MiRNAs and LncRNAs: Dual Roles in TGF-β Signaling-Regulated Metastasis in Lung Cancer
Abstract
:1. Introduction
2. Overview of miRNAs and lncRNAs
2.1. MiRNAs
2.2. LncRNAs
3. TGF-β Signaling Is Involved in Lung Cancer Metastasis
3.1. TGF-β Signaling Regulates Cell Migration and Invasion
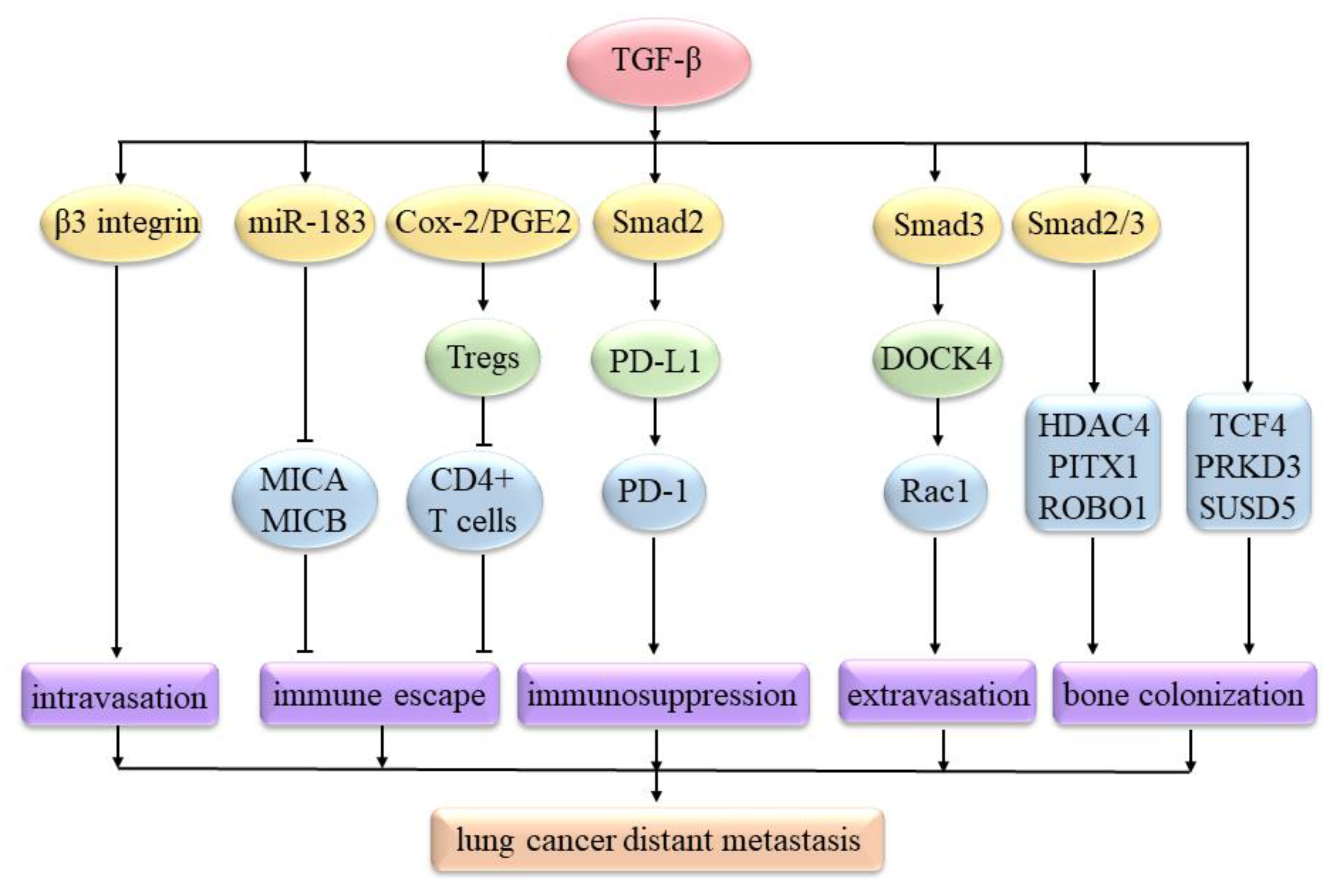
3.2. TGF-β Signaling Regulates Lung Cancer’s Distant Metastasis
4. Roles of miRNAs in TGF-β Signaling-Regulated Lung Cancer Metastasis
4.1. Positive Roles of miRNAs
4.1.1. MiR-93
4.1.2. MiR-128-3p
4.1.3. MiR-9
4.1.4. MiR-134 and miR-487b
4.1.5. MiR-330-3p
4.1.6. MiR-1246
4.1.7. MiR-9-5p
4.1.8. MiR-181b-5p
4.1.9. MiR-23a
4.2. Negative Roles of miRNAs
4.2.1. MiR-132
4.2.2. MiR-203 and miR-145
4.2.3. MiR-205
4.2.4. MiR-124
4.2.5. MiR-422a
4.2.6. MiR-196b
4.2.7. MiR-940
4.2.8. MiR-22
4.2.9. MiR-200 Family
4.2.10. MiR-145 and miR-497
4.2.11. MiR-149
4.2.12. MiR-134
4.2.13. MiR-133
4.2.14. MiR-29c
4.2.15. MiR-16
4.2.16. MiR-129
4.2.17. MiR-485-5p and miR-3127-5p
4.2.18. MiR-3607-3p
4.2.19. MiR-206 and miR-140
4.2.20. MiR-136
4.2.21. MiR-133a
4.2.22. MiR-143
4.2.23. MiR-886-3p
4.2.24. MiR-138
5. Roles of lncRNAs in TGF-β Signaling-Regulated Lung Cancer Metastasis
5.1. Positive Roles of lncRNAs
5.1.1. LncRNA HCP5
5.1.2. LncRNA NORAD
5.1.3. LncRNA XIST
5.1.4. LncRNA linc00673
5.1.5. LncRNA MEG3
5.1.6. LncRNA MEG8
5.1.7. LncRNA ATB
5.1.8. LncRNA TBILA
5.1.9. Lnc-MMP2-2
5.1.10. LncRNA AWPPH
5.1.11. LncRNA CASC11
5.2. Negative Roles of lncRNAs
5.2.1. LncRNA HAND2-AS1
5.2.2. LncRNA linc01186
5.2.3. LncRNA LINP1
5.2.4. LncRNA ANCR
5.2.5. LncRNA NKILA
6. MiRNA Sponges Participate in TGF-β Signaling-Regulated Lung Cancer Metastasis
6.1. CeRNA
6.1.1. CeRNA MYEOV
6.1.2. CeRNA HMGA2
6.2. CircRNA
7. Targeted Therapy
7.1. Curcumin
7.2. Carboplatin
7.3. DDP
7.4. DAC
8. Discussion and Conclusions
- ➣
- They can promote or suppress TGF-β/Smads pathways or other non-Smad pathways. MiR-93 [59] and NORAD [18] can promote the TGF-β/Smads signaling pathways. ATB can promote the AKT signal and JAK/STAT3 signal [113]. MiR-132 [77], miR-422a [17], miR-206, miR-140 [100], and HAND2-AS1 [119] can repress the TGF-β/Smads signaling pathways. MiR-133a [103] and NKILA [123] can repress the TGF-β/AKT and NF-κB/Snail pathway, respectively.
