Whey-Derived Peptides Interactions with ACE by Molecular Docking as a Potential Predictive Tool of Natural ACE Inhibitors
Abstract
:1. Introduction
2. Results
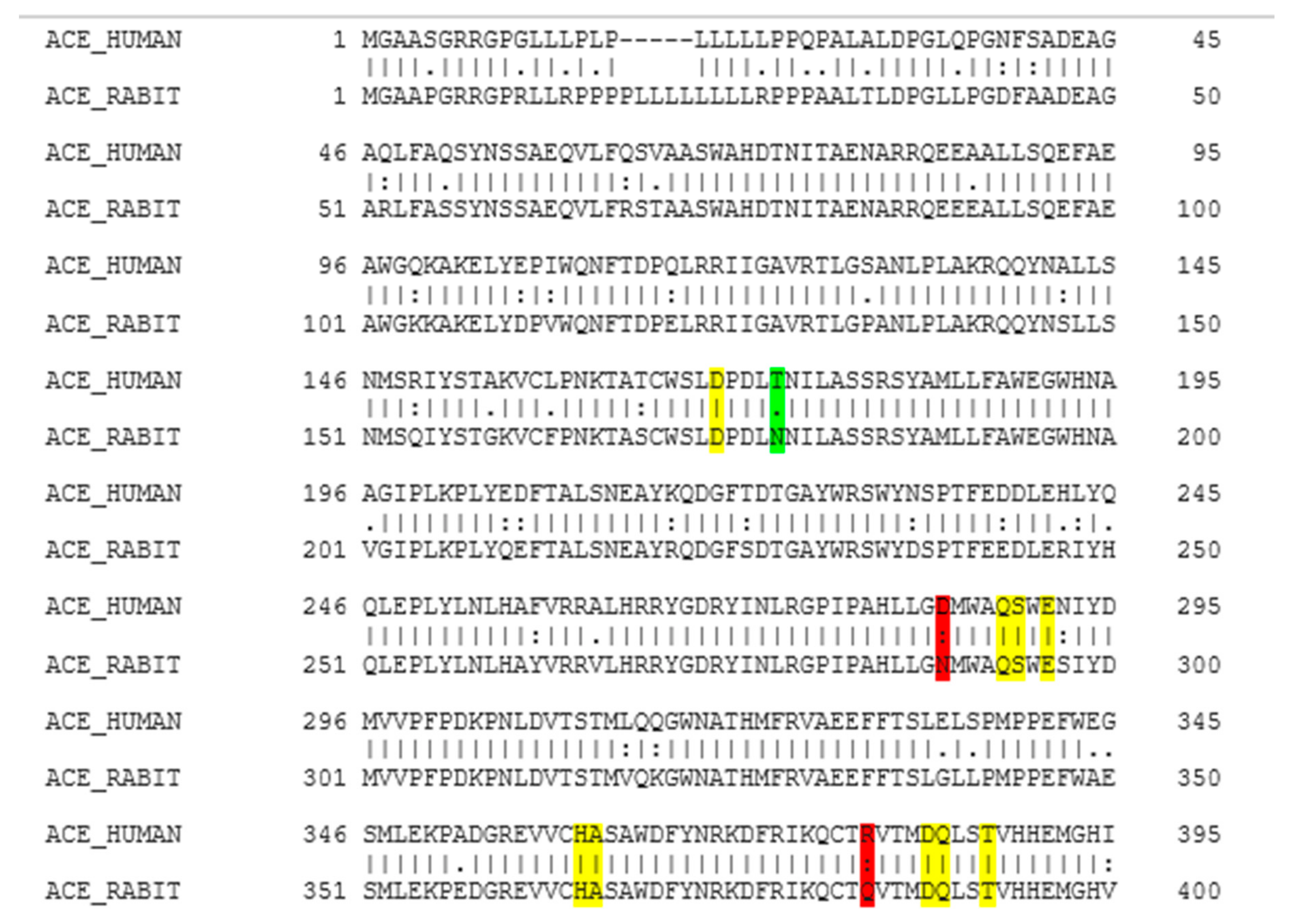
2.1. Molecular Homology between Human ACE and Rabbit ACE
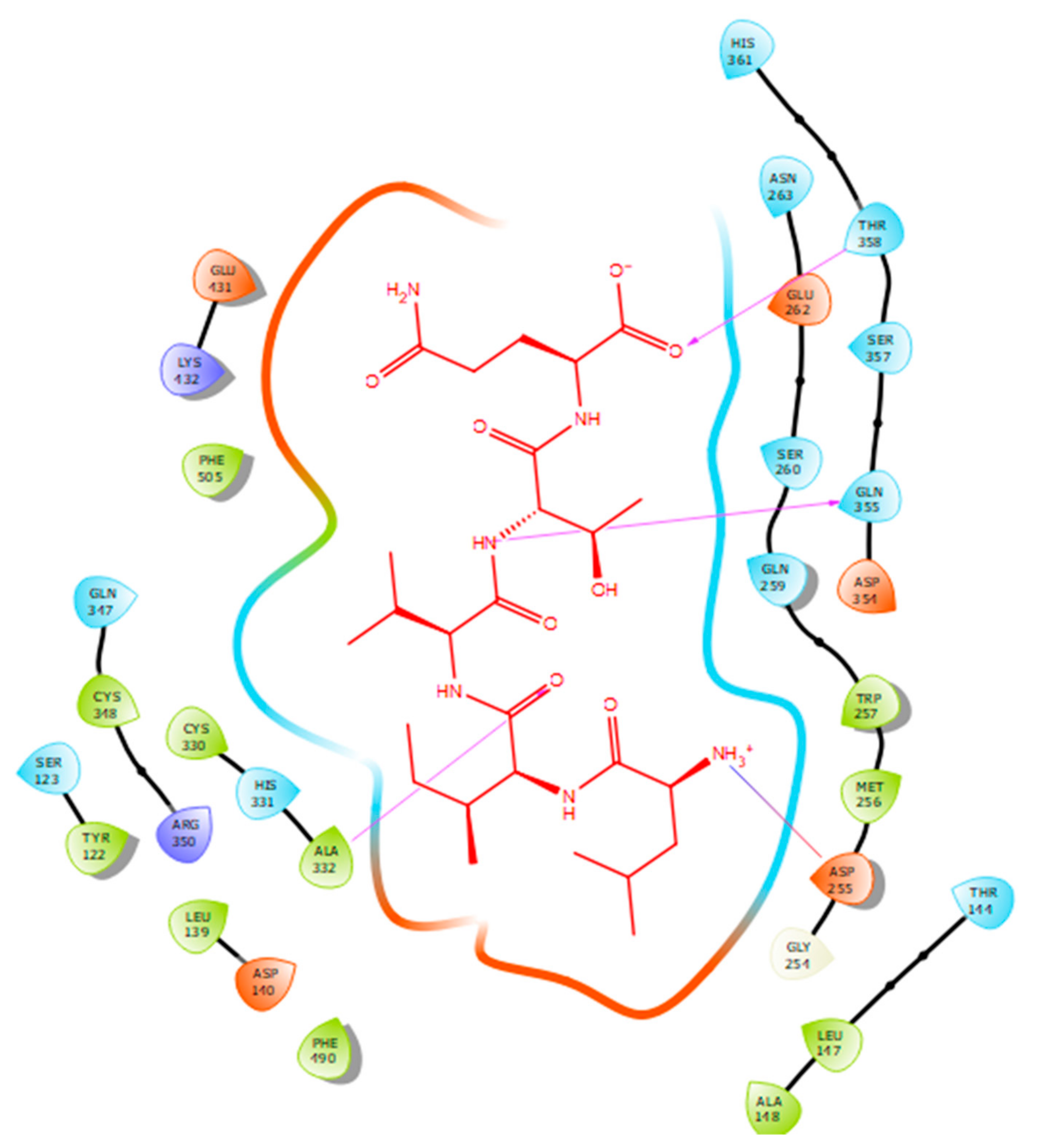
2.2. Molecular Docking
3. Discussion
4. Materials and Methods
4.1. Whey-Protein Derived Peptides
4.2. Homology between Human ACE and Rabbit ACE
4.3. Molecular Docking
4.3.1. Docking Validation
4.3.2. Docking Procedure
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Appendix A
References
- World Health Organization. Cardiovascular Diseases (CVDs): Fact Sheet No.317. 2015. Available online: http://www.who.int/mediacentre/factsheets/fs317/en/ (accessed on 20 October 2016).
- Celermajer, D.S.; Chow, C.K.; Marijon, E.; Anstey, N.M.; Woo, K.S. Cardiovascular disease in the developing world. J. Am. Coll. Cardiol. 2012, 60, 1207–1216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cannon, C.P. Cardiovascular disease and modifiable cardiometabolic risk factors. Clin. Cornerstone 2008, 9, 24–41. [Google Scholar] [CrossRef]
- Aluko, R.E. Antihypertensive Peptides from Food Proteins. Annu. Rev. Food Sci. Technol. 2015, 6, 235–262. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.Y.; Peng, C.; Jiao, R.; Wong, Y.M.; Yang, N.; Huang, Y. Anti-hypertensive Nutraceuticals and Functional Foods. J. Agric. Food Chem. 2009, 57, 4485–4499. [Google Scholar] [CrossRef]
- Donkor, O.N.; Henriksson, A.; Singh, T.K.; Vasiljevic, T.; Shah, N.P. ACE-inhibitory Activity of Probiotic Yoghurt. Int. Dairy J. 2007, 17, 1321–1331. [Google Scholar] [CrossRef]
- Acharya, K.R.; Sturrock, E.D.; Riordan, J.F.; Ehlers, M.R. Ace revisited: A new target for structure-based drug design. Nat. Rev. Drug Discov. 2003, 2, 891. [Google Scholar] [CrossRef]
- Li, G.H.; Le, G.W.; Shi, Y.H.; Shrestha, S. Angiotensin I–converting enzyme inhibitory peptides derived from food proteins and their physiological and pharmacological effects. Nutr. Res. 2004, 24, 469–486. [Google Scholar] [CrossRef]
- Wei, L.; Alhenc-Gelas, F.; Corvol, P.; Clauser, E. The two homologous domains of human angiotensin I-converting enzyme are both catalytically active. J. Biol. Chem. 1991, 266, 9002–9008. [Google Scholar]
- Sturrock, E.D.; Natesh, R.; Van Rooyen, J.M.; Acharya, K.R. Structure of angiotensin I-converting enzyme. Cell. Mol. Life Sci. 2004, 61, 2677–2686. [Google Scholar] [CrossRef]
- Natesh, R.; Schwager, S.L.; Sturrock, E.D.; Acharya, K.R. Crystal structure of the human angiotensin-converting enzyme–lisinopril complex. Nature 2003, 421, 551. [Google Scholar] [CrossRef] [Green Version]
