Identification of Potential Biomarkers of Radiation Exposure in Blood Cells by Capillary Electrophoresis Time-of-Flight Mass Spectrometry
Abstract
1. Introduction
2. Results
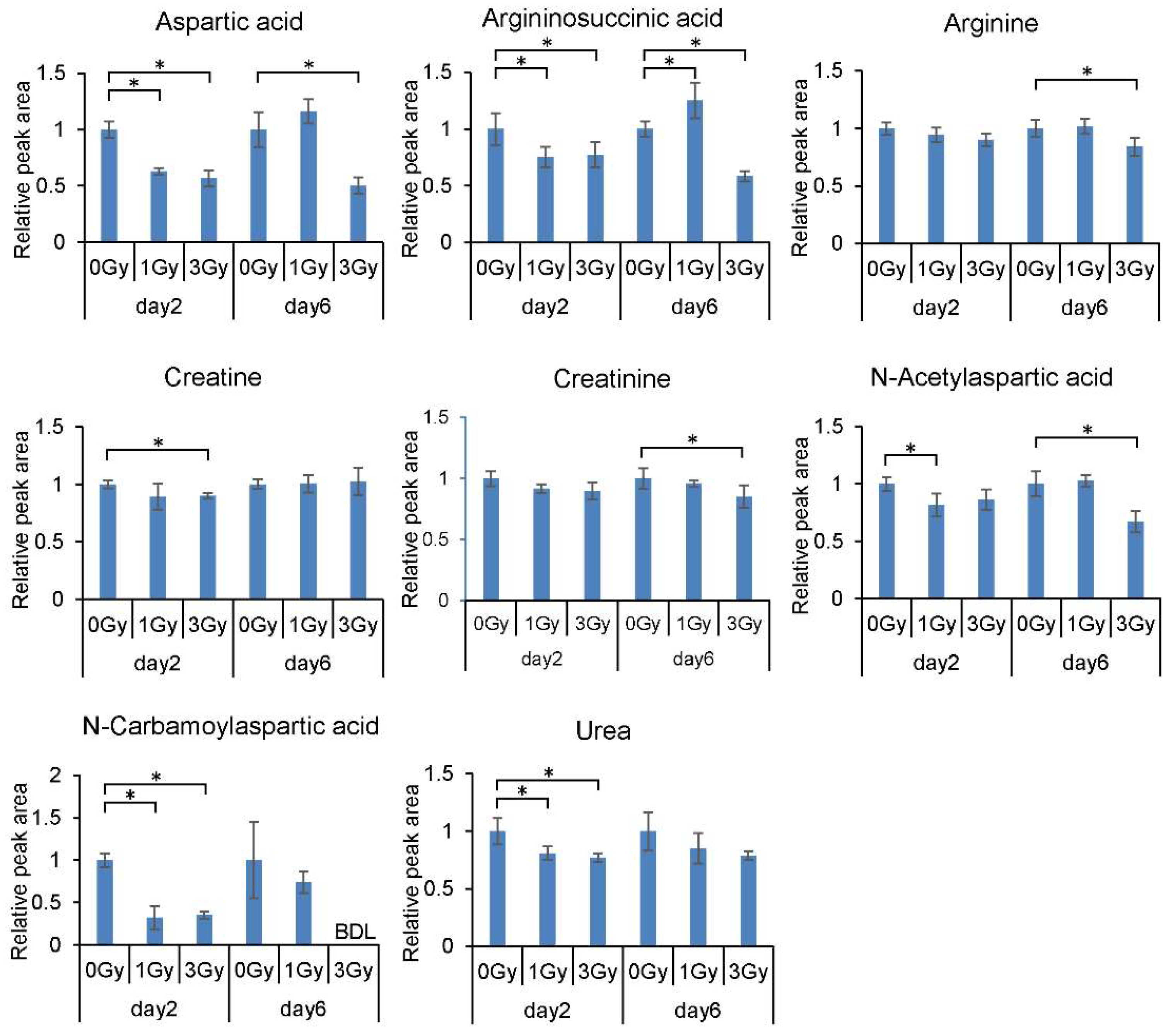
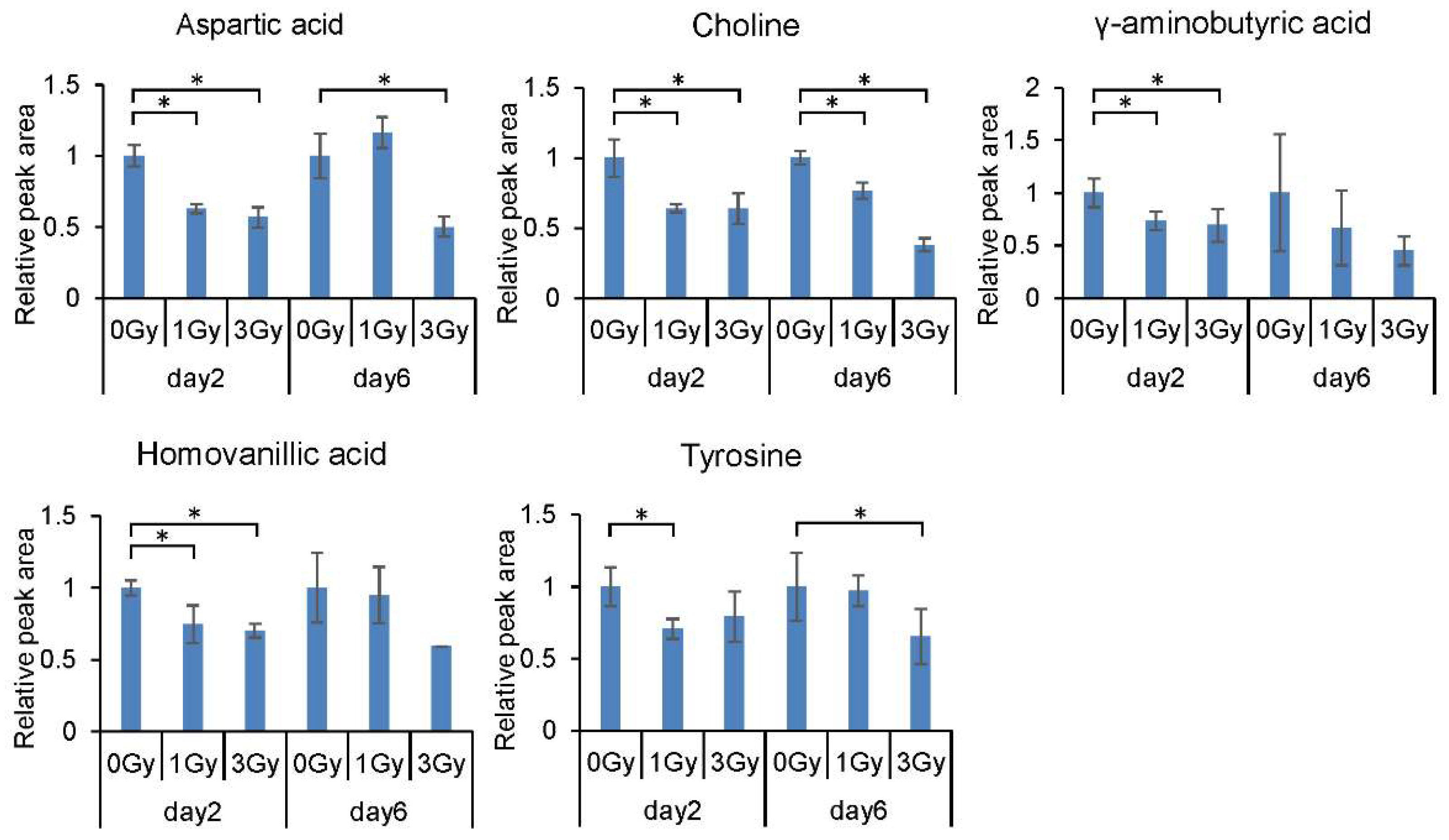
2.1. Changes in the Levels of Blood Cell Metabolites Following Exposure to Ionizing Radiation
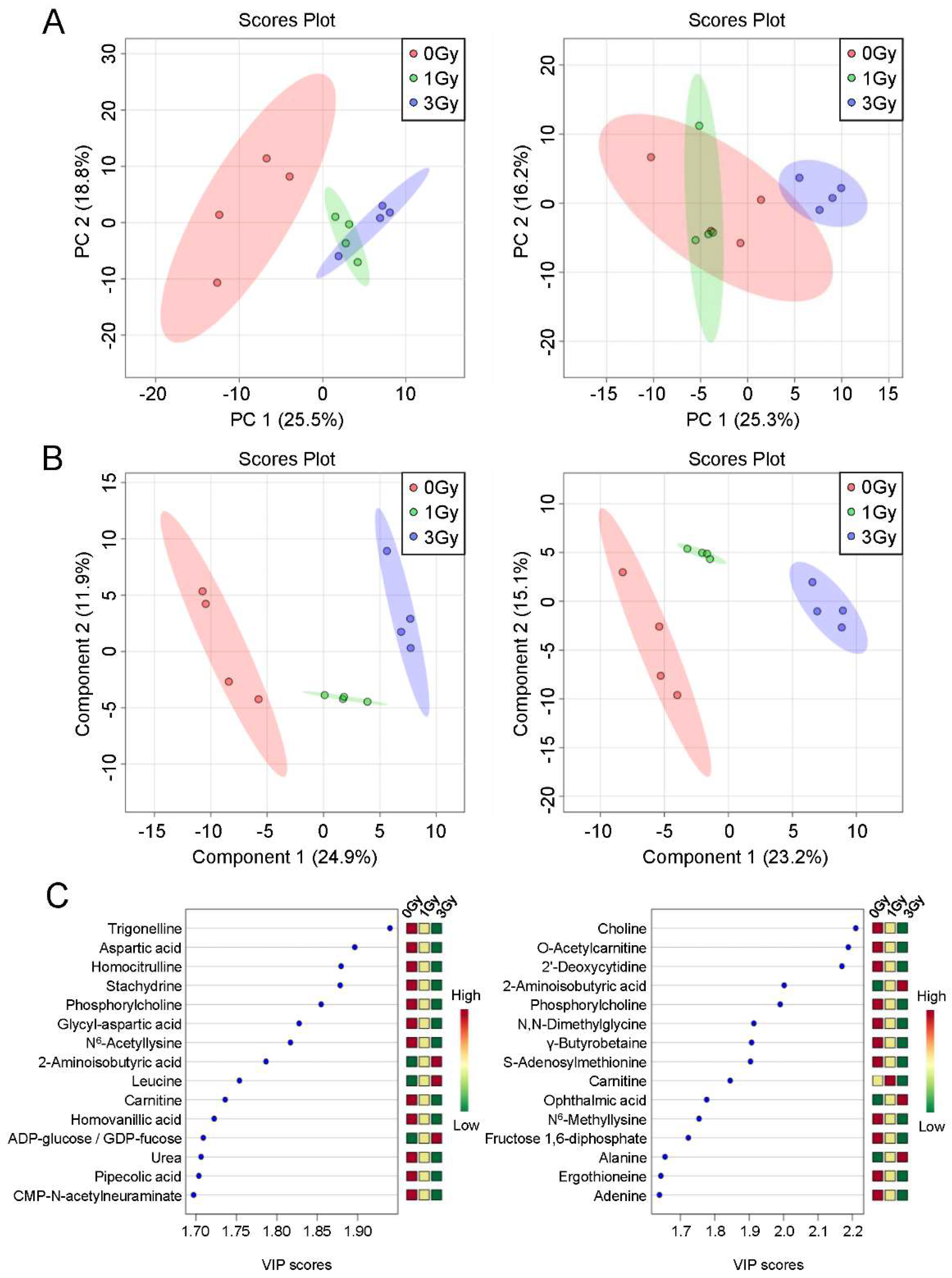
2.2. Multivariate Analysis of Blood Cell Metabolites Following Exposure to Ionizing Radiation
2.3. Establishment of a Potential Exposure Dose Prediction Panel
3. Discussion
4. Materials and Methods
4.1. Animals, Exposure, and Preparation of Blood Cells
4.2. Metabolite Extraction
4.3. Measurement of Blood Cell Metabolites by CE–TOFMS
4.4. Statistical Analysis
4.5. Ethical Considerations
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pernot, E.; Hall, J.; Baatout, S.; Benotmane, M.A.; Blanchardon, E.; Bouffler, S.; El Saghire, H.; Gomolka, M.; Guertler, A.; Harms-Ringdahl, M. Ionizing radiation biomarkers for potential use in epidemiological studies. Mutat. Res. 2012, 751, 258–286. [Google Scholar] [CrossRef] [PubMed]
- Williams, B.B.; Flood, A.B.; Demidenko, E.; Swartz, H.M. ROC analysis for evaluation of radiation biodosimetry technologies. Radiat. Prot. Dosim. 2016, 172, 145–151. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Simon, S.L.; Bouville, A. Long-term biodosimetry redux. Radiat. Prot. Dosim. 2016, 172, 244–247. [Google Scholar] [CrossRef] [PubMed]
- Oestreicher, U.; Samaga, D.; Ainsbury, E.; Antunes, A.C.; Baeyens, A.; Barrios, L.; Beinke, C.; Beukes, P.; Blakely, W.F.; Cucu, A. RENEB intercomparisons applying the conventional Dicentric Chromosome Assay (DCA). Int. J. Radiat. Biol. 2017, 93, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Moquet, J.; Barnard, S.; Rothkamm, K. Gamma-H2AX biodosimetry for use in large scale radiation incidents: Comparison of a rapid ‘96 well lyse/fix’protocol with a routine method. PeerJ 2014, 2, e282. [Google Scholar] [CrossRef] [PubMed]
- Williams, B.B.; Dong, R.; Flood, A.B.; Grinberg, O.; Kmiec, M.; Lesniewski, P.N.; Matthews, T.P.; Nicolalde, R.J.; Raynolds, T.; Salikhov, I.K. A deployable in vivo EPR tooth dosimeter for triage after a radiation event involving large populations. Radiat. Meas. 2011, 46, 772–777. [Google Scholar] [CrossRef]
- Grace, M.; McLeland, C.; Blakely, W. Real-time quantitative RT-PCR assay of GADD45 gene expression changes as a biomarker for radiation biodosimetry. Int. J. Radiat. Biol. 2002, 78, 1011–1021. [Google Scholar] [CrossRef]
- Sun, L.; Inaba, Y.; Sato, K.; Hirayama, A.; Tsuboi, K.; Okazaki, R.; Chida, K.; Moritake, T. Dose-dependent decrease in anti-oxidant capacity of whole blood after irradiation: A novel potential marker for biodosimetry. Sci. Rep. 2018, 8, 7425. [Google Scholar] [CrossRef]