- ➣
- ➣
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ADAM12 | a disintegrin and metalloprotease 12 |
| AKT | protein kinase B |
| ATB | activated by TGF-β |
| CCNE2 | cyclin E2 |
| ceRNA | competing endogenous RNA |
| circRNA | circular RNA |
| Cox-2 | cyclooxygenase-2 |
| cPP1 | catalytic subunit of protein phosphatase 1 |
| CSC | cancer stem cell |
| CTC | circulating tumor cell |
| DAC | Decitabine |
| DAP12 | DNAX activating protein 12 kDa |
| DDP | cisplatin |
| DOCK4 | dedicator of cytokinesis 4 |
| EC | endothelial cell |
| ECM | extracellular matrix |
| EED | embryonic ectoderm development |
| EMNs | exosome-mimetic nanosystems |
| EMT | epithelial-mesenchymal transition |
| ERK | extracellular signal-regulated kinase |
| EZH2 | enhancer of zeste homologue 2 |
| F-EMNs | Functionalization of EMNs with ITGα6β4 |
| FOXM1 | forkhead Box M1 |
| FOXQ1 | forkhead box Q1 |
| HCP5 | histocompatibility leukocyte antigen complex P5 |
| HDAC4 | histone deacetylase 4 |
| HGAL | human germinal center-associated lymphoma |
| IL-10 | interleukin-10 |
| JAB1 | c-Jun activation domain-binding protein 1 |
| JAK | Janus-activated kinase |
| LATS2 | large tumor suppressor homology 2 |
| LEC | lymphatic endothelial cell |
| LINP1 | lncRNA in nonhomologous end joining (NHEJ) pathway 1 |
| LMO7 | LIM-domain only protein 7 |
| lncRNA | long non-coding RNA |
| MAGI2 | membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 |
| MALAT1 | metastasis-associated lung adenocarcinoma transcript 1 |
| MAPK | mitogen-activated protein kinase |
| MICA | major histocompatibility complex class I chain-related A |
| miRNA | microRNA |
| mRNA | messenger RNA |
| MMP | matrix metalloprotease |
| MRE | miRNA response element |
| MTDH | metadherin |
| MT1-MMP | membrane type 1 MMP |
| MYEOV | myeloma overexpressed gene |
| nc-RNA | non-coding RNA |
| NEDD4L | neural precursor cell expressed developmentally downregulated gene 4-like |
| NF-κB | nuclear factor-kappa B |
| NK | natural killer |
| NKILA | NF-κB Interacting LncRNA |
| NSCLC | non-small cell lung cancer |
| PAI-1 | plasminogen activator inhibitor-1 |
| PD-1 | programmed cell death protein-1 |
| PD-L1 | programmed cell death ligand-1 |
| pFAK | phosphorylated focal adhesion kinase |
| PGE2 | prostaglandin E2 |
| PI3K | phosphatidylinositol-3-kinase |
| PITX1 | paired-like homeodomain 1 |
| PRC2 | polycomb repressive complex 2 |
| PRKD3 | protein kinase D 3 |
| PTX | paclitaxel |
| qRT-PCR | quantitative real-time polymerase chain reaction |
| ROBO1 | roundabout, axon guidance receptor, and homolog 1 |
| SCLC | small cell lung cancer |
| SMURF | Smad ubiquitin regulatory factor |
| SOX4 | sex-determining region Y-box 4 |
| SOX7 | SRY-Box 7 |
| Sp1 | specificity protein 1 |
| STAT3 | signal transducer and activator of transcription 3 |
| SULF2 | sulfatase 2 |
| TBILA | TGF-β-induced lncRNA |
| TCF4 | transcription factor 4 |
| TGF-β | transforming growth factor-β |
| TGFBR | TGF-β receptor |
| TIF1γ | transcriptional intermediary factor 1 γ |
| Tregs | regulatory T cells |
| USP15 | Ubiquitin specific protease 15 |
| UTR | untranslated region |
| XIST | X inactivate-specific transcript |
| YAP | yes-associated protein |
| ZEB | Zinc finger E-box-binding homeobox |
| Zo-1 | zonula occludens-1 |
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bender, E. Epidemiology: The dominant malignancy. Nature 2014, 513, S2. [Google Scholar] [CrossRef] [PubMed]
- Politi, K.; Herbst, R.S. Lung cancer in the era of precision medicine. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2015, 21, 2213–2220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, K.L.; Tsai, Y.M.; Lien, C.T.; Kuo, P.L.; Hung, A.J. The Roles of MicroRNA in Lung Cancer. Int. J. Mol. Sci. 2019, 20, 1611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riihimaki, M.; Hemminki, A.; Fallah, M.; Thomsen, H.; Sundquist, K.; Sundquist, J.; Hemminki, K. Metastatic sites and survival in lung cancer. Lung Cancer (Amst. Neth.) 2014, 86, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Morgensztern, D.; Ng, S.H.; Gao, F.; Govindan, R. Trends in stage distribution for patients with non-small cell lung cancer: A National Cancer Database survey. J. Thorac. Oncol. 2010, 5, 29–33. [Google Scholar] [CrossRef] [Green Version]
- Ettinger, D.S.; Wood, D.E.; Aisner, D.L.; Akerley, W.; Bauman, J.; Chirieac, L.R.; D’Amico, T.A.; DeCamp, M.M.; Dilling, T.J.; Dobelbower, M.; et al. Non-Small Cell Lung Cancer, Version 5.2017, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Canc. Netw. 2017, 15, 504–535. [Google Scholar] [CrossRef]
- Ettinger, D.S.; Wood, D.E.; Akerley, W.; Bazhenova, L.A.; Borghaei, H.; Camidge, D.R.; Cheney, R.T.; Chirieac, L.R.; D’Amico, T.A.; Dilling, T.J.; et al. NCCN Guidelines Insights: Non-Small Cell Lung Cancer, Version 4.2016. J. Natl. Compr. Canc. Netw. 2016, 14, 255–264. [Google Scholar] [CrossRef] [Green Version]
- Nicole, L.; Cappello, F.; Cappellesso, R.; VandenBussche, C.J.; Fassina, A. MicroRNA profiling in serous cavity specimens: Diagnostic challenges and new opportunities. Cancer Cytopathol. 2019, 127, 493–500. [Google Scholar] [CrossRef]
- Dykes, I.M.; Emanueli, C. Transcriptional and Post-transcriptional Gene Regulation by Long Non-coding RNA. Genom. Proteom. Bioinform. 2017, 15, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Sun, M.; Liu, H.; Yao, Y.; Song, Y. Long non-coding RNAs: A new frontier in the study of human diseases. Cancer Lett. 2013, 339, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Dutta, A. MicroRNAs in cancer. Annu. Rev. Pathol. 2009, 4, 199–227. [Google Scholar] [CrossRef] [PubMed]
- Guo, T.; Li, J.; Zhang, L.; Hou, W.; Wang, R.; Zhang, J.; Gao, P. Multidimensional communication of microRNAs and long non-coding RNAs in lung cancer. J. Cancer Res. Clin. Oncol. 2019, 145, 31–48. [Google Scholar] [CrossRef]
- Huang, Q. Predictive relevance of ncRNAs in non-small-cell lung cancer patients with radiotherapy: A review of the published data. Biomark. Med. 2018, 12, 1149–1159. [Google Scholar] [CrossRef]
- Li, W.Q.; Zhang, J.P.; Wang, Y.Y.; Li, X.Z.; Sun, L. MicroRNA-422a functions as a tumor suppressor in non-small cell lung cancer through SULF2-mediated TGF-beta/SMAD signaling pathway. Cell Cycle (Georget. Tex.) 2019, 18, 1727–1744. [Google Scholar] [CrossRef]
- Kawasaki, N.; Miwa, T.; Hokari, S.; Sakurai, T.; Ohmori, K.; Miyauchi, K.; Miyazono, K.; Koinuma, D. Long noncoding RNA NORAD regulates transforming growth factor-beta signaling and epithelial-to-mesenchymal transition-like phenotype. Cancer Sci. 2018, 109, 2211–2220. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Wan, L.; Liu, Z.; Xu, G.; Wang, S.; Su, Z.; Zhang, Y.; Zhang, C.; Liu, X.; Lei, Z. Long non-coding RNA XIST promotes TGF-β-induced epithelial-mesenchymal transition by regulating miR-367/141-ZEB2 axis in non-small-cell lung cancer. Cancer Lett. 2018, 418. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, G.; Cheng, Z.; Dai, L.; Jia, L.; Jing, X.; Wang, H.; Zhang, R.; Liu, M.; Jiang, T.; et al. Knockdown of LncRNA-XIST Suppresses Proliferation and TGF-beta1-Induced EMT in NSCLC Through the Notch-1 Pathway by Regulation of miR-137. Genet. Test. Mol. Biomark. 2018, 22, 333–342. [Google Scholar] [CrossRef]
- Dünker, N.; Krieglstein, K. Targeted mutations of transforming growth factor-beta genes reveal important roles in mouse development and adult homeostasis. Eur. J. Biochem. 2000, 267, 6982–6988. [Google Scholar] [CrossRef]
- Miyazono, K.; Katsuno, Y.; Koinuma, D.; Ehata, S.; Morikawa, M. Intracellular and extracellular TGF-beta signaling in cancer: Some recent topics. Front. Med. 2018, 12, 387–411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, D.-L.; Xiang, Y.-Y.; Ji, L.-j.; Lu, X.-J. Competing endogenous RNA interplay in cancer: Mechanism, methodology, and perspectives. Tumor Biol. 2015, 36, 479–488. [Google Scholar] [CrossRef] [PubMed]
