes modulates Alzheimer’s disease risk. Dement. Geriatr. Cogn. Disord. 2009, 27, 286–291. [Google Scholar] [CrossRef] [PubMed]
- Van Rensburg, S.J.; Carstens, M.E.; Potocnik, F.C.; Aucamp, A.K.; Taljaard, J.J. Increased frequency of the transferrin C2 subtype in Alzheimer’s disease. Neuroreport 1993, 4, 1269–1271. [Google Scholar] [CrossRef] [PubMed]
- Ciarlo, E.; Massone, S.; Penna, I.; Nizzari, M.; Gigoni, A.; Dieci, G.; Russo, C.; Florio, T.; Cancedda, R.; Pagano, A. An intronic ncRNA-dependent regulation of SORL1 expression affecting Aβ formation is upregulated in post-mortem Alzheimer’s disease brain samples. Dis. Models Mech. 2013, 6, 424–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacobsen, L.; Madsen, P.; Moestrup, S.K.; Lund, A.H.; Tommerup, N.; Nykjær, A.; Sottrup-Jensen, L.; Gliemann, J.; Petersen, C.M. Molecular characterization of a novel human hybrid-type receptor that binds the α2-macroglobulin receptor-associated protein. J. Biol. Chem. 1996, 271, 31379–31383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Motoi, Y.; Aizawa, T.; Haga, S.; Nakamura, S.; Namba, Y.; Ikeda, K. Neuronal localization of a novel mosaic apolipoprotein E receptor, LR11, in rat and human brain. Brais Res. 1999, 833, 209–215. [Google Scholar] [CrossRef]
- Rogaeva, E.; Meng, Y.; Lee, J.H.; Gu, Y.; Kawarai, T.; Zou, F.; Katayama, T.; Baldwin, C.T.; Cheng, R.; Hasegawa, H.; et al. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer’s disease. Nat. Genet. 2007, 39, 168–177. [Google Scholar] [CrossRef]
- Tan, L.; Yu, J.; Hu, N.; Tan, L. Non-coding RNAs in Alzheimer’s disease. Mol. Neurobiol. 2013, 47, 382–393. [Google Scholar] [CrossRef]
- Idda, M.L.; Munk, R.; Abdelmohsen, K.; Gorospe, M. Noncoding RNAs in Alzheimer’s disease. WIRES RNA 2017, 9, e1463. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Lee, Y.; Yeom, K.; Kim, Y.; Jin, H.; Kim, V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004, 18, 3016–3027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yi, R.; Qin, Y.; Macara, I.G.; Cullen, B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003, 17, 3011–3016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366. [Google Scholar] [CrossRef]
- Hutvágner, G.; McLachlan, J.; Pasquinelli, A.E.; Bálint, É.; Tuschl, T.; Zamore, P.D. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 2001, 293, 834–838. [Google Scholar] [CrossRef] [Green Version]
- Chendrimada, T.P.; Gregory, R.I.; Kumaraswamy, E.; Norman, J.; Cooch, N.; Nishikura, K.; Shiekhattar, R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 2005, 436, 740–744. [Google Scholar] [CrossRef]
- Yoda, M.; Kawamata, T.; Paroo, Z.; Ye, X.; Iwasaki, S.; Liu, Q.; Tomari, Y. ATP-dependent human RISC assembly pathways. Nat. Struct. Mol. Biol. 2010, 17, 17–23. [Google Scholar] [CrossRef] [Green Version]
- Khvorova, A.; Reynolds, A.; Jayasena, S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell 2003, 115, 209–216. [Google Scholar] [CrossRef] [Green Version]
- Amakiri, N.; Kubosumi, A.; Tran, J.; Reddy, P.H. Amyloid beta and microRNAs in Alzheimer’s disease. Front. Neurosci. 2019, 13, 430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brás, A.; Rodrigues, A.S.; Gomes, B.; Rueff, J. Down syndrome and microRNAs. Biomed. Rep. 2018, 8, 11–16. [Google Scholar] [PubMed]