- Sproull, M.; Camphausen, K. State-of-the-art advances in radiation biodosimetry for mass casualty events involving radiation exposure. Radiat. Res. 2016, 186, 423–435. [Google Scholar] [CrossRef]
- Abe, M.; Takahashi, M.; Takeuchi, K.; Fukuda, M. Studies on the significance of taurine in radiation injury. Radiat. Res. 1968, 33, 563–573. [Google Scholar] [CrossRef]
- Rist, M.J.; Roth, A.; Frommherz, L.; Weinert, C.H.; Krüger, R.; Merz, B.; Bunzel, D.; Mack, C.; Egert, B.; Bub, A. Metabolite patterns predicting sex and age in participants of the Karlsruhe Metabolomics and Nutrition (KarMeN) study. PLoS ONE 2017, 12, e0183228. [Google Scholar] [CrossRef] [PubMed]
- Nishiumi, S.; Kobayashi, T.; Kawana, S.; Unno, Y.; Sakai, T.; Okamoto, K.; Yamada, Y.; Sudo, K.; Yamaji, T.; Saito, Y. Investigations in the possibility of early detection of colorectal cancer by gas chromatography/triple-quadrupole mass spectrometry. Oncotarget 2017, 8, 17115. [Google Scholar] [CrossRef] [PubMed]
- Vlaanderen, J.; Janssen, N.; Hoek, G.; Keski-Rahkonen, P.; Barupal, D.; Cassee, F.; Gosens, I.; Strak, M.; Steenhof, M.; Lan, Q. The impact of ambient air pollution on the human blood metabolome. Environ. Res. 2017, 156, 341–348. [Google Scholar] [CrossRef] [PubMed]
- Hall, J.; Jeggo, P.A.; West, C.; Gomolka, M.; Quintens, R.; Badie, C.; Laurent, O.; Aerts, A.; Anastasov, N.; Azimzadeh, O. Ionizing radiation biomarkers in epidemiological studies–an update. Mutat. Res. 2017, 771, 59–84. [Google Scholar] [CrossRef]
- Pannkuk, E.L.; Laiakis, E.C.; Authier, S.; Wong, K.; Fornace, A.J., Jr. Targeted Metabolomics of Nonhuman Primate Serum after Exposure to Ionizing Radiation: Potential Tools for High-throughput Biodosimetry. RSC Adv. 2016, 6, 51192–51202. [Google Scholar] [CrossRef]
- Pannkuk, E.L.; Laiakis, E.C.; Mak, T.D.; Astarita, G.; Authier, S.; Wong, K.; Fornace, A.J., Jr. A Lipidomic and Metabolomic Serum Signature from Nonhuman Primates Exposed to Ionizing Radiation. Metabolomics 2016, 12, 80. [Google Scholar] [CrossRef]
- Laiakis, E.C.; Strassburg, K.; Bogumil, R.; Lai, S.; Vreeken, R.J.; Hankemeier, T.; Langridge, J.; Plumb, R.S.; Fornace, A.J., Jr.; Astarita, G. Metabolic phenotyping reveals a lipid mediator response to ionizing radiation. J. Proteome. Res. 2014, 13, 4143–4154. [Google Scholar] [CrossRef]
- Broin, P.Ó.; Vaitheesvaran, B.; Saha, S.; Hartil, K.; Chen, E.I.; Goldman, D.; Fleming, W.H.; Kurland, I.J.; Guha, C.; Golden, A. Intestinal microbiota-derived metabolomic blood plasma markers for prior radiation injury. Int. J. Radiat. Oncol. Biol. Phys. 2015, 91, 360–367. [Google Scholar] [CrossRef]
- Iizuka, D.; Yoshioka, S.; Kawai, H.; Izumi, S.; Suzuki, F.; Kamiya, K. Metabolomic screening using ESI-FT MS identifies potential radiation-responsive molecules in mouse urine. J. Radiat. Res. 2017, 58, 273–280. [Google Scholar] [CrossRef]