- Tzakos, A.G.; Galanis, A.S.; Spyroulias, G.A.; Cordopatis, P.; Manessi-Zoupa, E.; Gerothanassis, I.P. Structure–function discrimination of the N-and C-catalytic domains of human angiotensin-converting enzyme: Implications for Cl–activation and peptide hydrolysis mechanisms. Protein Eng. 2003, 16, 993–1003. [Google Scholar] [CrossRef] [Green Version]
- Korhonen, H.; Pihlanto, A. Technological options for the production of health-promoting proteins and peptides derived from milk and colostrum. Curr. Pharm. Des. 2007, 13, 829–843. [Google Scholar] [CrossRef]
- FitzGerald, R.J.; Murray, B.A.; Walsh, D.J. Hypotensive peptides from milk proteins. J. Nutr. 2004, 134, 980S–988S. [Google Scholar] [CrossRef] [Green Version]
- Miguel, M.; López-Fandino, R.; Ramos, M.; Aleixandre, A. Short-term effect of egg-white hydrolysate products on the arterial blood pressure of hypertensive rats. Br. J. Nutr. 2005, 94, 731–737. [Google Scholar] [CrossRef]
- Beltrami, L.; Zingale, L.C.; Carugo, S.; Cicardi, M. Angiotensin-converting enzyme inhibitor-related angioedema: How to deal with it. Expert Opin. Drug Saf. 2006, 5, 643–649. [Google Scholar] [CrossRef]
- Cicero, A.F.G.; Gerocarni, B.; Laghi, L.; Borghi, C. Blood pressure lowering effect of lactotripeptides assumed as functional foods: A meta-analysis of current available clinical trials. J. Hum. Hypertens. 2011, 25, 425. [Google Scholar] [CrossRef] [Green Version]
- Fekete, A.A.; Givens, D.I.; Lovegrove, J.A. The impact of milk proteins and peptides on blood pressure and vascular function: A review of evidence from human intervention studies. Nutr. Res. Rev. 2013, 26, 177–190. [Google Scholar] [CrossRef] [Green Version]
- Geleijnse, J.M.; Engberink, M.F. Lactopeptides and human blood pressure. Curr. Opin. Lipidol. 2010, 21, 58–63. [Google Scholar] [CrossRef]
- Martínez-Maqueda, D.; Miralles, B.; Recio, I.; Hernández-Ledesma, B. Antihypertensive peptides from food proteins: A review. Food Funct. 2012, 3, 350–361. [Google Scholar] [CrossRef]
- Meisel, H. Overview on milk protein-derived peptides. Int. Dairy J. 1998, 8, 363–373. [Google Scholar] [CrossRef]
- Kitts, D.D.; Weiler, K. Bioactive proteins and peptides from food sources. Applications of bioprocesses used in isolation and recovery. Curr. Pharm. Des. 2003, 9, 1309–1323. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Yamamoto, N.; Sakai, K.; Okubo, A.; Yamazaki, S.; Takano, T. Purification and characterization of angiotensin I-converting enzyme inhibitors from sour milk. J. Dairy Sci. 1995, 78, 777–783. [Google Scholar] [CrossRef]
- Ehlers, P.I.; Nurmi, L.; Turpeinen, A.M.; Korpela, R.; Vapaatalo, H. Casein-derived tripeptide Ile–Pro–Pro improves angiotensin-(1–7)-and bradykinin-induced rat mesenteric artery relaxation. Life Sci. 2011, 88, 206–211. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Rivera, L.; Martínez-Maqueda, D.; Cruz-Huerta, E.; Miralles, B.; Recio, I. Peptidomics for discovery, bioavailability and monitoring of dairy bioactive peptides. Food Res. Int. 2014, 63, 170–181. [Google Scholar] [CrossRef]