- Jonas, S.; Izaurralde, E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015, 16, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Croce, C.M. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009, 10, 704–714. [Google Scholar] [CrossRef]
- Stahlhut, C.; Slack, F.J. MicroRNAs and the cancer phenotype: Profiling, signatures and clinical implications. Genome Med. 2013, 5, 111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, J.; Zhan, Y.; Feng, J.; Luo, J.; Fan, S. MicroRNAs associated with therapy of non-small cell lung cancer. Int. J. Biol. Sci. 2018, 14, 390–397. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.; Li, H. miRNAs as biomarkers and for the early detection of non-small cell lung cancer (NSCLC). J. Thorac. Dis. 2018, 10, 3119–3131. [Google Scholar] [CrossRef]
- Osielska, M.A.; Jagodzinski, P.P. Long non-coding RNA as potential biomarkers in non-small-cell lung cancer: What do we know so far? Biomed. Pharmacother. Biomed. Pharmacother. 2018, 101, 322–333. [Google Scholar] [CrossRef]
- Sang, H.; Liu, H.; Xiong, P.; Zhu, M. Long non-coding RNA functions in lung cancer. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2015, 36, 4027–4037. [Google Scholar] [CrossRef]
- Hu, G.; Niu, F.; Humburg, B.A.; Liao, K.; Bendi, S.; Callen, S.; Fox, H.S.; Buch, S. Molecular mechanisms of long noncoding RNAs and their role in disease pathogenesis. Oncotarget 2018, 9, 18648–18663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, W.; Yuan, S.; Sun, Z.; Li, Y. Long noncoding and circular RNAs in lung cancer: Advances and perspectives. Epigenomics 2016, 8, 1275–1287. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ma, L.; Xu, F.; Zhai, W.; Dong, S.; Yin, L.; Liu, J.; Yu, Z. Role of long non-coding RNA in drug resistance in non-small cell lung cancer. Thorac. Cancer 2018, 9, 761–768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikushima, H.; Miyazono, K. TGFbeta signalling: A complex web in cancer progression. Nat. Rev. Cancer 2010, 10, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Tania, M.; Khan, M.A.; Fu, J. Epithelial to mesenchymal transition inducing transcription factors and metastatic cancer. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2014, 35, 7335–7342. [Google Scholar] [CrossRef]
- Pardali, K.; Moustakas, A. Actions of TGF-beta as tumor suppressor and pro-metastatic factor in human cancer. Biochim. Biophys. Acta 2007, 1775, 21–62. [Google Scholar] [CrossRef]
- Feng, H.; Lu, J.J.; Wang, Y.; Pei, L.; Chen, X. Osthole inhibited TGF beta-induced epithelial-mesenchymal transition (EMT) by suppressing NF-kappaB mediated Snail activation in lung cancer A549 cells. Cell Adhes. Migr. 2017, 11, 464–475. [Google Scholar] [CrossRef] [Green Version]
- Liu, R.-Y.; Zeng, Y.; Lei, Z.; Wang, L.; Yang, H.; Liu, Z.; Zhao, J.; Zhang, H.-T. JAK/STAT3 signaling is required for TGF-β-induced epithelial-mesenchymal transition in lung cancer cells. Int. J. Oncol. 2014, 44, 1643–1651. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.F.; Zhang, H.J.; Wang, H.B.; Zhu, J.; Zhou, W.Y.; Zhang, H.; Zhao, M.C.; Su, J.M.; Gao, W.; Zhang, L.; et al. Transforming growth factor-beta1 induces epithelial-to-mesenchymal transition in human lung cancer cells via PI3K/Akt and MEK/Erk1/2 signaling pathways. Mol. Biol. Rep. 2012, 39, 3549–3556. [Google Scholar] [CrossRef]
- Yang, J.; Weinberg, R.A. Epithelial-mesenchymal transition: At the crossroads of development and tumor metastasis. Dev. Cell 2008, 14, 818–829. [Google Scholar] [CrossRef] [Green Version]
- Gunaratne, A.; Di Guglielmo, G.M. Par6 is phosphorylated by aPKC to facilitate EMT. Cell Adhes. Migr. 2013, 7, 357–361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fong, Y.C.; Hsu, S.F.; Wu, C.L.; Li, T.M.; Kao, S.T.; Tsai, F.J.; Chen, W.C.; Liu, S.C.; Wu, C.M.; Tang, C.H. Transforming growth factor-beta1 increases cell migration and beta1 integrin up-regulation in human lung cancer cells. Lung Cancer (Amst. Neth.) 2009, 64, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Mise, N.; Savai, R.; Yu, H.; Schwarz, J.; Kaminski, N.; Eickelberg, O. Zyxin is a transforming growth factor-beta (TGF-beta)/Smad3 target gene that regulates lung cancer cell motility via integrin alpha5beta1. J. Biol. Chem. 2012, 287, 31393–31405. [Google Scholar] [CrossRef] [Green Version]
- Parekh, A.; Weaver, A.M. Regulation of invadopodia by mechanical signaling. Exp. Cell Res. 2016, 343, 89–95. [Google Scholar] [CrossRef] [Green Version]
- Eddy, R.J.; Weidmann, M.D.; Sharma, V.P.; Condeelis, J.S. Tumor Cell Invadopodia: Invasive Protrusions that Orchestrate Metastasis. Trends Cell Biol. 2017, 27, 595–607. [Google Scholar] [CrossRef]
- Pelaez, R.; Morales, X.; Salvo, E.; Garasa, S.; Ortiz de Solorzano, C.; Martinez, A.; Larrayoz, I.M.; Rouzaut, A. beta3 integrin expression is required for invadopodia-mediated ECM degradation in lung carcinoma cells. PLoS ONE 2017, 12, e0181579. [Google Scholar] [CrossRef] [Green Version]
- Reymond, N.; d’Agua, B.B.; Ridley, A.J. Crossing the endothelial barrier during metastasis. Nat. Rev. Cancer 2013, 13, 858–870. [Google Scholar] [CrossRef]
- Salvo, E.; Garasa, S.; Dotor, J.; Morales, X.; Pelaez, R.; Altevogt, P.; Rouzaut, A. Combined targeting of TGF-β1 and integrin β3 impairs lymph node metastasis in a mouse model of non-small-cell lung cancer. Mol. Cancer 2014, 13, 112. [Google Scholar] [CrossRef] [Green Version]
- Baratelli, F.; Lee, J.M.; Hazra, S.; Lin, Y.; Walser, T.C.; Schaue, D.; Pak, P.S.; Elashoff, D.; Reckamp, K.; Zhang, L.; et al. PGE(2) contributes to TGF-beta induced T regulatory cell function in human non-small cell lung cancer. Am. J. Transl. Res. 2010, 2, 356–367. [Google Scholar]
- David, J.M.; Dominguez, C.; McCampbell, K.K.; Gulley, J.L.; Schlom, J.; Palena, C. A novel bifunctional anti-PD-L1/TGF-beta Trap fusion protein (M7824) efficiently reverts mesenchymalization of human lung cancer cells. Oncoimmunology 2017, 6, e1349589. [Google Scholar] [CrossRef] [Green Version]
- Santarpia, M.; Gonzalez-Cao, M.; Viteri, S.; Karachaliou, N.; Altavilla, G.; Rosell, R. Programmed cell death protein-1/programmed cell death ligand-1 pathway inhibition and predictive biomarkers: Understanding transforming growth factor-beta role. Transl. Lung Cancer Res. 2015, 4, 728–742. [Google Scholar] [CrossRef] [PubMed]
- Trinh, T.L.; Kandell, W.M.; Donatelli, S.S.; Tu, N.; Tejera, M.M.; Gilvary, D.L.; Eksioglu, E.A.; Burnette, A.; Adams, W.A.; Liu, J.; et al. Immune evasion by TGFbeta-induced miR-183 repression of MICA/B expression in human lung tumor cells. Oncoimmunology 2019, 8, e1557372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miles, F.L.; Pruitt, F.L.; van Golen, K.L.; Cooper, C.R. Stepping out of the flow: Capillary extravasation in cancer metastasis. Clin. Exp. Metastasis 2008, 25, 305–324. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.R.; Tai, Y.; Jin, Y.; Hammell, M.C.; Wilkinson, J.E.; Roe, J.S.; Vakoc, C.R.; Van Aelst, L. TGF-beta/Smad signaling through DOCK4 facilitates lung adenocarcinoma metastasis. Genes Dev. 2015, 29, 250–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Popper, H.H. Progression and metastasis of lung cancer. Cancer Metastasis Rev. 2016, 35, 75–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vicent, S.; Luis-Ravelo, D.; Anton, I.; Garcia-Tunon, I.; Borras-Cuesta, F.; Dotor, J.; De Las Rivas, J.; Lecanda, F. A novel lung cancer signature mediates metastatic bone colonization by a dual mechanism. Cancer Res. 2008, 68, 2275–2285. [Google Scholar] [CrossRef] [Green Version]