- Tyburski, J.B.; Patterson, A.D.; Krausz, K.W.; Slavik, J.; Fornace, A.J., Jr.; Gonzalez, F.J.; Idle, J.R. Radiation metabolomics. 2. Dose- and time-dependent urinary excretion of deaminated purines and pyrimidines after sublethal gamma-radiation exposure in mice. Radiat. Res. 2009, 172, 42–57. [Google Scholar] [CrossRef]
- Laiakis, E.C.; Strawn, S.J.; Brenner, D.J.; Fornace, A.J., Jr. Assessment of Saliva as a Potential Biofluid for Biodosimetry: A Pilot Metabolomics Study in Mice. Radiat. Res. 2016, 186, 92–97. [Google Scholar] [CrossRef] [PubMed]
- Chaleckis, R.; Murakami, I.; Takada, J.; Kondoh, H.; Yanagida, M. Individual variability in human blood metabolites identifies age-related differences. Proc. Natl. Acad. Sci. USA 2016, 113, 4252–4259. [Google Scholar] [CrossRef] [PubMed]
- Teruya, T.; Chaleckis, R.; Takada, J.; Yanagida, M.; Kondoh, H. Diverse metabolic reactions activated during 58-hr fasting are revealed by non-targeted metabolomic analysis of human blood. Sci. Rep. 2019, 9, 854. [Google Scholar] [CrossRef] [PubMed]
- Yushmanov, V.E. Evaluation of radiation injury by 1H and 31P NMR of human urine. Magn. Reson. Med. 1994, 31, 48–52. [Google Scholar] [CrossRef] [PubMed]
- Pannkuk, E.L.; Laiakis, E.C.; Girgis, M.; Dowd, S.E.; Dhungana, S.; Nishita, D.; Bujold, K.; Bakke, J.; Gahagen, J.; Authier, S.; et al. Temporal Effects on Radiation Responses in Nonhuman Primates: Identification of Biofluid Small Molecule Signatures by Gas Chromatography(-)Mass Spectrometry Metabolomics. Metabolites 2019, 9, 98. [Google Scholar] [CrossRef] [PubMed]
- Pannkuk, E.L.; Laiakis, E.C.; Garcia, M.; Fornace, A.J., Jr.; Singh, V.K. Nonhuman Primates with Acute Radiation Syndrome: Results from a Global Serum Metabolomics Study after 7.2 Gy Total-Body Irradiation. Radiat. Res. 2018, 190, 576–583. [Google Scholar] [CrossRef]
- Ramautar, R.; Nevedomskaya, E.; Mayboroda, O.A.; Deelder, A.M.; Wilson, I.D.; Gika, H.G.; Theodoridis, G.A.; Somsen, G.W.; de Jong, G.J. Metabolic profiling of human urine by CE-MS using a positively charged capillary coating and comparison with UPLC-MS. Mol. Biosyst. 2011, 7, 194–199. [Google Scholar] [CrossRef]
- Stewart, F.; Akleyev, A.; Hauer-Jensen, M.; Hendry, J.; Kleiman, N.; Macvittie, T.; Aleman, B.; Edgar, A.; Mabuchi, K.; Muirhead, C. ICRP publication 118: ICRP statement on tissue reactions and early and late effects of radiation in normal tissues and organs–threshold doses for tissue reactions in a radiation protection context. Ann. ICRP 2012, 41, 1–322. [Google Scholar] [CrossRef]
- Carlisle, S.; Burchart, P.; Mitchel, R. Cancer and non-cancer risks in normal and cancer-prone Trp53 heterozygous mice exposed to high-dose radiation. Radiat. Res. 2010, 173, 40–48. [Google Scholar] [CrossRef]