- Majumder, K.; Wu, J. Molecular targets of antihypertensive peptides: Understanding the mechanisms of action based on the pathophysiology of hypertension. Int. J. Mol. Sci. 2014, 16, 256–283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Udenigwe, C.C.; Mohan, A. Mechanisms of food protein-derived antihypertensive peptides other than ACE inhibition. J. Funct. Foods 2014, 8, 45–52. [Google Scholar] [CrossRef]
- Masclee, G.M.; Coloma, P.M.; Kuipers, E.J.; Sturkenboom, M.C. Increased risk of microscopic colitis with use of proton pump inhibitors and non-steroidal anti-inflammatory drugs. Am. J. Gastroenterol. 2015, 110, 749–759. [Google Scholar] [CrossRef]
- Dallas, D.C.; Murray, N.M.; Gan, J. Proteolytic systems in milk: Perspectives on the evolutionary function within the mammary gland and the infant. J. Mammary Gland Biol. Neoplasia 2015, 20, 133–147. [Google Scholar] [CrossRef] [Green Version]
- Meisel, H.; FitzGerald, R.J. Biofunctional peptides from milk proteins: Mineral binding and cytomodulatory effects. Curr. Pharm. Des. 2003, 9, 1289–1296. [Google Scholar]
- Welderufael, F.T.; Gibson, T.; Methven, L.; Jauregi, P. Chemical characterisation and determination of sensory attributes of hydrolysates produced by enzymatic hydrolysis of whey proteins following a novel integrative process. Food Chem. 2012, 134, 1947–1958. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, Y.; Yamauchi, T.; Katsuda, T.; Yamaji, H.; Katoh, S. Angiotensin-I converting enzyme (ACE) inhibitory mechanism of tripeptides containing aromatic residues. J. Biosci. Bioeng. 2008, 106, 310–312. [Google Scholar] [CrossRef] [PubMed]
- Pan, D.; Guo, H.; Zhao, B.; Cao, J. The molecular mechanisms of interactions between bioactive peptides and angiotensin-converting enzyme. Bioorganic Med. Chem. Lett. 2011, 21, 3898–3904. [Google Scholar] [CrossRef] [PubMed]
- Udenigwe, C.C. Bioinformatics approaches, prospects and challenges of food bioactive peptide research. Trends Food Sci. Technol. 2014, 36, 137–143. [Google Scholar] [CrossRef]
- Tu, M.; Feng, L.; Wang, Z.; Qiao, M.; Shahidi, F.; Lu, W.; Du, M. Sequence analysis and molecular docking of antithrombotic peptides from casein hydrolysate by trypsin digestion. J. Funct. Foods 2017, 32, 313–323. [Google Scholar] [CrossRef]
- García-Mora, P.; Martín-Martínez, M.; Bonache, M.A.; González-Múniz, R.; Peñas, E.; Frias, J.; Martinez-Villaluenga, C. Identification, functional gastrointestinal stability and molecular docking studies of lentil peptides with dual antioxidant and angiotensin I converting enzyme inhibitory activities. Food Chem. 2017, 221, 464–472. [Google Scholar] [CrossRef] [Green Version]