- Luis-Ravelo, D.; Anton, I.; Zandueta, C.; Valencia, K.; Ormazabal, C.; Martinez-Canarias, S.; Guruceaga, E.; Perurena, N.; Vicent, S.; De Las Rivas, J.; et al. A gene signature of bone metastatic colonization sensitizes for tumor-induced osteolysis and predicts survival in lung cancer. Oncogene 2014, 33, 5090–5099. [Google Scholar] [CrossRef] [Green Version]
- Zhu, W.; He, J.; Chen, D.; Zhang, B.; Xu, L.; Ma, H.; Liu, X.; Zhang, Y.; Le, H. Expression of miR-29c, miR-93, and miR-429 as potential biomarkers for detection of early stage non-small lung cancer. PLoS ONE 2014, 9, e87780. [Google Scholar] [CrossRef]
- Qu, M.H.; Han, C.; Srivastava, A.K.; Cui, T.; Zou, N.; Gao, Z.Q.; Wang, Q.E. miR-93 promotes TGF-beta-induced epithelial-to-mesenchymal transition through downregulation of NEDD4L in lung cancer cells. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2016, 37, 5645–5651. [Google Scholar] [CrossRef]
- Wang, T.; Lv, M.; Shen, S.; Zhou, S.; Wang, P.; Chen, Y.; Liu, B.; Yu, L.; Hou, Y. Cell-free microRNA expression profiles in malignant effusion associated with patient survival in non-small cell lung cancer. PLoS ONE 2012, 7, e43268. [Google Scholar] [CrossRef] [Green Version]
- Cai, J.; Fang, L.; Huang, Y.; Li, R.; Xu, X.; Hu, Z.; Zhang, L.; Yang, Y.; Zhu, X.; Zhang, H.; et al. Simultaneous overactivation of Wnt/beta-catenin and TGFbeta signalling by miR-128-3p confers chemoresistance-associated metastasis in NSCLC. Nat. Commun. 2017, 8, 15870. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wu, Q.; Zhang, Y.; Zhang, H.N.; Wang, Y.B.; Wang, W. TGF-beta1-induced epithelial-mesenchymal transition in lung cancer cells involves upregulation of miR-9 and downregulation of its target, E-cadherin. Cell. Mol. Biol. Lett. 2017, 22, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, L.; Wang, W.; Ding, W.; Zhang, L. MiR-9 is involved in TGF-β1-induced lung cancer cell invasion and adhesion by targeting SOX7. J. Cell. Mol. Med. 2017, 21, 2000–2008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shoji, H.; Tsuchida, K.; Kishi, H.; Yamakawa, N.; Matsuzaki, T.; Liu, Z.; Nakamura, T.; Sugino, H. Identification and characterization of a PDZ protein that interacts with activin type II receptors. J. Biol. Chem. 2000, 275, 5485–5492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kitamura, K.; Seike, M.; Okano, T.; Matsuda, K.; Miyanaga, A.; Mizutani, H.; Noro, R.; Minegishi, Y.; Kubota, K.; Gemma, A. MiR-134/487b/655 cluster regulates TGF-β-induced epithelial-mesenchymal transition and drug resistance to gefitinib by targeting MAGI2 in lung adenocarcinoma cells. Mol. Cancer Ther. 2014, 13, 444–453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Shi, H.; Liu, B.; Li, J.; Liu, Y.; Yu, B. miR-330-3p controls cell proliferation by targeting early growth response 2 in non-small-cell lung cancer. Acta Biochim. Et Biophys. Sin. 2015, 47, 431–440. [Google Scholar] [CrossRef] [Green Version]
- Chen, G.; Ye, B. The key microRNAs regulated the development of non-small cell lung cancer by targeting TGF-beta-induced epithelial-mesenchymal transition. Comb. Chem. High Throughput Screen. 2019. [Google Scholar] [CrossRef]
- Yang, F.; Xiong, H.; Duan, L.; Li, Q.; Li, X.; Zhou, Y. MiR-1246 Promotes Metastasis and Invasion of A549 cells by Targeting GSK-3betaMediated Wnt/beta-Catenin Pathway. Cancer Res. Treat. 2019. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Hutter, D.; Liu, Y.; Wang, X.; Sheikh, M.S.; Chan, A.M.; Holbrook, N.J. Transforming growth factor-beta 1 suppresses serum deprivation-induced death of A549 cells through differential effects on c-Jun and JNK activities. J. Biol. Chem. 2000, 275, 18234–18242. [Google Scholar] [CrossRef] [Green Version]
- Pirozzi, G.; Tirino, V.; Camerlingo, R.; Franco, R.; La Rocca, A.; Liguori, E.; Martucci, N.; Paino, F.; Normanno, N.; Rocco, G. Epithelial to mesenchymal transition by TGFβ-1 induction increases stemness characteristics in primary non small cell lung cancer cell line. PLoS ONE 2011, 6, e21548. [Google Scholar] [CrossRef]
- Li, G.; Wu, F.; Yang, H.; Deng, X.; Yuan, Y. MiR-9-5p promotes cell growth and metastasis in non-small cell lung cancer through the repression of TGFBR2. Biomed. Pharmacother. Biomed. Pharmacother. 2017, 96, 1170–1178. [Google Scholar] [CrossRef] [PubMed]
- Leon, G.; MacDonagh, L.; Finn, S.P.; Cuffe, S.; Barr, M.P. Cancer stem cells in drug resistant lung cancer: Targeting cell surface markers and signaling pathways. Pharmacol. Ther. 2016, 158, 71–90. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Han, J.; Zhu, H.; Peng, L.; Chen, Z. miR181b5p mediates TGFbeta1-induced epithelial-to-mesenchymal transition in non-small cell lung cancer stem-like cells derived from lung adenocarcinoma A549 cells. Int. J. Oncol. 2017, 51, 158–168. [Google Scholar] [CrossRef] [Green Version]
- Cao, M.; Seike, M.; Soeno, C.; Mizutani, H.; Kitamura, K.; Minegishi, Y.; Noro, R.; Yoshimura, A.; Cai, L.; Gemma, A. MiR-23a regulates TGF-beta-induced epithelial-mesenchymal transition by targeting E-cadherin in lung cancer cells. Int. J. Oncol. 2012, 41, 869–875. [Google Scholar] [CrossRef] [Green Version]
- Soung, Y.H.; Nguyen, T.; Cao, H.; Lee, J.; Chung, J. Emerging roles of exosomes in cancer invasion and metastasis. Bmb. Rep. 2016, 49, 18–25. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Kim, T.Y.; Lee, M.S.; Mun, J.Y.; Ihm, C.; Kim, S.A. Exosome cargo reflects TGF-beta1-mediated epithelial-to-mesenchymal transition (EMT) status in A549 human lung adenocarcinoma cells. Biochem. Biophys. Res. Commun. 2016, 478, 643–648. [Google Scholar] [CrossRef]
- Zhang, J.X.; Zhai, J.F.; Yang, X.T.; Wang, J. MicroRNA-132 inhibits migration, invasion and epithelial-mesenchymal transition by regulating TGFβ1/Smad2 in human non-small cell lung cancer. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 3793–3801. [Google Scholar]
- Hu, H.; Xu, Z.; Li, C.; Xu, C.; Lei, Z.; Zhang, H.T.; Zhao, J. MiR-145 and miR-203 represses TGF-beta-induced epithelial-mesenchymal transition and invasion by inhibiting SMAD3 in non-small cell lung cancer cells. Lung Cancer (Amst. Neth.) 2016, 97, 87–94. [Google Scholar] [CrossRef]
- Zeng, Y.; Zhu, J.; Shen, D.; Qin, H.; Lei, Z.; Li, W.; Huang, J.A.; Liu, Z. Repression of Smad4 by miR205 moderates TGF-beta-induced epithelial-mesenchymal transition in A549 cell lines. Int. J. Oncol. 2016, 49, 700–708. [Google Scholar] [CrossRef] [Green Version]
- Zu, L.; Xue, Y.; Wang, J.; Fu, Y.; Wang, X.; Xiao, G.; Hao, M.; Sun, X.; Wang, Y.; Fu, G.; et al. The feedback loop between miR-124 and TGF-β pathway plays a significant role in non-small cell lung cancer metastasis. Carcinogenesis 2016, 37, 333–343. [Google Scholar] [CrossRef] [Green Version]
- Wu, L.; Hu, B.; Zhao, B.; Liu, Y.; Yang, Y.; Zhang, L.; Chen, J. Circulating microRNA-422a is associated with lymphatic metastasis in lung cancer. Oncotarget 2017, 8, 42173–42188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, X.; Meng, L.; Sun, H.; Li, Z.; Zhang, X.; Hua, S. MicroRNA-196b Inhibits Cell Growth and Metastasis of Lung Cancer Cells by Targeting Runx2. Cell. Physiol. Biochem. 2017, 43, 757–767. [Google Scholar] [CrossRef] [PubMed]
- Jiang, K.; Zhao, T.; Shen, M.; Zhang, F.; Duan, S.; Lei, Z.; Chen, Y. MiR-940 inhibits TGF-beta-induced epithelial-mesenchymal transition and cell invasion by targeting Snail in non-small cell lung cancer. J. Cancer 2019, 10, 2735–2744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, K.; Li, X.Y.; Wang, Z.M.; Han, Z.F.; Zhao, Y.H. MiR-22 inhibits lung cancer cell EMT and invasion through targeting Snail. Eur. Rev. Med Pharmacol. Sci. 2017, 21, 3598–3604. [Google Scholar]
- Oktyabri, D.; Tange, S.; Terashima, M.; Ishimura, A.; Suzuki, T. EED regulates epithelial-mesenchymal transition of cancer cells induced by TGF-β. Biochem. Biophys. Res. Commun. 2014, 453, 124–130. [Google Scholar] [CrossRef] [Green Version]