- Flood, A.B.; Ali, A.N.; Boyle, H.K.; Du, G.; Satinsky, V.A.; Swarts, S.G.; Williams, B.B.; Demidenko, E.; Schreiber, W.; Swartz, H.M. Evaluating the Special Needs of The Military for Radiation Biodosimetry for Tactical Warfare Against Deployed Troops: Comparing Military to Civilian Needs for Biodosimetry Methods. Health Phys. 2016, 111, 169–182. [Google Scholar] [CrossRef][Green Version]
- Pannkuk, E.L.; Laiakis, E.C.; Gill, K.; Jain, S.K.; Mehta, K.Y.; Nishita, D.; Bujold, K.; Bakke, J.; Gahagen, J.; Authier, S.; et al. Liquid Chromatography-Mass Spectrometry-Based Metabolomics of Nonhuman Primates after 4 Gy Total Body Radiation Exposure: Global Effects and Targeted Panels. J. Proteome Res. 2019, 18, 2260–2269. [Google Scholar] [CrossRef] [PubMed]
- Bryk, A.H.; Wiśniewski, J.R. Quantitative analysis of human red blood cell proteome. J. Proteome Res. 2017, 16, 2752–2761. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, Y.; Itoh, N.; Hayakawa, M.; Habuchi, Y.; Inoue, R.; Chen, Z.-H.; Cao, J.; Cynshi, O.; Niki, E. Lipid peroxidation in mice fed a choline-deficient diet as evaluated by total hydroxyoctadecadienoic acid. Nutrition 2006, 22, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Pannkuk, E.L.; Laiakis, E.C.; Authier, S.; Wong, K.; Fornace, A.J., Jr. Gas chromatography/mass spectrometry metabolomics of urine and serum from nonhuman primates exposed to ionizing radiation: Impacts on the tricarboxylic acid cycle and protein metabolism. J. Proteome Res. 2017, 16, 2091–2100. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, N.; Shinoda, K.; Sato, H.; Sasaki, K.; Suzuki, M.; Yamaki, K.; Fujimori, T.; Yamamoto, H.; Osei-Hyiaman, D.; Ohashi, Y. Plasma metabolome analysis of patients with major depressive disorder. Psychiatry Clin. Neurosci. 2018, 72, 349–361. [Google Scholar] [CrossRef]
- Petty, F.; Kramer, G.L.; Gullion, C.M.; Rush, A.J. Low plasma γ-aminobutyric acid levels in male patients with depression. Biol. Psychiatry 1992, 32, 354–363. [Google Scholar] [CrossRef]
- Trivedi, R.; Khan, A.R.; Rana, P.; Haridas, S.; Hemanth Kumar, B.; Manda, K.; Rathore, R.K.; Tripathi, R.P.; Khushu, S. Radiation-induced early changes in the brain and behavior: Serial diffusion tensor imaging and behavioral evaluation after graded doses of radiation. J. Neurosci. Res. 2012, 90, 2009–2019. [Google Scholar] [CrossRef]
- Brown, P.D.; Pugh, S.; Laack, N.N.; Wefel, J.S.; Khuntia, D.; Meyers, C.; Choucair, A.; Fox, S.; Suh, J.H.; Roberge, D. Memantine for the prevention of cognitive dysfunction in patients receiving whole-brain radiotherapy: A randomized, double-blind, placebo-controlled trial. Neuro-Oncology 2013, 15, 1429–1437. [Google Scholar] [CrossRef]
- Ohashi, Y.; Hirayama, A.; Ishikawa, T.; Nakamura, S.; Shimizu, K.; Ueno, Y.; Tomita, M.; Soga, T. Depiction of metabolome changes in histidine-starved Escherichia coli by CE-TOFMS. Mol. BioSyst. 2008, 4, 135–147. [Google Scholar] [CrossRef]