- Guo, M.; Chen, X.; Wu, Y.; Zhang, L.; Huang, W.; Yuan, Y.; Wei, D. Angiotensin I-converting enzyme inhibitory peptides from Sipuncula (Phascolosoma esculenta): Purification, identification, molecular docking and antihypertensive effects on spontaneously hypertensive rats. Process Biochem. 2017, 63, 84–95. [Google Scholar] [CrossRef]
- Sangsawad, P.; Choowongkomon, K.; Kitts, D.D.; Chen, X.M.; Li-Chan, E.C.; Yongsawatdigul, J. Transepithelial transport and structural changes of chicken angiotensin I-converting enzyme (ACE) inhibitory peptides through Caco-2 cell monolayers. J. Funct. Foods 2018, 45, 401–408. [Google Scholar] [CrossRef]
- Shi, L.; Wu, T.; Sheng, N.; Yang, L.; Wang, Q.; Liu, R.; Wu, H. Characterization of angiotensin-I converting enzyme inhibiting peptide from Venerupis philippinarum with nano-liquid chromatography in combination with orbitrap mass spectrum detection and molecular docking. J. Ocean Univ. China 2017, 16, 473–478. [Google Scholar] [CrossRef]
- Wang, X.; Chen, H.; Fu, X.; Li, S.; Wei, J. A novel antioxidant and ACE inhibitory peptide from rice bran protein: Biochemical characterization and molecular docking study. Lwt-Food Sci. Technol. 2017, 75, 93–99. [Google Scholar] [CrossRef]
- Wu, Q.; Du, J.; Jia, J.; Kuang, C. Production of ACE inhibitory peptides from sweet sorghum grain protein using alcalase: Hydrolysis kinetic, purification and molecular docking study. Food Chem. 2016, 199, 140–149. [Google Scholar] [CrossRef]
- Yu, Z.; Chen, Y.; Zhao, W.; Li, J.; Liu, J.; Chen, F. Identification and molecular docking study of novel angiotensin-converting enzyme inhibitory peptides from Salmo salar using in silico methods. J. Sci. Food Agric. 2018. [Google Scholar] [CrossRef] [PubMed]
- Ashok, N.R.; Aparna, H. Empirical and bioinformatic characterization of buffalo (Bubalus bubalis) colostrum whey peptides & their angiotensin I-converting enzyme inhibition. Food Chem. 2017, 228, 582–594. [Google Scholar] [PubMed]
- Lin, K.; Zhang, L.W.; Han, X.; Cheng, D.Y. Novel angiotensin I-converting enzyme inhibitory peptides from protease hydrolysates of Qula casein: Quantitative structure-activity relationship modeling and molecular docking study. J. Funct. Foods 2017, 32, 266–277. [Google Scholar] [CrossRef]
- Nongonierma, A.B.; FitzGerald, R.J. Strategies for the discovery and identification of food protein-derived biologically active peptides. Trends Food Sci. Technol. 2017. [Google Scholar] [CrossRef] [Green Version]
- Soubrier, F.; Alhenc-Gelas, F.; Hubert, C.; Allegrini, J.; John, M.; Tregear, G.; Corvol, P. Two putative active centers in human angiotensin I-converting enzyme revealed by molecular cloning. Proc. Natl. Acad. Sci. USA 1998, 85, 9386–9390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cozier, G.E.; Schwager, S.L.; Sharma, R.K.; Chibale, K.; Sturrock, E.D.; Acharya, K.R. Crystal structures of sampatrilat and sampatrilat-Asp in complex with human ACE–a molecular basis for domain selectivity. Febs J. 2018, 285, 1477–1490. [Google Scholar] [CrossRef] [Green Version]