- Tange, S.; Oktyabri, D.; Terashima, M.; Ishimura, A.; Suzuki, T. JARID2 is involved in transforming growth factor-β-induced epithelial-mesenchymal transition of lung and colon cancer cell lines. PLoS ONE 2014, 9, e115684. [Google Scholar] [CrossRef]
- Yin, Q.; Han, Y.; Zhu, D.; Li, Z.; Shan, S.; Jin, W.; Lu, Q.; Ren, T. miR-145 and miR-497 suppress TGF-β-induced epithelial-mesenchymal transition of non-small cell lung cancer by targeting MTDH. Cancer Cell Int. 2018, 18, 105. [Google Scholar] [CrossRef]
- Ke, Y.; Zhao, W.; Xiong, J.; Cao, R. miR-149 Inhibits Non-Small-Cell Lung Cancer Cells EMT by Targeting FOXM1. Biochem. Res. Int. 2013, 2013, 506731. [Google Scholar] [CrossRef]
- Li, J.; Wang, Y.; Luo, J.; Fu, Z.; Ying, J.; Yu, Y.; Yu, W. miR-134 inhibits epithelial to mesenchymal transition by targeting FOXM1 in non-small cell lung cancer cells. Febs Lett. 2012, 586, 3761–3765. [Google Scholar] [CrossRef] [Green Version]
- Xiao, B.; Liu, H.; Gu, Z.; Ji, C. Expression of microRNA-133 inhibits epithelial-mesenchymal transition in lung cancer cells by directly targeting FOXQ1. Arch. De Bronconeumol. 2016, 52, 505–511. [Google Scholar] [CrossRef]
- Zhang, H.W.; Wang, E.W.; Li, L.X.; Yi, S.H.; Li, L.C.; Xu, F.L.; Wang, D.L.; Wu, Y.Z.; Nian, W.Q. A regulatory loop involving miR-29c and Sp1 elevates the TGF-beta1 mediated epithelial-to-mesenchymal transition in lung cancer. Oncotarget 2016, 7, 85905–85916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yue, Z.; Jin, S.; Yang, C.; Levine, A.J.; Heintz, N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl. Acad. Sci. USA 2003, 100, 15077–15082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qu, X.; Yu, J.; Bhagat, G.; Furuya, N.; Hibshoosh, H.; Troxel, A.; Rosen, J.; Eskelinen, E.L.; Mizushima, N.; Ohsumi, Y.; et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Investig. 2003, 112, 1809–1820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Zhang, Y.; Wu, Q.; Wang, Y.B.; Wang, W. miR-16 mimics inhibit TGF-beta1-induced epithelial-to-mesenchymal transition via activation of autophagy in non-small cell lung carcinoma cells. Oncol. Rep. 2018, 39, 247–254. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Li, C.; Bi, H.; Bai, S.; Zhao, L.; Zhang, J.; Qi, C. Targeting ZEB2 By microRNA-129 In Non-Small Cell Lung Cancer Suppresses Cell Proliferation, Invasion and Migration Via Regulating Wnt/β-Catenin Signaling Pathway and Epithelial-Mesenchymal Transition. Oncotargets Ther. 2019, 12, 9165–9175. [Google Scholar] [CrossRef] [Green Version]
- Bin, C.; Xiaofeng, H.; Wanzi, X. The effect of microRNA-129 on the migration and invasion in NSCLC cells and its mechanism. Exp. Lung Res. 2018, 44, 280–287. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.S.; Zheng, Y.L.; Li, C.; Ding, C.; Xu, C.; Zhao, J. MicroRNA-485-5p suppresses growth and metastasis in non-small cell lung cancer cells by targeting IGF2BP2. Life Sci. 2018, 199, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Sun, Y.; Wu, Y.; Tang, D.; Ding, X.; Xu, W.; Su, B.; Gao, W. Downregulation of miR-3127-5p promotes epithelial-mesenchymal transition via FZD4 regulation of Wnt/β-catenin signaling in non-small-cell lung cancer. Mol. Carcinog. 2018, 57, 842–853. [Google Scholar] [CrossRef]
- Gao, P.; Wang, H.; Yu, J.; Zhang, J.; Yang, Z.; Liu, M.; Niu, Y.; Wei, X.; Wang, W.; Li, H.; et al. miR-3607-3p suppresses non-small cell lung cancer (NSCLC) by targeting TGFBR1 and CCNE2. PLoS Genet. 2018, 14, e1007790. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.X.; Yan, Y.F.; Liu, Y.M.; Li, Y.J.; Zhang, H.H.; Pang, M.; Hu, J.X.; Zhao, W.; Xie, N.; Zhou, L.; et al. Smad3-related miRNAs regulated oncogenic TRIB2 promoter activity to effectively suppress lung adenocarcinoma growth. Cell Death Dis. 2016, 7, e2528. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Sempere, L.F.; Ouyang, H.; Memoli, V.A.; Andrew, A.S.; Luo, Y.; Demidenko, E.; Korc, M.; Shi, W.; Preis, M.; et al. MicroRNA-31 functions as an oncogenic microRNA in mouse and human lung cancer cells by repressing specific tumor suppressors. J. Clin. Investig. 2010, 120, 1298–1309. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Liu, L.; Cai, J.; Wu, J.; Guan, H.; Zhu, X.; Yuan, J.; Chen, S.; Li, M. Targeting Smad2 and Smad3 by miR-136 suppresses metastasis-associated traits of lung adenocarcinoma cells. Oncol. Res. 2013, 21, 345–352. [Google Scholar] [CrossRef]
- Wang, L.K.; Hsiao, T.H.; Hong, T.M.; Chen, H.Y.; Kao, S.H.; Wang, W.L.; Yu, S.L.; Lin, C.W.; Yang, P.C. MicroRNA-133a suppresses multiple oncogenic membrane receptors and cell invasion in non-small cell lung carcinoma. PLoS ONE 2014, 9, e96765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, T.; Hu, C.; Yang, H.; Cao, L.; An, J. Transforming growth factor-β-induced miR143 expression in regulation of non-small cell lung cancer cell viability and invasion capacity in vitro and in vivo. Int. J. Oncol. 2014, 45, 1977–1988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, J.; Song, Y.; Bi, N.; Shen, J.; Liu, W.; Fan, J.; Sun, G.; Tong, T.; He, J.; Shi, Y.; et al. DNA methylation-mediated repression of miR-886-3p predicts poor outcome of human small cell lung cancer. Cancer Res. 2013, 73, 3326–3335. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Li, T.; Han, L.; Qin, P.; Wu, Z.; Xu, B.; Gao, Q.; Song, Y. TGFbeta1-induced down-regulation of microRNA-138 contributes to epithelial-mesenchymal transition in primary lung cancer cells. Biochem. Biophys. Res. Commun. 2018, 496, 1169–1175. [Google Scholar] [CrossRef]
- Jiang, L.; Wang, R.; Fang, L.; Ge, X.; Chen, L.; Zhou, M.; Zhou, Y.; Xiong, W.; Hu, Y.; Tang, X.; et al. HCP5 is a SMAD3-responsive long non-coding RNA that promotes lung adenocarcinoma metastasis via miR-203/SNAI axis. Theranostics 2019, 9, 2460–2474. [Google Scholar] [CrossRef]
- Chen, T.; Qin, S.; Gu, Y.; Pan, H.; Bian, D. Long non-coding RNA NORAD promotes the occurrence and development of non-small cell lung cancer by adsorbing MiR-656-3p. Mol. Genet. Genom. Med. 2019, 7, e757. [Google Scholar] [CrossRef] [Green Version]
- Lu, W.; Zhang, H.; Niu, Y.; Wu, Y.; Sun, W.; Li, H.; Kong, J.; Ding, K.; Shen, H.M.; Wu, H.; et al. Long non-coding RNA linc00673 regulated non-small cell lung cancer proliferation, migration, invasion and epithelial mesenchymal transition by sponging miR-150-5p. Mol. Cancer 2017, 16, 118. [Google Scholar] [CrossRef]
- Terashima, M.; Tange, S.; Ishimura, A.; Suzuki, T. MEG3 Long Noncoding RNA Contributes to the Epigenetic Regulation of Epithelial-Mesenchymal Transition in Lung Cancer Cell Lines. J. Biol. Chem. 2017, 292, 82–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terashima, M.; Ishimura, A.; Wanna-Udom, S.; Suzuki, T. MEG8 long noncoding RNA contributes to epigenetic progression of the epithelial-mesenchymal transition of lung and pancreatic cancer cells. J. Biol. Chem. 2018, 293, 18016–18030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, L.; Wu, T.; He, P.; Zhang, J.L.; Wu, W. LncRNA ATB promotes the proliferation and metastasis of lung cancer via activation of the p38 signaling pathway. Oncol. Lett. 2018, 16, 3907–3912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, Y.; Luo, X.; Ding, X.; Cui, S.; Guo, C. LncRNA ATB promotes proliferation and metastasis in A549 cells by down-regulation of microRNA-494. J. Cell. Biochem. 2018, 119, 6935–6942. [Google Scholar] [CrossRef] [PubMed]
- Ke, L.; Xu, S.B.; Wang, J.; Jiang, X.L.; Xu, M.Q. High expression of long non-coding RNA ATB indicates a poor prognosis and regulates cell proliferation and metastasis in non-small cell lung cancer. Clin. Transl. Oncol. 2017, 19, 599–605. [Google Scholar] [CrossRef]