- Ooga, T.; Sato, H.; Nagashima, A.; Sasaki, K.; Tomita, M.; Soga, T.; Ohashi, Y. Metabolomic anatomy of an animal model revealing homeostatic imbalances in dyslipidaemia. Mol. Biosyst. 2011, 7, 1217–1223. [Google Scholar] [CrossRef]
- Sugimoto, M.; Wong, D.T.; Hirayama, A.; Soga, T.; Tomita, M. Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics 2010, 6, 78–95. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Wishart, D.S.; Xia, J. Using metaboanalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef] [PubMed]

| Metabolite | Category † | Day 2 | Day 6 | ||
|---|---|---|---|---|---|
| Fold Change | Fold Change | ||||
| 1 Gy/0 Gy | 3 Gy/0 Gy | 1 Gy/0 Gy | 3 Gy/0 Gy | ||
| Increased (38 metabolites) | |||||
| 2-Aminobutyric acid | 1.58 * | 1.7 * | 1.27 * | 1.45 * | |
| 3-Phosphoglyceric acid | Sugar metabolism | 1.33 | 1.71 * | 1.05 | 0.74 |
| 5-Oxohexanoic acid | 1.48 * | 1.21 | 1.37 * | 1.22 | |
| ADP-glucose | Nucleotide sugars, Nucleotide sugars, Vascular | 1.22 | 1.42 * | 1.08 | 1.07 |
| GDP-fucose | |||||
| ADP-ribose | 0.48 | 0.87 | 1.1 | 1.83 * | |
| Alanine | Cytokine, Hormone, Hemocyte, Renal disease, Uremic toxin, Sugar metabolism | 1.04 | 1.06 | 1.12 | 1.3 * |
| ATP | Purine bases, Sugar metabolism | 1.22 | 1.46 * | 1.09 | 0.81 |
| Cumic acid | N.D. | N.D. | 1.4 * | 1.1 | |
| Decanoic acid | Lipid, Fatty Acid metabolism | 1.25 | 1.01 | 1.38 * | 1.06 |
| Flavin mononucleotide | 1.62 | 2.08 * | 1.29 * | 0.78 | |
| Glutamine | Hemocyte | 0.99 | 1.15 * | 1.05 | 1 |
| Glutathione (GSH) | Anti-oxidant, Lipid Fatty Acid metabolism, Methylglyoxal | 0.83 | 1.67 * | 0.92 | 0.95 |
| GTP | Purine bases | 1.38 | 1.76 * | 1.24 | 0.81 |
| Heptanoic acid | Lipid, Fatty Acid metabolism | 1.51 | 1 | 1.4 * | 1.17 |
| Hexanoic acid | Lipid, Fatty Acid metabolism | 1.33 | 1.06 | 1.21 * | 1.02 |
| Histidine | 1.06 | 1.18 * | 1.01 | 1.02 | |
| Isoleucine | Essential amino acid, Lipid, Fatty Acid metabolism, Liver disease, Sugar metabolism | 1.03 | 1.27 * | 1.12 | 1.01 |
| Isobutyric acid | Apoptosis, Carcinogenesis, Cell function, Lipid, Fatty Acid metabolism | 1.67 * | 1.11 | 1.08 | 1.12 |
| Butyric acid | |||||
| Isovaleric acid | Lipid, Fatty Acid metabolism | 1.57 * | 1.16 | 1.19 | 1 |
| Valeric acid | |||||
| Leucine | Essential amino acid, Insulin, Lipid, Fatty Acid metabolism, Liver disease, Protein metabolism | 1.11 | 1.33 * | 1.17 | 1.19 * |
| Malonylcarnitine | 1.02 | 1.16 | 1.31 * | 0.85 | |
| Octanoic acid | Lipid, Fatty Acid metabolism | 1.02 | 0.94 | 1.15 * | 0.98 |
| Octanoylcarnitine | 0.53 | 0.81 | 1.31 * | N.D. | |
| Ophthalmic acid | 1.11 | 1.16 | 1.15 | 1.34 * | |
| p-Toluic acid | 1.7 * | 1.12 | N.D. | N.D. | |
| m-Toluic acid | |||||
| o-Toluic acid | |||||
| Pelargonic acid | Lipid, Fatty Acid metabolism | 1.33 | 1.04 | 1.22 * | 0.99 |
| Phenylalanine | Catecholamines and Derivatives, Essential amino acid | 1.04 | 1.17 * | 1.06 | 0.92 |