- Tu, M.; Wang, C.; Chen, C.; Zhang, R.; Liu, H.; Lu, W.; Du, M. Identification of a novel ACE-inhibitory peptide from casein and evaluation of the inhibitory mechanisms. Food Chem. 2018, 256, 98–104. [Google Scholar] [CrossRef]
- Ling, Y.; Sun, L.P.; Zhuang, Y.L. Preparation and identification of novel inhibitory Angiotensin-I-converting enzyme peptides from tilapia skin gelatin hydrolysates: Inhibition kinetics and molecular docking. Food Funct. 2018, 9, 5251–5259. [Google Scholar] [CrossRef]
- Venn, R.F.; Barnard, G.; Kaye, B.; Macrae, P.V.; Saunders, K.C. Clinical analysis of sampatrilat, a combined renal endopeptidase and angiotensin-converting enzyme inhibitor: II: Assay in the plasma and urine of human volunteers by dissociation enhanced lanthanide fluorescence immunoassay (DELFIA). J. Pharm. Biomed. Anal. 1998, 16, 883–892. [Google Scholar] [CrossRef]
- Wallis, E.J.; Ramsay, L.E.; Hettiarachchi, J. Combined inhibition of neutral endopeptidase and angiotensin-converting enzyme by sampatrilat in essential hypertension. Clin. Pharmacol. Ther. 1998, 64, 439–449. [Google Scholar] [CrossRef]
- Natesh, R.; Schwager, S.L.; Evans, H.R.; Sturrock, E.D.; Acharya, K.R. Structural details on the binding of antihypertensive drugs captopril and enalaprilat to human testicular angiotensin I-converting enzyme. Biochemistry 2004, 43, 8718–8724. [Google Scholar] [CrossRef] [PubMed]
- Vercruysse, L.; Van Camp, J.; Morel, N.; Rougé, P.; Herregods, G.; Smagghe, G. Ala-Val-Phe and Val-Phe: ACE inhibitory peptides derived from insect protein with antihypertensive activity in spontaneously hypertensive rats. Peptides 2010, 31, 482–488. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.L.; Zhang, S.S.; Wei, W.A.N.G.; Feng, F.Q.; Shan, W.G. A novel angiotensin I converting enzyme inhibitory peptide from the milk casein: Virtual screening and docking studies. Agric. Sci. China 2011, 10, 463–467. [Google Scholar] [CrossRef]
- Gonzalez-Gonzalez, C.; Gibson, T.; Jauregi, P. Novel probiotic-fermented milk with angiotensin I-converting enzyme inhibitory peptides produced by Bifidobacterium bifidum MF 20/5. Int. J. Food Microbiol. 2013, 167, 131–137. [Google Scholar] [CrossRef]
- Jain, R.K. Molecular regulation of vessel maturation. Nat. Med. 2003, 9, 685. [Google Scholar] [CrossRef]
- Schrodinger LLC. Version 1.8, The PyMOL Molecular Graphics System; Technical Report; Schrödinger LLC: New York, NY, USA, 2015. [Google Scholar]
- PDBeFold - Structure Similarity, Embl-Ebi. 2019. Available online: http://www.ebi.ac.uk/msd-srv/ssm/cgi-bin/ssmserver (accessed on 2 December 2019).