- Paul, P.; Chakraborty, A.; Sarkar, D.; Langthasa, M.; Rahman, M.; Bari, M.; Singha, R.K.S.; Malakar, A.K.; Chakraborty, S. Interplay between miRNAs and human disease. J. Cell. Physiol. 2018, 233, 2007–2018. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.; Agarwala, P.; Jayaraj, G.G.; Gargallo, R.; Maiti, S. The RNA stem-loop to G-quadruplex equilibrium controls mature microRNA production inside the cell. Biochemistry 2015, 54, 7067–7078. [Google Scholar] [CrossRef] [Green Version]
- Arachchilage, G.M.; Dassanayake, A.C.; Basu, S. A potassium ion-dependent RNA structural switch regulates pre-miRNA 92b maturation. Chem. Biol. 2015, 22, 262–272. [Google Scholar]
- Williamson, J.R.; Raghuraman, M.K.; Cech, T.R. Monovalent cation-induced structure of telomeric DNA: The G-quartet model. Cell 1989, 59, 871–880. [Google Scholar] [CrossRef]
- Williamson, J.R. G-quartet structures in telomeric DNA. Annu. Rev. Biophys. Biomol. Struct. 1994, 23, 703–730. [Google Scholar] [CrossRef]
- Ghanbari, M.; Ikram, M.A.; de Looper, H.W.J.; Hofman, A.; Erkeland, S.J.; Franco, O.H.; Dehghan, A. Genome-wide identification of microRNA-related variants associated with risk of Alzheimer’s disease. Sci. Rep. 2016, 6, 28387. [Google Scholar] [CrossRef] [Green Version]
- Balasubramanian, S.; Neidle, S. G-quadruplex nucleic acids as therapeutic targets. Curr. Opin. Chem. Biol. 2009, 13, 345–353. [Google Scholar] [CrossRef] [Green Version]
- Fürtig, B.; Richter, C.; Wöhnert, J.; Schwalbe, H. NMR spectroscopy of RNA. ChemBioChem 2003, 4, 936–962. [Google Scholar] [CrossRef]
- Joachimi, A.; Benz, A.; Hartig, J.S. A comparison of DNA and RNA quadruplex structures and stabilities. Bioorg. Med. Chem. 2009, 19, 6811–6815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyoshi, D.; Nakao, A.; Sugimoto, N. Structural transition from antiparallel to parallel G-quadruplex of d(G4T4G4) induced by Ca2+. Nucleic Acids Res. 2003, 31, 1156–1163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ranjbar, B.; Gill, P. Circular dichroism techniques: biomolecular and nanostructural analyses—A review. Chem. Biol. Drug Des. 2009, 74, 101–120. [Google Scholar] [CrossRef] [PubMed]
- Mergny, J.L.; Phan, A.T.; Lacroix, L. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998, 435, 74–78. [Google Scholar] [CrossRef] [Green Version]
- Menon, L.; Mader, S.A.; Mihailescu, M. Fragile X mental retardation protein interactions with the microtubule associated protein 1B RNA. RNA 2008, 14, 1644–1655. [Google Scholar] [CrossRef] [Green Version]
- Hardin, C.C.; Perry, A.G.; White, K. Thermodynamic and kinetic characterization of the dissociation and assembly of quadruplex nucleic acids. Biopolymers 2001, 56, 147–194. [Google Scholar] [CrossRef]
- Hendry, P.; Hannan, G. Detection and quantitation of unlabeled nucleic acids in polyacrylamide gels. BioTechniques 1996, 20, 258–264. [Google Scholar] [CrossRef]
- Arthanari, H.; Basu, S.; Kawano, T.L.; Bolton, P.H. Fluorescent dyes specific for quadruplex DNA. Nucleic Acids Res. 1998, 26, 3724–3728. [Google Scholar] [CrossRef] [Green Version]
- Glemarec, C.; Kufel, J.; Földesi, A.; Maltseva, T.; Sandström, A.; Kirsebom, L.A.; Chattopadhyaya, J. The NMR structure of 31mer RNA domain of Escherichia coli RNAse P RNA using its non-uniformly deuterium labelled counterpart [the ‘NMR-window’ concept]. Nucleic Acids Res. 1996, 24, 2022–2035. [Google Scholar] [CrossRef] [Green Version]
- Shankar, N.; Kennedy, S.D.; Chen, G.; Krugh, T.R.; Turner, D.H. The NMR structure of an internal loop from 23S ribosomal RNA differs from its structure in crystals of 50S ribosomal subunits. Biochemistry 2006, 45, 11776–11789. [Google Scholar] [CrossRef] [Green Version]