| Phosphoenolpyruvic acid | Sugar metabolism | 1.45 | 1.9 * | 1.13 | 0.75 |
| S-Methylcysteine | Anti-oxidant, Sugar metabolism | 0.97 | 1.17 * | N.D. | N.D. |
| Sedoheptulose 7-phosphate | 1.44 | 1.24 | 1.19 | 1.63* | |
| Thiaproline | 1.13 | 1.52 * | 1.15 | 1.09 | |
| Tyramine | Catecholamines and Derivatives, Nervous system | N.D. | N.D. | 1.47 * | 1.17 |
| UDP-N-acetylgalactosamine-1 | 1.22 * | 1.27 * | 1.09 | 0.93 | |
| UDP-N-acetylglucosamine-1 | |||||
| Undecanoic acid | Lipid, Fatty Acid metabolism | N.D. | N.D. | 1.56 * | 1.09 |
| Valine | Essential amino acid, Liver disease, Nervous system, Sugar metabolism | 1.06 | 1.25 * | 1.06 | 1.02 |
| XA0002 (unknown peak) | 0.93 | 0.97 | 0.92 | 1.16 * | |
| XC0132 (unknown peak) | 1.13 | 1.26 * | N.D. | N.D. | |
| γ-Glutamyl-cysteine | 1.03 | 2.06 * | 0.83 | 0.87 | |
| Decreased (61 metabolites) | |||||
| 2-Hydroxy-4-methylvaleric acid | Maple syrup urine disease | 0.63 * | 0.6 * | 0.78 | 0.8 |
| 2’-Deoxycytidine | Pyrimidine bases | 0.57 * | 0.74 * | 0.73 * | 0.46 * |
| 3-Indoxylsulfuric acid | Renal disease, Uremic toxin, Vascular | 0.76 | 0.51 * | 0.61 | 0.78 |
| 5’-Deoxy-5’-methylthioadenosine | Purine bases | 0.84 | 0.55 * | 0.9 | 0.89 |
| 7,8-Dihydrobiopterin | Vascular | 0.41 * | 0.39 * | 0.99 | 0.3 * |
| Acetoacetamide | 0.8 | 0.65 * | N.D. | N.D. | |
| Adenine | Purine bases, Salvage pathway, Purine bases, Salvage pathway | 1.32 | 1.09 | 0.91 | 0.71 * |
| Arginine | Guanidino compounds | 0.94 | 0.9 | 1.02 | 0.84 * |
| Aspartic acid | Nervous system, Neuropsychiatric disorder, Sugar metabolism | 0.63 * | 0.57 * | 1.16 | 0.5 * |
| Betaine | Osmolytes, Renal disease, Uremic toxin, Transmethyration | 0.79 * | 0.76 * | 1.11 | 0.83 |
| Betonicine | 0.71 * | 0.69 * | 0.86 | N.D. | |
| Butyrylcarnitine | 0.73 | 0.63 * | 0.75 | 0.93 | |
| Carnitine | Lipid, Fatty Acid metabolism, Liver disease, Cardiac disease | 0.74 * | 0.72 * | 1.02 | 0.55 * |
| Choline | Lipid, Fatty Acid metabolism, Transmethyration | 0.64 * | 0.64 * | 0.77 * | 0.38 * |
| Citrulline | 0.87 | 0.8 * | 1.04 | 0.92 * | |
| CMP-N-acetylneuraminate | Nucleotide sugars | 0.45 * | 0.44 * | 1.01 | 0.95 |
| Creatine | Cell function | 0.89 | 0.9 * | 1.01 | 1.03 |
| Creatinine | Protein metabolism, Renal disease, Uremic toxin, | 0.92 | 0.9 | 0.95 | 0.85 * |
| Ectoine | 0.98 | 0.61 * | 0.84 | 0.67 | |
| Ergothioneine | Anti-oxidant, Oxidative stress | 0.98 | 1.03 | 0.95 | 0.69 * |
| Ethanolamine | 0.79 * | 0.89 | 1.08 | 0.59 | |
| Fructose 1,6-diphosphate | Sugar metabolism | 0.75 | 0.92 | 0.87 | 0.4 * |
| γ-Aminobutyric acid | Nervous system, Sugar metabolism | 0.73 * | 0.69 * | 0.67 | 0.45 |
| Glycine | 0.84 * | 0.92 | 1.13 | 0.85 | |
| Glycyl-aspartic acid | 0.46 * | 0.41 * | 1.3 | 0.36 * | |
| Glycerol 3-phosphate | Sugar metabolism | 0.62 * | 0.69 * | 1.05 | 0.91 |
| Hippuric acid | 0.98 | 0.57 * | 0.63 | 1.2 | |