- Lu, Z.; Li, Y.; Che, Y.; Huang, J.; Sun, S.; Mao, S.; Lei, Y.; Li, N.; Sun, N.; He, J. The TGFβ-induced lncRNA TBILA promotes non-small cell lung cancer progression in vitro and in vivo via cis-regulating HGAL and activating S100A7/JAB1 signaling. Cancer Lett. 2018, 432, 156–168. [Google Scholar] [CrossRef]
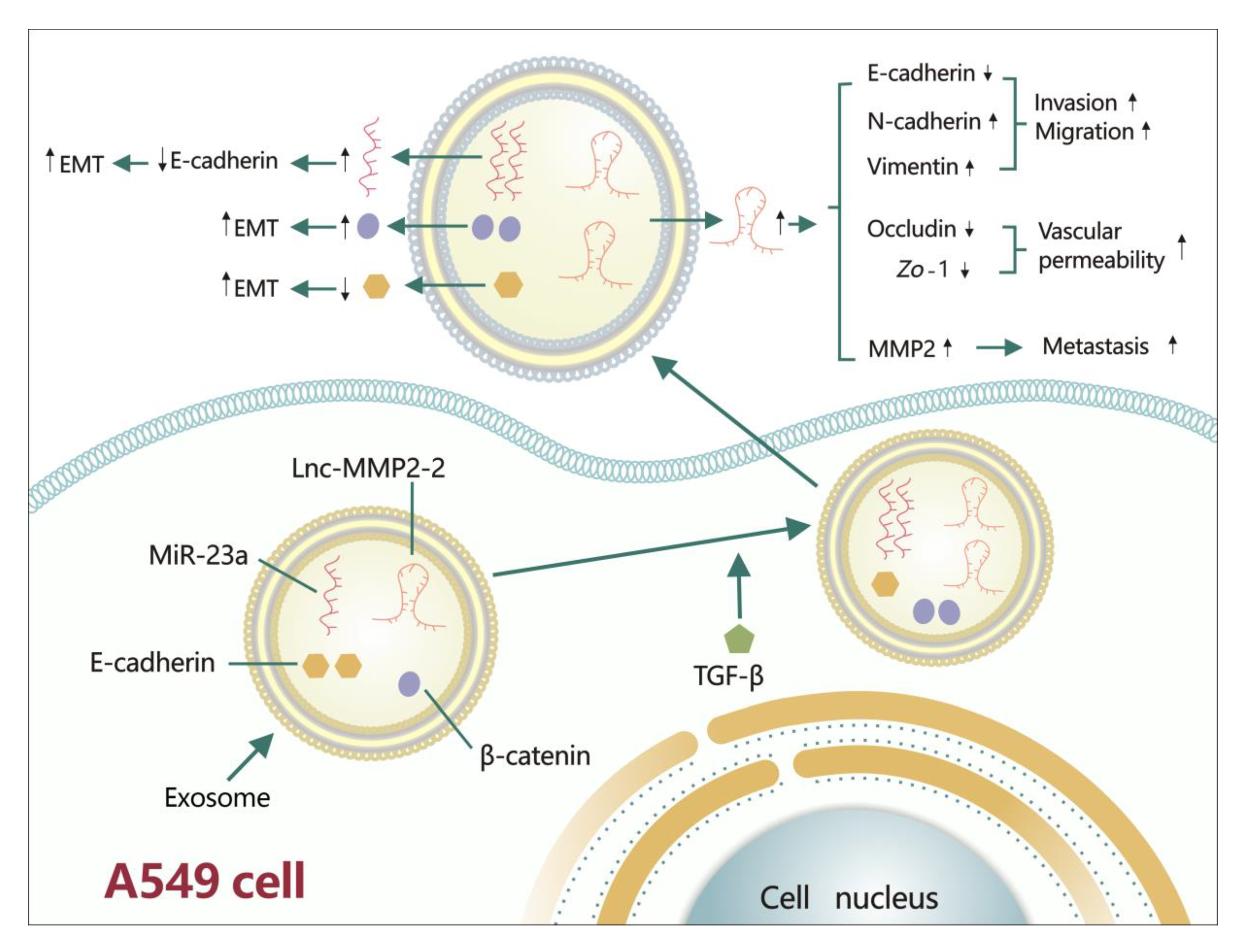
- Wu, D.M.; Deng, S.H.; Liu, T.; Han, R.; Zhang, T.; Xu, Y. TGF-β-mediated exosomal lnc-MMP2-2 regulates migration and invasion of lung cancer cells to the vasculature by promoting MMP2 expression. Cancer Med. 2018, 7, 5118–5129. [Google Scholar] [CrossRef]
- Tang, L.; Wang, T.; Zhang, Y.; Zhang, J.; Zhao, H.; Wang, H.; Wu, Y.; Liu, K. Long Non-Coding RNA AWPPH Promotes Postoperative Distant Recurrence in Resected Non-Small Cell Lung Cancer by Upregulating Transforming Growth Factor beta 1 (TGF-β1). Med Sci. Monit. Int. Med J. Exp. Clin. Res. 2019, 25, 2535–2541. [Google Scholar] [CrossRef]
- Fu, Y.; Zhang, P.; Nan, H.; Lu, Y.; Zhao, J.; Yang, M.; Song, Q. LncRNA CASC11 promotes TGF-beta1, increases cancer cell stemness and predicts postoperative survival in small cell lung cancer. Gene 2019, 704, 91–96. [Google Scholar] [CrossRef]
- Miao, F.; Chen, J.; Shi, M.; Song, Y.; Chen, Z.; Pang, L. LncRNA HAND2-AS1 inhibits non-small cell lung cancer migration, invasion and maintains cell stemness through the interactions with TGF-β1. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef] [Green Version]
- Hao, Y.; Yang, X.; Zhang, D.; Luo, J.; Chen, R. Long noncoding RNA LINC01186, regulated by TGF-beta/SMAD3, inhibits migration and invasion through Epithelial-Mesenchymal-Transition in lung cancer. Gene 2017, 608, 1–12. [Google Scholar] [CrossRef]
- Zhang, C.; Hao, Y.; Wang, Y.; Xu, J.; Teng, Y.; Yang, X. TGF-beta/SMAD4-Regulated LncRNA-LINP1 Inhibits Epithelial-Mesenchymal Transition in Lung Cancer. Int. J. Biol. Sci. 2018, 14, 1715–1723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Lan, F.; Xia, Y. lncRA ANCR Inhibits Non-Small Cell Lung Cancer Cell Migration and Invasion by Inactivating TGF-beta Pathway. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2018, 24, 6002–6009. [Google Scholar] [CrossRef]
- Lu, Z.; Li, Y.; Wang, J.; Che, Y.; Sun, S.; Huang, J.; Chen, Z.; He, J. Long non-coding RNA NKILA inhibits migration and invasion of non-small cell lung cancer via NF-κB/Snail pathway. J. Exp. Clin. Cancer Res. 2017, 36, 54. [Google Scholar] [CrossRef]
- Iqbal, M.A.; Arora, S.; Prakasam, G.; Calin, G.A.; Syed, M.A. MicroRNA in lung cancer: Role, mechanisms, pathways and therapeutic relevance. Mol. Asp. Med. 2019, 70, 3–20. [Google Scholar] [CrossRef]
- Ergun, S.; Oztuzcu, S. Oncocers: ceRNA-mediated cross-talk by sponging miRNAs in oncogenic pathways. Tumour Biol. 2015, 36, 3129–3136. [Google Scholar] [CrossRef]
- Panda, A.C. Circular RNAs Act as miRNA Sponges. Adv. Exp. Med. Biol. 2018, 1087, 67–79. [Google Scholar] [CrossRef]
- Chen, Y.; Wei, S.; Wang, X.; Zhu, X.; Han, S. Progress in research on the role of circular RNAs in lung cancer. World J. Surg. Oncol. 2018, 16, 215. [Google Scholar] [CrossRef]
- Hu, W.; Bi, Z.-Y.; Chen, Z.-L.; Liu, C.; Li, L.-L.; Zhang, F.; Zhou, Q.; Zhu, W.; Song, Y.-Y.-Y.; Zhan, B.-T.; et al. Emerging landscape of circular RNAs in lung cancer. Cancer Lett. 2018, 427, 18–27. [Google Scholar] [CrossRef]
- Fang, L.; Wu, S.; Zhu, X.; Cai, J.; Wu, J.; He, Z.; Liu, L.; Zeng, M.; Song, E.; Li, J.; et al. MYEOV functions as an amplified competing endogenous RNA in promoting metastasis by activating TGF-β pathway in NSCLC. Oncogene 2019, 38, 896–912. [Google Scholar] [CrossRef]
- Kumar, M.S.; Armenteros-Monterroso, E.; East, P.; Chakravorty, P.; Matthews, N.; Winslow, M.M.; Downward, J. HMGA2 functions as a competing endogenous RNA to promote lung cancer progression. Nature 2014, 505, 212–217. [Google Scholar] [CrossRef]
- He, W.; Dorn, D.C.; Erdjument-Bromage, H.; Tempst, P.; Moore, M.A.; Massague, J. Hematopoiesis controlled by distinct TIF1gamma and Smad4 branches of the TGFbeta pathway. Cell 2006, 125, 929–941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Tong, X.; Zhou, Z.; Wang, S.; Lei, Z.; Zhang, T.; Liu, Z.; Zeng, Y.; Li, C.; Zhao, J.; et al. Circular RNA hsa_circ_0008305 (circPTK2) inhibits TGF-β-induced epithelial-mesenchymal transition and metastasis by controlling TIF1gamma in non-small cell lung cancer. Mol. Cancer 2018, 17, 140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhan, J.W.; Jiao, D.M.; Wang, Y.; Song, J.; Wu, J.H.; Wu, L.J.; Chen, Q.Y.; Ma, S.L. Integrated microRNA and gene expression profiling reveals the crucial miRNAs in curcumin anti-lung cancer cell invasion. Thorac. Cancer 2017, 8, 461–470. [Google Scholar] [CrossRef]
- Scagliotti, G.V. Pemetrexed plus carboplatin or oxaliplatin in advanced non-small cell lung cancer. Semin. Oncol. 2005, 32, S5–S8. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Tu, H.B.; Wu, L.; Liu, M.; Jiang, G.N. MicroRNA-21 Regulates Non-Small Cell Lung Cancer Cell Invasion and Chemo-Sensitivity through SMAD7. Cell. Physiol. Biochem. 2016, 38, 2152–2162. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Chen, X.; Meng, Q.; Jing, H.; Lu, H.; Yang, Y.; Cai, L.; Zhao, Y. MiR-181b regulates cisplatin chemosensitivity and metastasis by targeting TGFbetaR1/Smad signaling pathway in NSCLC. Sci. Rep. 2015, 5, 17618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Z.; Yin, J.; Fu, W.; Mo, Y.; Pan, Y.; Dai, L.; Huang, H.; Li, S.; Zhao, J. MiRNA 17 family regulates cisplatin-resistant and metastasis by targeting TGFbetaR2 in NSCLC. PLoS ONE 2014, 9, e94639. [Google Scholar] [CrossRef] [Green Version]
- Zhang, N.; Liu, Y.; Wang, Y.; Zhao, M.; Tu, L.; Luo, F. Decitabine reverses TGF-beta1-induced epithelial-mesenchymal transition in non-small-cell lung cancer by regulating miR-200/ZEB axis. Drug Des. Dev. Ther. 2017, 11, 969–983. [Google Scholar] [CrossRef] [Green Version]
- Tong, W.; Han, T.C.; Wang, W.; Zhao, J. LncRNA CASC11 promotes the development of lung cancer through targeting microRNA-302/CDK1 axis. Eur. Rev. Med Pharmacol. Sci. 2019, 23, 6539–6547. [Google Scholar] [CrossRef]
- Donatelli, S.S.; Zhou, J.-M.; Gilvary, D.L.; Eksioglu, E.A.; Chen, X.; Cress, W.D.; Haura, E.B.; Schabath, M.B.; Coppola, D.; Wei, S.; et al. TGF-β-inducible microRNA-183 silences tumor-associated natural killer cells. Proc. Natl. Acad. Sci. USA 2014, 111, 4203–4208. [Google Scholar] [CrossRef] [Green Version]
- Weber, D.G.; Johnen, G.; Casjens, S.; Bryk, O.; Pesch, B.; Jockel, K.H.; Kollmeier, J.; Bruning, T. Evaluation of long noncoding RNA MALAT1 as a candidate blood-based biomarker for the diagnosis of non-small cell lung cancer. BMC Res. Notes 2013, 6, 518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, L.; Dou, Y.; Sui, Z.; Cheng, H.; Liu, X.; Wang, Q.; Gao, P.; Qu, Y.; Xu, M. Upregulated miRNA-182-5p expression in tumor tissue and peripheral blood samples from patients with non-small cell lung cancer is associated with downregulated Caspase 2 expression. Exp. Ther. Med. 2020, 19, 603–610. [Google Scholar] [CrossRef]
- Tang, H.; Zhao, L.; Li, M.; Li, T.; Hao, Y. Investigation of LINC00342 as a poor prognostic biomarker for human patients with non-small cell lung cancer. J. Cell. Biochem. 2019, 120, 5055–5061. [Google Scholar] [CrossRef] [PubMed]
- Kwok, H.-H.; Ning, Z.; Chong, P.W.-C.; Wan, T.S.-K.; Ng, M.H.-L.; Ho, G.Y.F.; Ip, M.S.-M.; Lam, D.C.-L. Transfer of Extracellular Vesicle-Associated-RNAs Induces Drug Resistance in ALK-Translocated Lung Adenocarcinoma. Cancers 2019, 11, 104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, H.; Zhou, J.; Mei, S.; Wu, D.; Mu, Z.; Chen, B.; Xie, Y.; Ye, Y.; Liu, J. Circulating exosomal microRNA-96 promotes cell proliferation, migration and drug resistance by targeting LMO7. J. Cell. Mol. Med. 2017, 21, 1228–1236. [Google Scholar] [CrossRef] [PubMed]
