A Unique Regulation Region in the 3′ UTR of HLA-G with a Promising Potential
Abstract
:1. Introduction
2. Results
2.1. Screening for RBPs That Bind the Unique Region in the 3′ UTR of HLA-G
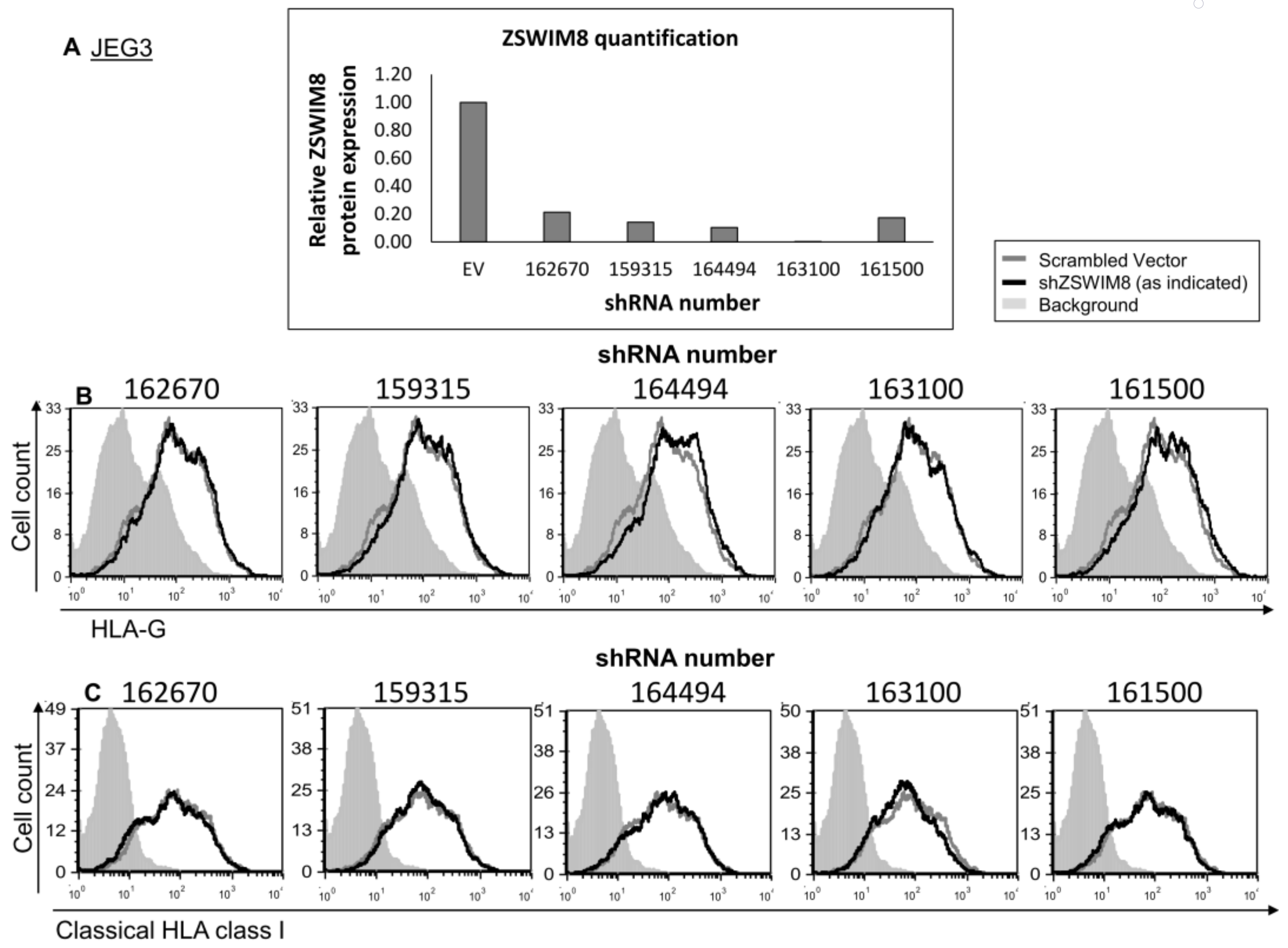
2.2. DDX47 and ZSWIM8 RBPs Do Not Alter HLA-G Expression
2.3. miR-1301 Is Not Involved in HLA-G Regulation
2.4. Dual Expression of miR-1301 and DDX47 Does Not Affect HLA-G Protein Expression
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. RNA Affinity Purification and Mass Spectrometry
4.3. Generation of Lentivirus, Knockdown, Overexpression
4.4. RNA Extraction and cDNA Preparation
4.5. Quantitative Real-Time PCR
4.6. Western Blot Analyses
4.7. Flow Cytometry
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Niederkorn, J.Y. Immune escape mechanisms of intraocular tumors. Prog. Retin. Eye Res. 2009, 28, 329–347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pratheek, B.M.; Nayak, T.K.; Sahoo, S.S.; Mohanty, P.K.; Chattopadhyay, S.; Chakraborty, N.G.; Chattopadhyay, S. Mammalian non-classical major histocompatibility complex I and its receptors: Important contexts of gene, evolution, and immunity. Indian J. Hum. Genet. 2014, 20, 129–141. [Google Scholar] [PubMed] [Green Version]
- Carosella, E.D.; Rouas-Freiss, N.; Roux, D.T.; LeMoreau, P.; Le Maoult, J. HLA-G. An Immune Checkpoint Molecule. In Advances in Immunology; Academic Press Inc.: Cambridge, MA, USA, 2015; Volume 127, pp. 33–144. [Google Scholar]
- Clements, C.S.; Kjer-Nielsen, L.; McCluskey, J.; Rossjohn, J. Structural Studies on HLA-G: Implications for Ligand and Receptor Binding. Hum. Immunol. 2007, 68, 220–226. [Google Scholar] [CrossRef] [PubMed]
- LeMaoult, J.; Zafaranloo, K.; Le Banff, C.; Carosella, E.D. HLA-G up-regulates ILT2, ILT3, ILT4, and KIR2DL4 in antigen presenting cells, NK cells, and T cells. FASEB J. 2005, 19, 662–664. [Google Scholar] [CrossRef]
- Sheu, J.; Shih, I.M. HLA-G and Immune Evasion in Cancer Cells. J. Formos. Med. Assoc. 2010, 109, 248–257. [Google Scholar] [CrossRef] [Green Version]
- Rebmann, V.; Da Silva Nardi, F.; Wagner, B.; Horn, P.A. HLA-G as a tolerogenic molecule in transplantation and pregnancy. J. Immunol. Res. 2014, 2014, 297073. [Google Scholar] [CrossRef]
- Gamliel, M.; Goldman-Wohl, D.; Isaacson, B.; Gur, C.; Stein, N.; Yamin, R.; Berger, M.; Grunewald, M.; Keshet, E.; Rais, Y.; et al. Trained Memory of Human NK Cells Enhances Their Function in Subsequent Pregnancies. Immunity 2018, 48, 951–962.e5. [Google Scholar] [CrossRef] [Green Version]
- Manaster, I.; Mandelboim, O. The unique properties of human NK cells in the uterine mucosa. Placenta 2008, 29 (Suppl. A), S60–S66. [Google Scholar] [CrossRef]