| Homocitrulline | 0.84 | 0.65 * | 0.76 * | 0.79 | |
| Homovanillic acid | Catecholamines and Derivatives, Dopamine related substances, Nervous system | 0.74 * | 0.7 * | 0.95 | 0.6 |
| Imidazolelactic acid | 0.64 * | 0.85 | 0.85 | 0.71 | |
| Isethionic acid | 0.63 * | 0.63 * | 0.87 | 0.87 | |
| Methionine sulfoxide | Oxidative stress | 0.9 | 0.67 * | 0.95 | 1.13 |
| N-Acetylaspartic acid | N-Acetylated compounds, Nervous system | 0.82 * | 0.86 | 1.03 | 0.67 * |
| N-Acetylgalactosamine | Sugar metabolism | 0.91 | 0.99 | 1.08 | 0.69 * |
| N-Acetylmannosamine | |||||
| N-Acetylglucosamine | |||||
| N-Carbamoylaspartic acid | 0.32 * | 0.35 * | 0.74 | N.D. | |
| N-Methylproline | 0.94 | 0.77 * | 0.73 | 0.53 | |
| N,N-Dimethylglycine | Methylated compounds, Oxidative stress, Transmethyration | 0.96 | 0.87 | 0.9 | 0.8 * |
| N6-Acetyllysine | Apoptosis, Cell function, DNA damage, N-Acetylated compounds | 0.74 * | 0.55 * | 0.78 | 0.75 * |
| N6-Methyllysine | Methylated compounds, Transmethyration | 0.97 | 1.02 | 0.95 | 0.78 * |
| O-Acetylcarnitine | Lipid, Fatty Acid metabolism, Nervous system | 0.89 | 0.94 | 0.85 * | 0.61 * |
| Phosphorylcholine | Liver disease | 0.4 * | 0.32 * | 0.82 | 0.34 * |
| Pipecolic acid | Liver disease | 0.81 * | 0.77 * | 0.86 | 0.75 |
| Pyruvic acid | Ketosis, Lipid, Fatty Acid metabolism, Protein metabolism, Sugar metabolism | 0.72 * | 0.91 | 1.19 | 0.96 |
| Ribulose 5-phosphate | 0.58 * | 0.69 | 0.89 | 0.75 | |
| S-Adenosylmethionine | Liver disease, Neuropsychiatric disorder, Oxidative stress, Transmethyration | 0.94 | 1 | 0.94 | 0.87 * |
| S-Lactoylglutathione | Methylglyoxal | 0.58 * | 0.76 | 0.68 | 0.57 |
| Symmetric dimethylarginine | Methylated compounds, Protein metabolism, Renal disease, Uremic toxin, Transmethyration, Vascular | 0.91 | 1 | 1.05 | 0.6 * |
| Spermidine | Polyamines | 0.3 * | 0.41 * | 1.18 | 0.11 * |
| Stachydrine | Osmolytes, Renal disease, Uremic toxin, Transmethyration | 0.73 * | 0.61 * | 0.98 | 0.83 |
| Thymidine | Pyrimidine bases | 0.56 * | 0.77 * | 0.88 | 0.43 * |
| Trigonelline | 0.83 * | 0.62 * | 0.88 | 0.96 | |
| Trimethylamine N-oxide | Osmolytes, Renal disease, Uremic toxin | 0.69 | 0.55 * | 1.52 | 1.36 |
| Tyrosine | Catecholamines and Derivatives | 0.71 * | 0.79 | 0.97 | 0.65 * |
| Urea | Protein metabolism | 0.81 * | 0.77 * | 0.85 | 0.78 |
| XA0013 (unknown peak) | 0.65 | 0.42 * | N.D. | N.D. | |
| XA0033 (unknown peak) | 0.45 * | 0.39 * | N.D. | N.D. | |
| XA0055 (unknown peak) | 0.73 | 0.91 | 0.79 | 0.43 * | |
| XC0040 (unknown peak) | 0.77 * | 0.78 * | 0.83 | 0.68 * | |
| XC0061 (unknown peak) | 0.59 * | 0.57 * | 1.16 | 0.41 * | |
| β-Alanine | 0.93 | 0.79 | 1.05 | 0.57 * | |
| γ-Butyrobetaine | 0.76 * | 0.78 * | 0.78 | 0.36* | |
| Increased and decreased (one metabolite) | |||||
| Argininosuccinic acid | Renal disease, Uremic toxin | 0.75 * | 0.77 * | 1.25 * | 0.58 * |