- Świtlik, W.Z.; Karbownik, M.S.; Suwalski, M.; Kozak, J.; Szemraj, J. Serum miR-210-3p as a Potential Noninvasive Biomarker of Lung Adenocarcinoma: A Preliminary Study. Genet. Test. Mol. Biomark. 2019, 23, 353–358. [Google Scholar] [CrossRef]
- Li, N.; Feng, X.B.; Tan, Q.; Luo, P.; Jing, W.; Zhu, M.; Liang, C.; Tu, J.; Ning, Y. Identification of Circulating Long Noncoding RNA Linc00152 as a Novel Biomarker for Diagnosis and Monitoring of Non-Small-Cell Lung Cancer. Dis. Markers 2017, 2017, 7439698. [Google Scholar] [CrossRef] [Green Version]
- Lv, P.; Yang, S.; Liu, W.; Qin, H.; Tang, X.; Wu, F.; Liu, Z.; Gao, H.; Liu, X. Circulating plasma lncRNAs as novel markers of EGFR mutation status and monitors of epidermal growth factor receptor-tyrosine kinase inhibitor therapy. Thorac. Cancer 2020, 11, 29–40. [Google Scholar] [CrossRef]
- Wang, W.; Ding, M.; Duan, X.; Feng, X.; Wang, P.; Jiang, Q.; Cheng, Z.; Zhang, W.; Yu, S.; Yao, W.; et al. Diagnostic Value of Plasma MicroRNAs for Lung Cancer Using Support Vector Machine Model. J. Cancer 2019, 10, 5090–5098. [Google Scholar] [CrossRef]
- Hojbjerg, J.A.; Ebert, E.B.F.; Clement, M.S.; Winther-Larsen, A.; Meldgaard, P.; Sorensen, B. Circulating miR-30b and miR-30c predict erlotinib response in EGFR-mutated non-small cell lung cancer patients. Lung Cancer (Amst. Neth.) 2019, 135, 92–96. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.L.; Zhang, Z.L.; Zhu, X.B.; Xu, L.; Lu, P.; Xu, M.; Liu, W.J.; Zhang, X.Y.; Yao, H.M.; Ye, X.W. Low plasma miR-25 expression is a favorite prognosis factor in non-small cell lung cancer. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 5251–5259. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, H.; Gao, X.; Wei, F.; Zhang, X.; Su, Y.; Wang, C.; Li, H.; Ren, X. Identification of a three-miRNA signature as a blood-borne diagnostic marker for early diagnosis of lung adenocarcinoma. Oncotarget 2016, 7, 26070–26086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, C.; Su, J.; Zhan, M.; Stass, S.A.; Jiang, F. Sputum long non-coding RNA biomarkers for diagnosis of lung cancer. Cancer Biomark. Sect. A Dis. Mark. 2019, 26, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Leng, Q.; Lin, Y.; Ma, J.; Jiang, F.; Lee, C.-J.; Fang, H.; Jiang, F. Integrating Circulating Immunological and Sputum Biomarkers for the Early Detection of Lung Cancer. Biomark Cancer 2018, 10. [Google Scholar] [CrossRef]
- Wang, W.W.; Zhou, X.L.; Song, Y.J.; Yu, C.H.; Zhu, W.G.; Tong, Y.S. Combination of long noncoding RNA MALAT1 and carcinoembryonic antigen for the diagnosis of malignant pleural effusion caused by lung cancer. Oncotargets Ther. 2018, 11, 2333–2344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hydbring, P.; De Petris, L.; Zhang, Y.; Brandén, E.; Koyi, H.; Novak, M.; Kanter, L.; Hååg, P.; Hurley, J.; Tadigotla, V.; et al. Exosomal RNA-profiling of pleural effusions identifies adenocarcinoma patients through elevated miR-200 and LCN2 expression. Lung Cancer (Amst. Neth.) 2018, 124, 45–52. [Google Scholar] [CrossRef]
- Kim, J.E.; Eom, J.S.; Kim, W.Y.; Jo, E.J.; Mok, J.; Lee, K.; Kim, K.U.; Park, H.K.; Lee, M.K.; Kim, M.H. Diagnostic value of microRNAs derived from exosomes in bronchoalveolar lavage fluid of early-stage lung adenocarcinoma: A pilot study. Thorac. Cancer 2018, 9, 911–915. [Google Scholar] [CrossRef]
- Wu, T.; Hu, H.; Zhang, T.; Jiang, L.; Li, X.; Liu, S.; Zheng, C.; Yan, G.; Chen, W.; Ning, Y.; et al. miR-25 Promotes Cell Proliferation, Migration, and Invasion of Non-Small-Cell Lung Cancer by Targeting the LATS2/YAP Signaling Pathway. Oxid. Med. Cell Longev. 2019, 2019, 9719723. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Wu, X. MiR-21-5p promotes the progression of non-small-cell lung cancer by regulating the expression of SMAD7. Oncotargets Ther. 2018, 11, 8445–8454. [Google Scholar] [CrossRef] [Green Version]
- Hetta, H.F.; Zahran, A.M.; Shafik, E.A.; El-Mahdy, R.I.; Mohamed, N.A.; Nabil, E.E.; Esmaeel, H.M.; Alkady, O.A.; Elkady, A.; Mohareb, D.A.; et al. Circulating miRNA-21 and miRNA-23a Expression Signature as Potential Biomarkers for Early Detection of Non-Small-Cell Lung Cancer. Microrna 2019, 8, 206–215. [Google Scholar] [CrossRef]
- Liu, C.; Li, H.; Jia, J.; Ruan, X.; Liu, Y.; Zhang, X. High Metastasis-Associated Lung Adenocarcinoma Transcript 1 (MALAT1) Expression Promotes Proliferation, Migration, and Invasion of Non-Small Cell Lung Cancer via ERK/Mitogen-Activated Protein Kinase (MAPK) Signaling Pathway. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2019, 25, 5143–5149. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Wang, K.; Huang, X.; Zhao, Z.; Zhao, Z. LncRNA MALAT1 contributes to non-small cell lung cancer progression via modulating miR-200a-3p/programmed death-ligand 1 axis. Int. J. Immunopathol. Pharm. 2019, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, R.; Xia, Y.; Wang, Z.; Zheng, J.; Chen, Y.; Li, X.; Wang, Y.; Ming, H. Serum long non coding RNA MALAT-1 protected by exosomes is up-regulated and promotes cell proliferation and migration in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2017, 490, 406–414. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.-P.; Wang, Y.-Q.; Weng, W.-W.; Nie, W.; Wu, Y.; Deng, Y.; Wei, P.; Xu, M.-D.; Wang, C.-F. Linc00152 promotes Cancer Cell Proliferation and Invasion and Predicts Poor Prognosis in Lung adenocarcinoma. J. Cancer 2017, 8, 2042–2050. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.F.; Kong, J.L.; Zou, S.C.; Gao, H.; Wang, F.; Qin, S.M.; Wang, W. LncRNA LINC00342 regulated cell growth and metastasis in non-small cell lung cancer via targeting miR-203a-3p. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 7408–7418. [Google Scholar] [CrossRef]
- Stieber, P.; Hasholzner, U.; Bodenmüller, H.; Nagel, D.; Sunder-Plassmann, L.; Dienemann, H.; Meier, W.; Fateh-Moghadam, A. CYFRA 21-1. A new marker in lung cancer. Cancer 1993, 72, 707–713. [Google Scholar] [CrossRef]
- de Miguel-Pérez, D.; Bayarri-Lara, C.I.; Ortega, F.G.; Russo, A.; Moyano Rodriguez, M.J.; Alvarez-Cubero, M.J.; Maza Serrano, E.; Lorente, J.A.; Rolfo, C.; Serrano, M.J. Post-Surgery Circulating Tumor Cells and AXL Overexpression as New Poor Prognostic Biomarkers in Resected Lung Adenocarcinoma. Cancers 2019, 11, 1750. [Google Scholar] [CrossRef] [Green Version]
- Economopoulou, P.; Georgoulias, V.; Kotsakis, A. Classifying circulating tumor cells to monitor cancer progression. Exp. Rev. Mol. Diagn. 2017, 17, 153–165. [Google Scholar] [CrossRef]
- Park, G.; Son, B.; Kang, J.; Lee, S.; Jeon, J.; Kim, J.-H.; Yi, G.-R.; Youn, H.; Moon, C.; Nam, S.Y.; et al. LDR-Induced miR-30a and miR-30b Target the PAI-1 Pathway to Control Adverse Effects of NSCLC Radiotherapy. Mol. Ther. 2019, 27, 342–354. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Shen, Z.; Zheng, Y.; Wang, S.; Mao, W. Radiotherapy induced Lewis lung cancer cell apoptosis via inactivating β-catenin mediated by upregulated HOTAIR. Int. J. Clin. Exp. Pathol. 2015, 8, 7878–7886. [Google Scholar]
- Xu, J.; Su, C.; Zhao, F.; Tao, J.; Hu, D.; Shi, A.; Pan, J.; Zhang, Y. Paclitaxel promotes lung cancer cell apoptosis via MEG3-P53 pathway activation. Biochem. Biophys. Res. Commun. 2018, 504, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Yuwen, D.; Chen, J.; Zheng, B.; Gao, J.; Fan, M.; Xue, W.; Wang, Y.; Li, W.; Shu, Y.; et al. Exosomal Transfer Of Cisplatin-Induced miR-425-3p Confers Cisplatin Resistance In NSCLC Through Activating Autophagy. Int. J. Nanomed. 2019, 14, 8121–8132. [Google Scholar] [CrossRef] [Green Version]
- D’Almeida, O.; Mothar, O.; Bondzie, E.A.; Lieumo, Y.; Tagne, L.; Gupta, S.; Volkert, T.; Levine, S.; Tagne, J.-B. Encapsulated miR-200c and Nkx2.1 in a nuclear/mitochondria transcriptional regulatory network of non-metastatic and metastatic lung cancer cells. BMC Cancer 2019, 19, 136. [Google Scholar] [CrossRef]