- Hanna, J.; Goldman-Wohl, D.; Hamani, Y.; Avraham, I.; Greenfield, C.; Natanson-Yaron, S.; Prus, D.; Cohen-Daniel, L.; Arnon, T.I.; Manaster, I.; et al. Decidual NK cells regulate key developmental processes at the human fetal-maternal interface. Nat. Med. 2006, 12, 1065–1074. [Google Scholar] [CrossRef]
- Seidel, E.; Glasner, A.; Mandelboim, O. Virus-mediated inhibition of natural cytotoxicity receptor recognition. Cell. Mol. Life Sci. 2012, 69, 3911–3920. [Google Scholar] [CrossRef]
- Koch, J.; Steinle, A.; Watzl, C.; Mandelboim, O. Activating natural cytotoxicity receptors of natural killer cells in cancer and infection. Trends Immunol. 2013, 34, 182–191. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Oksvold, P.; Fagerberg, L.; Lundberg, E.; Jonasson, K.; Forsberg, M.; Zwahlen, M.; Kampf, C.; Wester, K.; Hober, S.; et al. Towards a knowledge-based Human Protein Atlas. Nat. Biotechnol. 2010, 28, 1248–1250. [Google Scholar] [CrossRef] [PubMed]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar]
- Glisovic, T.; Bachorik, J.L.; Yong, J.; Dreyfuss, G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008, 582, 1977–1986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moore, M.J.; Proudfoot, N.J. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 2009, 136, 688–700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sonenberg, N.; Hinnebusch, A.G. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 2009, 136, 731–745. [Google Scholar] [CrossRef] [Green Version]
- Martin, K.C.; Ephrussi, A. mRNA localization: gene expression in the spatial dimension. Cell 2009, 136, 719–730. [Google Scholar] [CrossRef] [Green Version]
- Nachmani, D.; Gutschner, T.; Reches, A.; Diederichs, S.; Mandelboim, O. RNA-binding proteins regulate the expression of the immune activating ligand MICB. Nat. Commun. 2014, 5, 4186. [Google Scholar] [CrossRef] [Green Version]
- Von Brandenstein, M.; Bernhart, S.H.; Pansky, A.; Richter, C.; Kohl, T.; Deckert, M.; Heidenreich, A.; Stadler, P.F.; Montesinos-Rongen, M.; Fries, J.W.U. Beyond the 3′ UTR binding-microRNA-induced protein truncation via DNA binding. Oncotarget 2018, 9, 32855–32867. [Google Scholar] [CrossRef]
- Szostak, E.; Gebauer, F. Translational control by 3′-UTR-binding proteins. Brief. Funct. Genomics 2013, 12, 58–65. [Google Scholar] [CrossRef]
- Matoulkova, E.; Michalova, E.; Vojtesek, B.; Hrstka, R. The role of the 3′ untranslated region in post-transcriptional regulation of protein expression in mammalian cells. RNA Biol. 2012, 9, 563–576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwich, E.; Rebmann, V.; Michita, R.T.; Rohn, H.; Voncken, J.W.; Horn, P.A.; Kimmig, R.; Kasimir-Bauer, S.; Buderath, P. HLA-G 3′ untranslated region variants +3187G/G, +3196G/G and +3035T define diametrical clinical status and disease outcome in epithelial ovarian cancer. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Reches, A.; Nachmani, D.; Berhani, O.; Duev-Cohen, A.; Shreibman, D.; Ophir, Y.; Seliger, B.; Mandelboim, O. HNRNPR Regulates the Expression of Classical and Nonclassical MHC Class I Proteins. J. Immunol. 2016, 196, 4967–4976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988, 16, 10881–10890. [Google Scholar] [CrossRef]