- Serra, M.J.; Baird, J.D.; Dale, T.; Fey, B.L.; Retatagos, K.; Westhof, E. Effects of magnesium ions on the stabilization of RNA oligomers of defined structures. RNA 2002, 8, 307–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ke, S.; Yang, Z.; Yang, F.; Wang, X.; Tan, J.; Liao, B. Long noncoding RNA NEAT1 aggravates Aβ-induced neuronal damage by targeting miR-107 in Alzheimer’s disease. Yonsei Med. J. 2019, 60, 640–650. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Ling, S.; Sun, W.; Liu, T.; Li, Y.; Zhong, G.; Zhao, D.; Zhang, P.; Song, J.; Jin, X.; et al. Circulating microRNAs correlated with the level of coronary artery calcification in symptomatic patients. Sci. Rep. 2015, 5, 16099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, H.; Yu, C.; Zhang, H.; Zhang, S.; Yu, W.; Yang, Y.; Chen, Q. Exosomal miR-1229 derived from colorectal cancer cells promotes angiogenesis by targeting HIPK2. Int. J. Biol. Macromol. 2019, 132, 470–477. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.; Zheng, H.; Liu, X.; Zhang, W.; Zhu, J.; Wu, G.; Cao, L.; Song, J.; Wu, S.; Song, L. MicroRNA-1229 overexpression promotes cell proliferation and tumorigenicity and activates Wnt/β-catenin signaling in breast cancer. Oncotarget 2016, 7, 24076–24087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milligan, J.F.; Uhlenbeck, O.C. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 1989, 180, 51–62. [Google Scholar] [PubMed]
- Piotto, M.; Saudek, V.; Sklenár, V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR 1992, 2, 661–665. [Google Scholar] [CrossRef]





| pre-miRNA-1229_WT GQ | 5′ GGGUAGGGUUUGGGGGAGAGCGUGGGCUGGGGUUCAGGG ACA 3′ |
| pre-miRNA-1229_WT FL | 5′ GGUGGGUAGGGUUUGGGGGAGAGCGUGGGCUGGGG UUCAGGGACACCCUCUCACCACUGCCCUCCCACAG 3′ |
| pre-miRNA-1229_SNP GQ | 5′ GGGUAGGGUUUGGGGGAGAGUGUGGGCUGGGGUUCAGGG ACA 3′ |
| pre-miRNA-1229_SNP FL | 5′ GGUGGGUAGGGUUUGGGGGAGAGUGUGGGCUGGGG UUCAGGGACACCCUCUCACCACUGCCCUCCCACAG 3′ |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Imperatore, J.A.; Then, M.L.; McDougal, K.B.; Mihailescu, M.R. Characterization of a G-Quadruplex Structure in Pre-miRNA-1229 and in Its Alzheimer’s Disease-Associated Variant rs2291418: Implications for miRNA-1229 Maturation. Int. J. Mol. Sci. 2020, 21, 767. https://doi.org/10.3390/ijms21030767
Imperatore JA, Then ML, McDougal KB, Mihailescu MR. Characterization of a G-Quadruplex Structure in Pre-miRNA-1229 and in Its Alzheimer’s Disease-Associated Variant rs2291418: Implications for miRNA-1229 Maturation. International Journal of Molecular Sciences. 2020; 21(3):767. https://doi.org/10.3390/ijms21030767
Chicago/Turabian StyleImperatore, Joshua A., McKenna L. Then, Keefe B. McDougal, and Mihaela Rita Mihailescu. 2020. "Characterization of a G-Quadruplex Structure in Pre-miRNA-1229 and in Its Alzheimer’s Disease-Associated Variant rs2291418: Implications for miRNA-1229 Maturation" International Journal of Molecular Sciences 21, no. 3: 767. https://doi.org/10.3390/ijms21030767
APA StyleImperatore, J. A., Then, M. L., McDougal, K. B., & Mihailescu, M. R. (2020). Characterization of a G-Quadruplex Structure in Pre-miRNA-1229 and in Its Alzheimer’s Disease-Associated Variant rs2291418: Implications for miRNA-1229 Maturation. International Journal of Molecular Sciences, 21(3), 767. https://doi.org/10.3390/ijms21030767