- Rana, S.; Zöller, M. Exosome target cell selection and the importance of exosomal tetraspanins: A hypothesis. Biochem. Soc. Trans. 2011, 39, 559–562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoshino, A.; Costa-Silva, B.; Shen, T.-L.; Rodrigues, G.; Hashimoto, A.; Tesic Mark, M.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srivastava, A.; Amreddy, N.; Razaq, M.; Towner, R.; Zhao, Y.D.; Ahmed, R.A.; Munshi, A.; Ramesh, R. Exosomes as Theranostics for Lung Cancer. Adv. Cancer Res. 2018, 139, 1–33. [Google Scholar] [CrossRef] [PubMed]
- Mo, D.; Yang, D.; Xiao, X.; Sun, R.; Huang, L.; Xu, J. MiRNA-145 suppresses lung adenocarcinoma cell invasion and migration by targeting N-cadherin. Biotechnol. Lett. 2017, 39, 701–710. [Google Scholar] [CrossRef]
- Vázquez-Ríos, A.J.; Molina-Crespo, Á.; Bouzo, B.L.; López-López, R.; Moreno-Bueno, G.; de la Fuente, M. Exosome-mimetic nanoplatforms for targeted cancer drug delivery. J. Nanobiotechnol. 2019, 17, 85. [Google Scholar] [CrossRef] [Green Version]
- Anderson, C.F.; Grimmett, M.E.; Domalewski, C.J.; Cui, H. Inhalable nanotherapeutics to improve treatment efficacy for common lung diseases. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2020, 12, e1586. [Google Scholar] [CrossRef]
- Garbuzenko, O.B.; Kuzmov, A.; Taratula, O.; Pine, S.R.; Minko, T. Strategy to enhance lung cancer treatment by five essential elements: Inhalation delivery, nanotechnology, tumor-receptor targeting, chemo- and gene therapy. Theranostics 2019, 9, 8362–8376. [Google Scholar] [CrossRef]
- Wu, D.; Wang, W.; He, X.; Jiang, M.; Lai, C.; Hu, X.; Xi, J.; Wang, M. Biofabrication of nano copper oxide and its aptamer bioconjugate for delivery of mRNA 29b to lung cancer cells. Mater. Sci. Eng. C Mater. Biol. Appl. 2019, 97, 827–832. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Guan, X.; Tu, Y.; Zheng, S.; Long, J.; Li, S.; Qi, C.; Xie, X.; Zhang, H.; Zhang, Y. MicroRNA-29b attenuates non-small cell lung cancer metastasis by targeting matrix metalloproteinase 2 and PTEN. J. Exp. Clin. Cancer Res. Cr 2015, 34, 59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sessa, R.; Hata, A. Role of microRNAs in lung development and pulmonary diseases. Pulm. Circ. 2013, 3, 315–328. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Nc-RNA | Cell Lines | Roles | References |
|---|---|---|---|
| miR-93 | A549, H1650 | miR-93→NEDD4L↓→TGF-β/Smad2 signal↑→EMT↑ | [59] |
| miR-128-3p | A549 | miR-128-3p→SMURF2↓, cPP1↓→TGF-β signal↑→EMT↑, metastasis↑, chemoresistance↑ | [61] |
| miR-9 | A549 | miR-9→E-cadherin↓→EMT↑ | [62] |
| miR-23a | A549 | miR-23a→E-cadherin↓→EMT↑ | [74] |
| miR-134, miR-487b | A549 | miR-134, miR-487b→MAGI2↓→EMT↑ | [65] |
| miR-330-3p | NCI-H1975 | miR-330-3p→E-cadherin↓, vimentin↑→EMT↑ | [67] |
| miR-1246 | A549 | miR-1246→E-cadherin↓, vimentin↑, TGF-β↑→EMT↑ | [68] |
| miR-9 | A549, HCC827 | miR-9→SOX7↓→invasion↑, adhesion↑ | [63] |
| miR-9-5p | A549, SK-MES-1 | miR-9-5p→TGFBR2↓→invasion↑, migration↑ | [71] |
| miR-181b-5p | A549 | miR-181b-5p→E-cadherin↓→EMT↑→invasion↑, metastasis↑ | [73] |
| lncRNA HCP5 | A549 | HCP5→miR-203↓→Snail↑, Slug↑→EMT↑, invasion↑ | [107] |
| lncRNA NORAD | A549 | NORAD→Smad3/importin β1 complex formation↑→TGF-β/Smad2/3 signal↑→SNAI1↑, FN1↑→EMT↑ | [18] |
| lncRNA XIST | A549 | XIST→miR-137↓→EMT↑ | [20] |
| XIST→miR-367↓, miR-141↓→ZEB2↑→EMT↑ | [19] | ||
| lncRNA linc00673 | A549, H1975 | linc00673→miR-150-5p↓→ZEB1↑→EMT↑, invasion↑, migration↑ | [109] |
| lncRNA MEG3 | A549, LC-2/ad | MEG3→CDH1↓, miR-200a↓, miR-200c↓, ZEB1↑, ZEB2↑→EMT↑ | [110] |
| lncRNA MEG8 | A549 | MEG8→miR-34a↓, miR-203↓→SNAI1↑, SNAI2↑→EMT↑ | [111] |
| lncRNA ATB | A549, HCC827 | ATB→E-cadherin↓, N-cadherin↑→migration↑ | [112] |
| A549 | ATB→miR-494↓→AKT signal↑, JAK/STAT3 signal↑→EMT↑ | [113] | |
| lncRNA TBILA | A549 | TBILA→HGAL↑, RhoA↑, S100A7/ JAB1 signal↑→invasion↑, migration↑ | [115] |
| lnc-MMP2-2 | A549 | lnc-MMP2-2→E-cadherin↓, N-cadherin↑, vimentin↑→invasion↑, migration↑ | [116] |
| lnc-MMP2-2→occludin↓, Zo-1↓→vascular permeability↑ | |||
| lnc-MMP2-2→MMP2↑→metastasis↑ | |||
| lncRNA AWPPH | NCI-H1993, NCI-H2170 | AWPPH→TGF-β1↑→invasion↑, migration↑ | [117] |
| lncRNA CASC11 | SHP-77, DMS79 | CASC11→TGF-β1↑→stemness↑ | [118] |
| Nc-RNA | Cell Lines | Roles | References |
|---|---|---|---|
| miR-132 | A549 | miR-132→TGF-β1/Smad2 signal↓→EMT↓, invasion↓, migration↓ | [77] |
| miR-203, miR-145 | A549, 95C | miR-203, miR-145→Smad3↓→EMT↓, invasion↓ | [78] |
| miR-124 | A549 | miR-124→Smad4↓→EMT↓ | [80] |
| miR-205 | NCI-H1975 | miR-205→E-cadherin↑, vimentin↓→EMT↓ | [67] |
| A549 | miR-205→Smad4↓→EMT↓ | [79] | |
| miR-422a | H522 | miR-422a→SULF2↓→TGF-β1/Smad2/3 signal↓→EMT↓ | [17] |
| miR-196b | A549 | miR-196b→N-cadherin↓, vimentin↓, ZEB1↓, Snail↓→EMT↓ | [82] |
| miR-940 | A549, H226 | miR-940→Snail↓→EMT↓ | [83] |
| miR-22 | Anip973, AGZY83-a | miR-22→Snail↓→EMT↓ | [84] |
| miR-200a, miR-200c | A549 | miR-200a, miR-200c→ZEB1↓, ZEB2↓→EMT↓ | [85,86] |
| miR-145, miR-497 | A549, H1299 | miR-145, miR-497→MTDH↓→EMT↓,invasion↓, migration↓ | [87] |
| miR-149 | A549 | miR-149→FOXM1↓→EMT↓ | [88] |
| miR-134 | A549 | miR-134→FOXM1↓→EMT↓ | [89] |
| miR-133 | A549, HCC827 | miR-133→FOXQ1↓→TGF-β↓→EMT↓ | [90] |
| miR-29c | 95C, A549 | miR-29c→Sp1↓→EMT↓ | [91] |
| miR-16 | A549 | miR-16→autophagy↑→EMT↓ | [94] |
| miR-129 | A549 | miR-129→SOX4↓→EMT↓ | [96] |
| miR-485-5p | A549 | miR-485-5p→E-cadherin↑, vimentin↓→EMT↓ | [97] |
| miR-3127-5p | A549, H1299 | miR-3127-5p→E-cadherin↑, vimentin↓→EMT↓ | [98] |
| miR-3607-3p | H157, H292 | miR-3607-3p→TGFBR1↓, CCNE2↓→ invasion↓, migration↓ | [99] |
| miR-206, miR-140 | A549 | miR-206, miR-140→TGF-β1/Smad3 signal↓→invasion↓, migration↓, metastasis↓ | [100] |
| miR-136 | A549, Calu-3 | miR-136→Smad2↓, Smad3↓→invasion↓, migration↓ | [102] |
| miR-133a | A549 | miR-133a→TGF-β/TGFBR1 signal↓→AKT signal↓→invasion↓, migration↓ | [103] |
| miR-143 | A549 | miR-143→Smad3↓, CD44↓, K-Ras↓→invasion↓, migration↓ | [104] |
| miR-886-3p | NCI-H446 | miR-886-3p→TGF-β1↓→invasion↓, migration↓ | [105] |
| miR-138 | LC006, LC021 | miR-138→EMT↓→CSCs formation↓ | [106] |
| lncRNA HAND2-AS1 | H1581, H1993 | HAND2-AS1→TGF-β1/Smad2/3 signal↓→invasion↓, migration↓, stemness↓ | [119] |
| lncRNA linc01186 | A549 | linc01186→EMT↓→invasion↓, migration↓ | [120] |
| lncRNA LINP1 | A549 | LINP1→E-cadherin↑, vimentin↓, Snail↓→EMT↓ | [121] |
| lncRNA ANCR | NCI-H23, NCI-H522 | ANCR→TGF-β1↓→invasion↓, migration↓ | [122] |
| lncRNA NKILA | A549, H226 | NKILA→NF-κB↓→Snail↓→EMT↓→invasion↓, migration↓ | [123] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lai, X.-N.; Li, J.; Tang, L.-B.; Chen, W.-T.; Zhang, L.; Xiong, L.-X. MiRNAs and LncRNAs: Dual Roles in TGF-β Signaling-Regulated Metastasis in Lung Cancer. Int. J. Mol. Sci. 2020, 21, 1193. https://doi.org/10.3390/ijms21041193
Lai X-N, Li J, Tang L-B, Chen W-T, Zhang L, Xiong L-X. MiRNAs and LncRNAs: Dual Roles in TGF-β Signaling-Regulated Metastasis in Lung Cancer. International Journal of Molecular Sciences. 2020; 21(4):1193. https://doi.org/10.3390/ijms21041193
Chicago/Turabian StyleLai, Xing-Ning, Jun Li, Li-Bo Tang, Wen-Tong Chen, Lei Zhang, and Li-Xia Xiong. 2020. "MiRNAs and LncRNAs: Dual Roles in TGF-β Signaling-Regulated Metastasis in Lung Cancer" International Journal of Molecular Sciences 21, no. 4: 1193. https://doi.org/10.3390/ijms21041193
APA StyleLai, X.-N., Li, J., Tang, L.-B., Chen, W.-T., Zhang, L., & Xiong, L.-X. (2020). MiRNAs and LncRNAs: Dual Roles in TGF-β Signaling-Regulated Metastasis in Lung Cancer. International Journal of Molecular Sciences, 21(4), 1193. https://doi.org/10.3390/ijms21041193