| Metabolite | Coefficient | Standard Error | T-Value | p-Value |
|---|---|---|---|---|
| Two days after irradiation (R2 = 1; p < 0.001; F-value = 4.76 × 1010) | ||||
| Constant | 1.333 | 0.000000065 | 20,573,133 | <0.001 |
| Trigonelline | −0.731 | 0.000000704 | −1,037,888 | <0.001 |
| γ-Glutamyl-cysteine | 0.842 | 0.000000709 | 1,186,369 | <0.001 |
| Kynurenine | −0.382 | 0.000000265 | −1,441,436 | <0.001 |
| Isethionic acid | −0.286 | 0.000000521 | −548,634 | <0.001 |
| UDP-glucuronic acid | −0.181 | 0.000000773 | −234,621 | <0.001 |
| Hypotaurine | −0.042 | 0.000000209 | −202,910 | <0.001 |
| N6-Acetyllysine | 0.029 | 0.000001165 | 24,655 | <0.001 |
| NADPH_divalent | 0.005 | 0.000000767 | 6794 | <0.001 |
| S-Methylcysteine | −0.0002 | 0.000000627 | −333 | 0.0019 |
| Adenine | −0.00005 | 0.000000725 | −72 | 0.0089 |
| Six days after irradiation (R2 = 1; p < 0.001; F-value = 1.1 × 1011) | ||||
| Constant | 1.333 | 0.00000120 | 1,109,219 | <0.001 |
| Choline | −1.180 | 0.00000631 | −186,890 | <0.001 |
| Dihydroxyacetone phosphate | −0.115 | 0.00000407 | −28,344 | <0.001 |
| Histamine | 0.095 | 0.00000211 | 45,188 | <0.001 |
| Glycerophosphocholine | −0.129 | 0.00000559 | −23,197 | <0.001 |
| Ornithine | 0.092 | 0.00000387 | 23,692 | <0.001 |
| Fructose 1,6-diphosphate | 0.063 | 0.00000397 | 15,820 | <0.001 |
| Ethanolamine | −0.017 | 0.00000413 | −4103 | <0.001 |
| Methionine sulfoxide | 0.004 | 0.00000381 | 1075 | <0.001 |
| Threonic acid | 0.002 | 0.00000447 | 346 | 0.0018 |
| Spermidine | −0.0004 | 0.00000684 | −67 | 0.0094 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, L.; Inaba, Y.; Kanzaki, N.; Bekal, M.; Chida, K.; Moritake, T. Identification of Potential Biomarkers of Radiation Exposure in Blood Cells by Capillary Electrophoresis Time-of-Flight Mass Spectrometry. Int. J. Mol. Sci. 2020, 21, 812. https://doi.org/10.3390/ijms21030812
Sun L, Inaba Y, Kanzaki N, Bekal M, Chida K, Moritake T. Identification of Potential Biomarkers of Radiation Exposure in Blood Cells by Capillary Electrophoresis Time-of-Flight Mass Spectrometry. International Journal of Molecular Sciences. 2020; 21(3):812. https://doi.org/10.3390/ijms21030812
Chicago/Turabian StyleSun, Lue, Yohei Inaba, Norie Kanzaki, Mahesh Bekal, Koichi Chida, and Takashi Moritake. 2020. "Identification of Potential Biomarkers of Radiation Exposure in Blood Cells by Capillary Electrophoresis Time-of-Flight Mass Spectrometry" International Journal of Molecular Sciences 21, no. 3: 812. https://doi.org/10.3390/ijms21030812
APA StyleSun, L., Inaba, Y., Kanzaki, N., Bekal, M., Chida, K., & Moritake, T. (2020). Identification of Potential Biomarkers of Radiation Exposure in Blood Cells by Capillary Electrophoresis Time-of-Flight Mass Spectrometry. International Journal of Molecular Sciences, 21(3), 812. https://doi.org/10.3390/ijms21030812