- Wang, R.; Lu, Y.; Wang, S. Comparative evaluation of 11 scoring functions for molecular docking. J. Med. Chem. 2003, 46, 2287–2303. [Google Scholar] [CrossRef]
- Sharma, R.; Dhingra, N.; Patil, S. CoMFA, CoMSIA, HQSAR and molecular docking analysis of ionone-based chalcone derivatives as antiprostate cancer activity. Indian J. Pharm. Sci. 2016, 78, 54. [Google Scholar] [CrossRef] [Green Version]
- Ai, Y.; Wang, S.T.; Sun, P.H.; Song, F.J. Combined 3D-QSAR modeling and molecular docking studies on Pyrrole-Indolin-2-ones as Aurora A Kinase inhibitors. Int. J. Mol. Sci. 2011, 12, 1605–1624. [Google Scholar] [CrossRef] [Green Version]
- Lan, P.; Chen, W.N.; Chen, W.M. Molecular modeling studies on imidazo [4, 5-b] pyridine derivatives as Aurora A kinase inhibitors using 3D-QSAR and docking approaches. Eur. J. Med. Chem. 2011, 46, 77–94. [Google Scholar] [CrossRef]
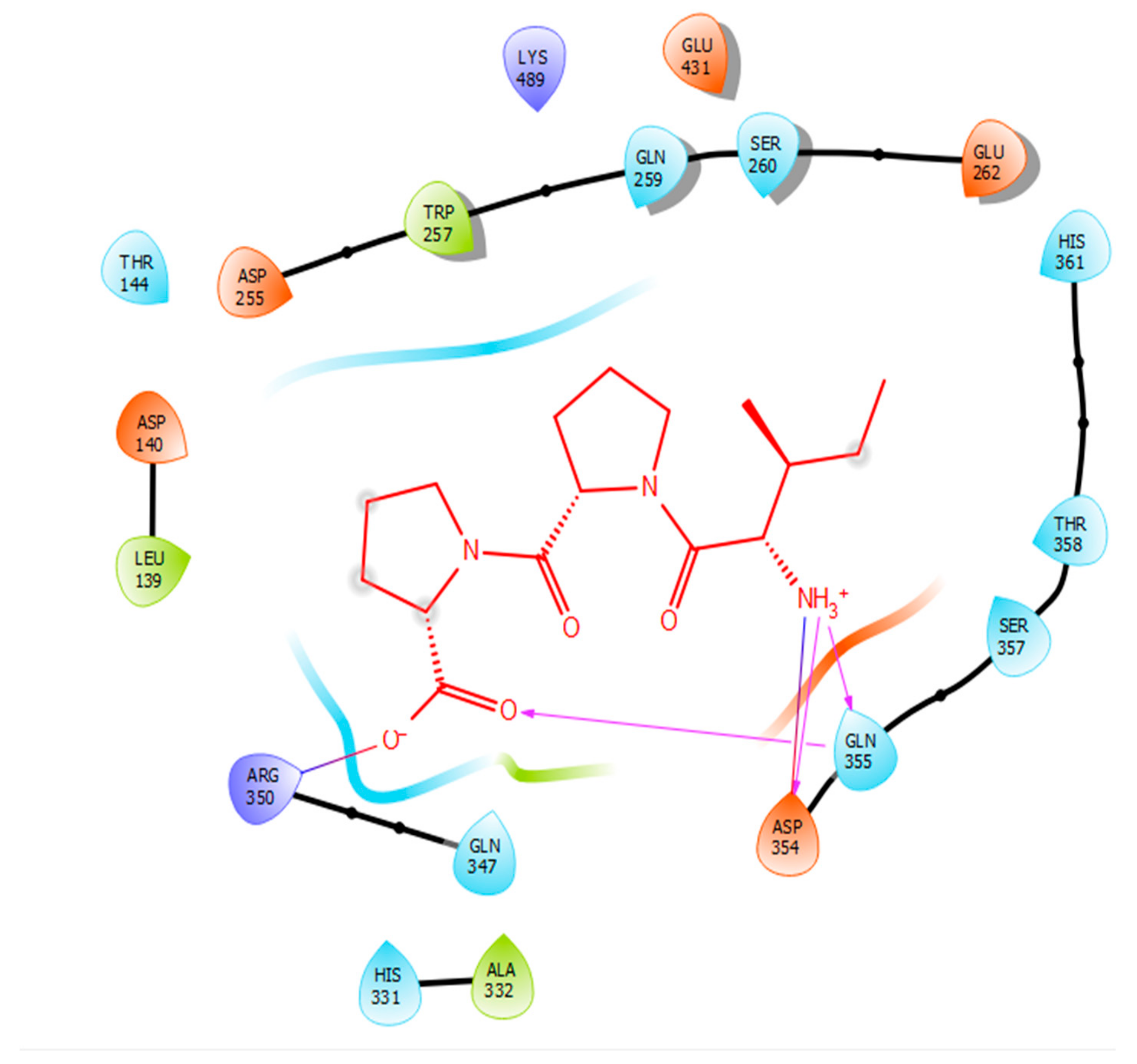

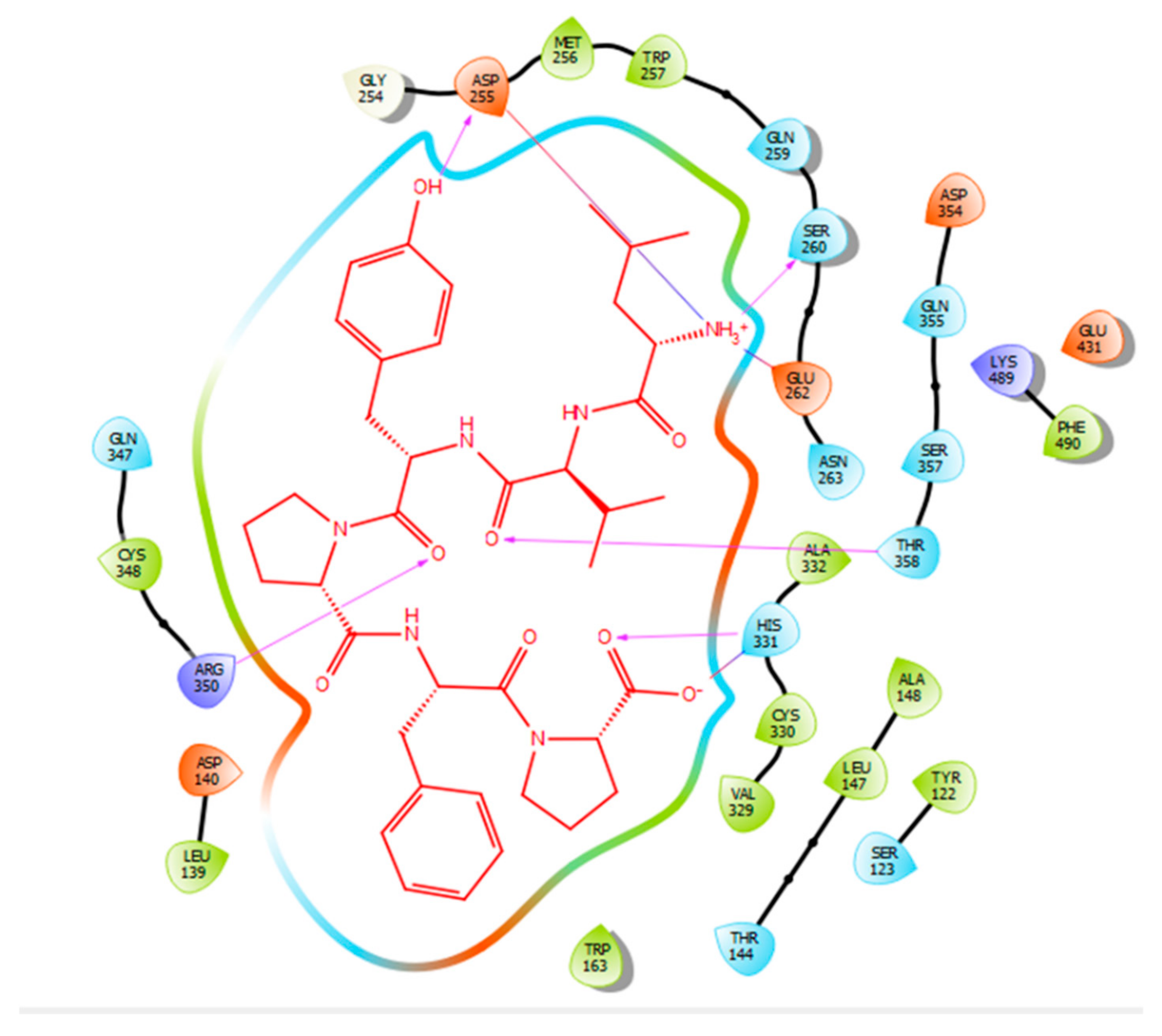
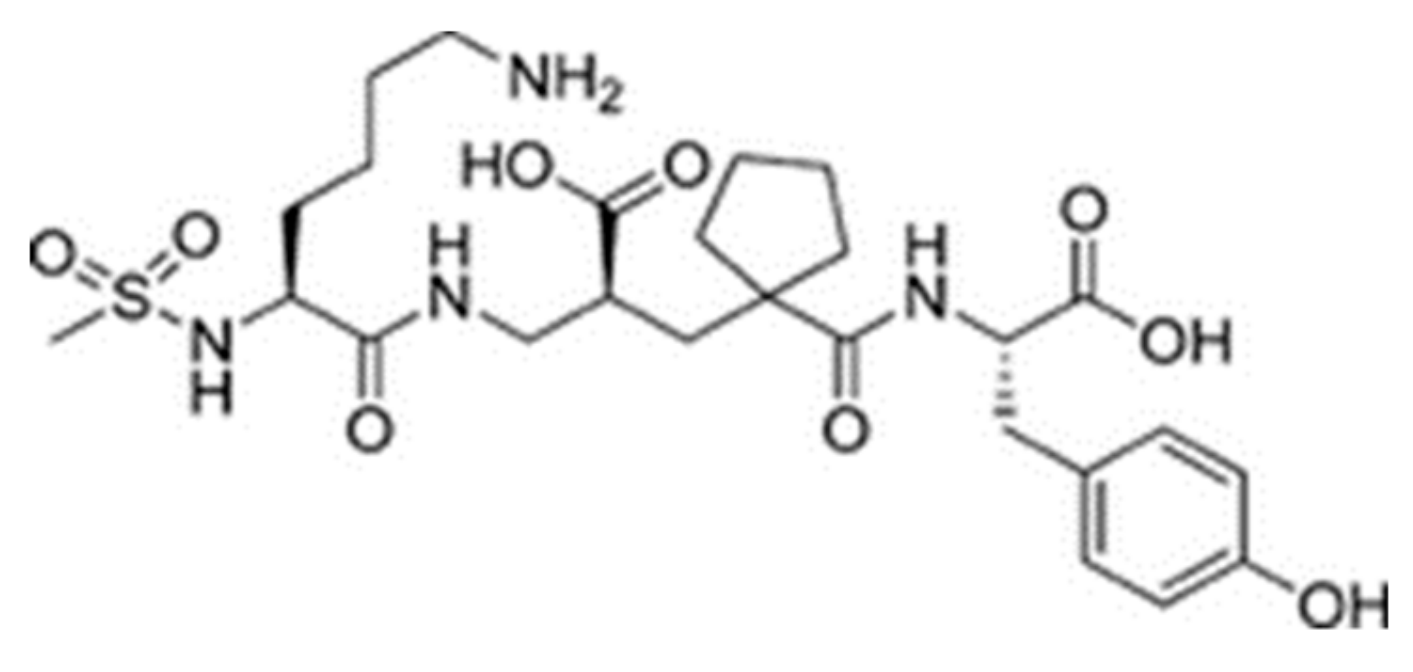
| Protein 69FV | Ligand IPP | ||
|---|---|---|---|
| Residue | Atom Name | Interaction Type | Distance (Å) |
| NH2 Arg 350 | O− (Pro) | Salt bridge | 2.86 |