- Kovats, S.; Main, E.K.; Librach, C.; Stubblebine, M.; Fisher, S.J.; DeMars, R. A class I antigen, HLA-G, expressed in human trophoblasts. Science 1990, 248, 220–223. [Google Scholar] [CrossRef]
- The UniProt Consortium UniProt: A hub for protein information. Nucleic Acids Res. 2014, 43, D204–D212.
- Weedon-Fekjae, R.M.S.; Sheng, Y.; Sugulle, M.; Johnsen, G.M.; Herse, F.; Redman, C.W.; Lyle, R.; Dechend, R.; Staff, A.C. Placental miR-1301 is dysregulated in early-onset preeclampsia and inversely correlated with maternal circulating leptin. Placenta 2014, 35, 709–717. [Google Scholar] [CrossRef]
- Nevalainen, J.; Skarp, S.; Savolainen, E.R.; Ryynänen, M.; Järvenpää, J. Intrauterine growth restriction and placental gene expression in severe preeclampsia, comparing early-onset and late-onset forms. J. Perinat. Med. 2017, 45, 869–877. [Google Scholar] [CrossRef]
- Krüger, J.; Rehmsmeier, M. RNAhybrid: MicroRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006, 34, 451–454. [Google Scholar] [CrossRef]
- Hämmerle, M.; Gutschner, T.; Uckelmann, H.; Ozgur, S.; Fiskin, E.; Gross, M.; Skawran, B.; Geffers, R.; Longerich, T.; Breuhahn, K.; et al. Posttranscriptional destabilization of the liver-specific long noncoding RNA HULC by the IGF2 mRNA-binding protein 1 (IGF2BP1). Hepatology 2013, 58, 1703–1712. [Google Scholar] [CrossRef]
- Stern-Ginossar, N.; Elefant, N.; Zimmermann, A.; Wolf, D.G.; Saleh, N.; Biton, M.; Horwitz, E.; Prokocimer, Z.; Prichard, M.; Hahn, G.; et al. Host immune system gene targeting by a viral miRNA. Science 2007, 317, 376–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]






© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reches, A.; Berhani, O.; Mandelboim, O. A Unique Regulation Region in the 3′ UTR of HLA-G with a Promising Potential. Int. J. Mol. Sci. 2020, 21, 900. https://doi.org/10.3390/ijms21030900
Reches A, Berhani O, Mandelboim O. A Unique Regulation Region in the 3′ UTR of HLA-G with a Promising Potential. International Journal of Molecular Sciences. 2020; 21(3):900. https://doi.org/10.3390/ijms21030900
Chicago/Turabian StyleReches, Adi, Orit Berhani, and Ofer Mandelboim. 2020. "A Unique Regulation Region in the 3′ UTR of HLA-G with a Promising Potential" International Journal of Molecular Sciences 21, no. 3: 900. https://doi.org/10.3390/ijms21030900
APA StyleReches, A., Berhani, O., & Mandelboim, O. (2020). A Unique Regulation Region in the 3′ UTR of HLA-G with a Promising Potential. International Journal of Molecular Sciences, 21(3), 900. https://doi.org/10.3390/ijms21030900