| OD2 Asp 354 | NH3+ (Ile) | Hydrogen bond | 1.86 |
| OE1 Gln 355 | NH3+ (Ile) | Hydrogen bond | 1.81 |
| NE2 Gln 355 | O- (Pro) | Hydrogen bond | 2.04 |
| Protein 69FV | Ligand IIAE | ||
|---|---|---|---|
| Residue | Atom Name | Interaction Type | Distance (Å) |
| OD2 Asp 140 | NH (Ile) | Salt bridge | 2.57 |
| OG1 Thr 144 | N (Ile) | Hydrogen bond | 2.15 |
| NE2 Gln 259 | O (Glu) | Hydrogen bond | 2.2 |
| OG1 Thr 358 | O (Glu) | Hydrogen bond | 1.88 |
| Protein 69FV | Ligand LIVTQ | ||
|---|---|---|---|
| Residue | Atom Name | Interaction Type | Distance (Å) |
| OD2 Asp 255 | NH3+ (Leu) | Salt bridge | 4.79 |
| N Ala 332 | O (Ile) | Hydrogen bond | 2.56 |
| OE1 Gln 355 | N (Thr) | Hydrogen bond | 1.85 |
| OG1 Thr 358 | O- (Gln) | Hydrogen bond | 1.88 |
| Protein 69FV | Ligand LVYPFP | ||
|---|---|---|---|
| Residue | Atom Name | Interaction Type | Distance (Å) |
| OD2 Asp 255 | OH (Tyr) | Hydrogen bond | 2.12 |
| OG Ser 260 | NH3+ (Leu) | Hydrogen bond | 1.89 |
| OE2 Glu 262 | NH3+ (Leu) | Salt bridge | 3.58 |
| ND1 His 331 | O (Pro) | Hydrogen bond | 1.92 |
| ND1 His 331 | O (Pro) | Salt bridge | 2.71 |
| NH2 Arg 350 | O (Tyr) | Hydrogen bond | 2.61 |
| OG1 Thr 358 | O (Valine) | Hydrogen bond | 1.75 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chamata, Y.; Watson, K.A.; Jauregi, P. Whey-Derived Peptides Interactions with ACE by Molecular Docking as a Potential Predictive Tool of Natural ACE Inhibitors. Int. J. Mol. Sci. 2020, 21, 864. https://doi.org/10.3390/ijms21030864
Chamata Y, Watson KA, Jauregi P. Whey-Derived Peptides Interactions with ACE by Molecular Docking as a Potential Predictive Tool of Natural ACE Inhibitors. International Journal of Molecular Sciences. 2020; 21(3):864. https://doi.org/10.3390/ijms21030864
Chicago/Turabian StyleChamata, Yara, Kimberly A. Watson, and Paula Jauregi. 2020. "Whey-Derived Peptides Interactions with ACE by Molecular Docking as a Potential Predictive Tool of Natural ACE Inhibitors" International Journal of Molecular Sciences 21, no. 3: 864. https://doi.org/10.3390/ijms21030864
APA StyleChamata, Y., Watson, K. A., & Jauregi, P. (2020). Whey-Derived Peptides Interactions with ACE by Molecular Docking as a Potential Predictive Tool of Natural ACE Inhibitors. International Journal of Molecular Sciences, 21(3), 864. https://doi.org/10.3390/ijms21030864