Transcriptome Profiling Analysis Reveals Co-Regulation of Hormone Pathways in Foxtail Millet during Sclerospora graminicola Infection
Abstract
:1. Introduction
2. Results
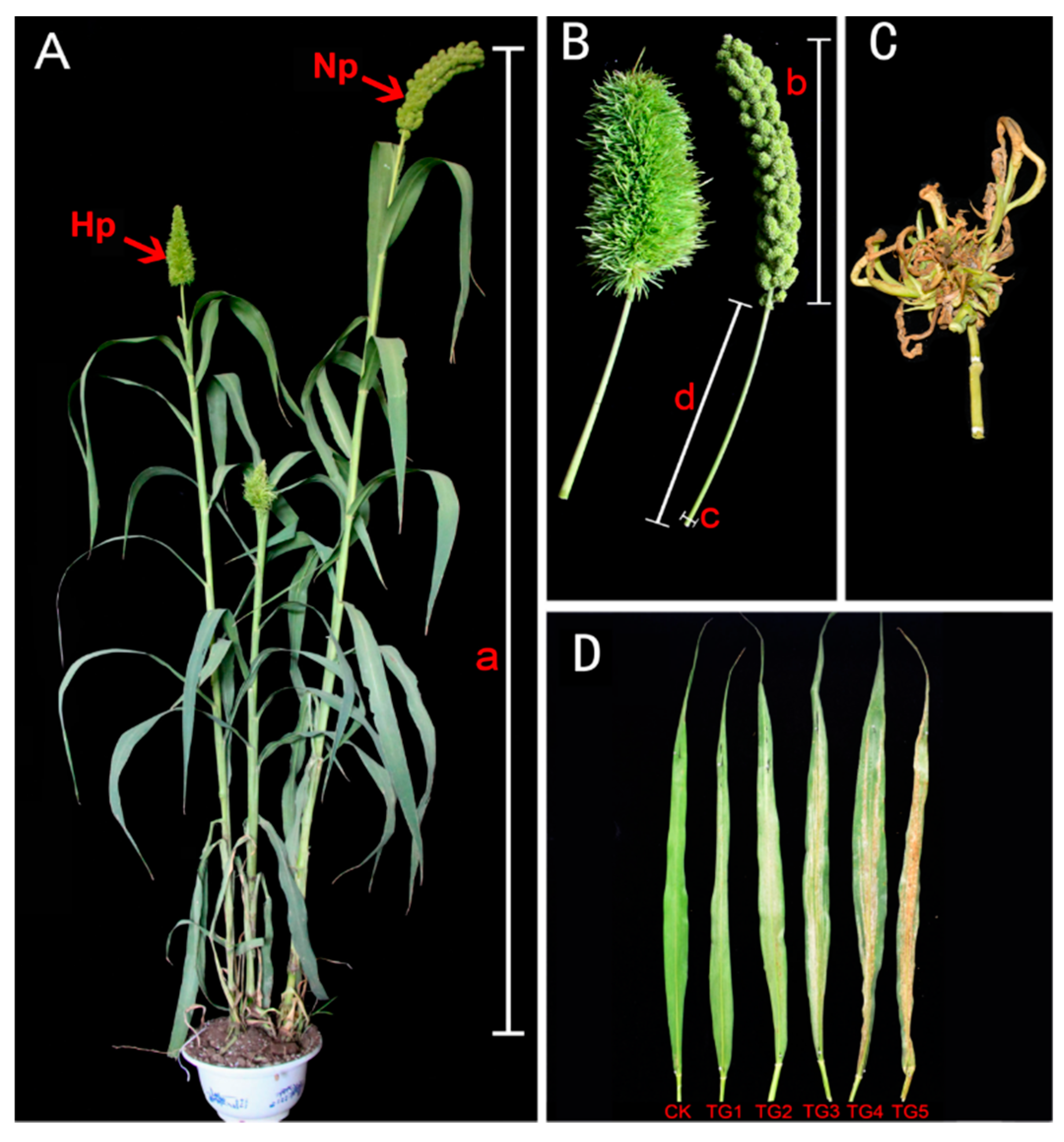
2.1. S. graminicola Caused Dwarfing and “Hedgehog Panicle” in Foxtail Millet
2.2. Identification of the Hormone-Associated Genes
2.3. Hierarchical Clustering Analysis of the Expression Patterns of Hormone-associated Genes
2.4. Identification of Hormone-associated Genes at Different Stages
2.5. S. graminicola Alters the Accumulation of Gene Transcripts in Hormone Biosynthesis Pathways in Foxtail Millet
2.6. S. graminicola Alters Transcripts of Genes in the Hormone Signal Transduction Pathway in Foxtail Millet and Verification by Quantitative Real-Time PCR
3. Discussion
4. Materials and Methods
4.1. Foxtail Millet Varieties, Inoculation, and Planting
4.2. RNA Extraction, Library Construction, and Sequencing
4.3. Bioinformatic Analysis of Transcriptome Data
4.4. Quantitative Real-Time PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| S.graminicola | Sclerospora graminicola (Sacc.) Schroeter |
| GA | Gibberellin |
| IAA | Auxin |
| ABA | Abscisic acid |
| CTK | Cytokinin |
| ETH | Ethylene |
| SA | Salicylic acid |
| JA | Jasmonic acid |
| BR | Brassinosteroids |
| IPA | Indole-3-pyruvic acid |
| IAOx | Indole acetaldehyde oxime |
| TAA1 | L-tryptophan---pyruvate aminotransferase |
| YUC | Indole-3-pyruvate monooxygenase |
| AAO1_2 | Indole-3-acetaldehyde oxidase |
| ALDH | Acetaldehyde dehydrogenase |
| CPS | Ent-copalyl diphosphate synthetase |
| KS | Ent-kaurene synthase |
| KO | Ent-kaurene oxidase |
| NCED | 9-cis-epoxycarotenoid dioxygenase |
| CYP707A | ABA 8-hydroxylase |
| GH3 | Gretchen hagen3 |
| AUX/IAA | IAA/indole-3-acetic acid |
| SAUR | Small IAA up RNA |
| TIR1 | Transport inhibitor response 1 |
| TF3 | Phytochrome-interacting factor 3 |
| TF4 | Phytochrome-interacting factor 4 |
| PYR/PYL | ABA receptor |
| PP2C | Protein phosphatase 2C |
| ABF | ABA responsive element binding factor |
| GID1 | Gibberellin receptor 1 |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
References
- Obayashi, M.; Hiraka, Y.; Abe, A.; Yaegashi, H.; Natsume, S.; Kikuchi, H.; Takagi, H.; Saitoh, H.; Win, J.; Kamoun, S.; et al. Genome analysis of the foxtail millet pathogen Sclerospora graminicola reveals the complex effector repertoire of graminicolous downy mildews. BMC Genom. 2017, 18, 897. [Google Scholar]
- Lavanya, S.N.; Udayashankar, A.C.; Raj, S.N.; Mohan, C.D.; Gupta, V.K.; Tarasatyavati, C.; Srivastava, R.; Nayaka, S.C. Lipopolysaccharide-induced priming enhances NO-mediated activation of defense responses in pearl millet challenged with Sclerospora graminicola. 3 Biotech 2018, 8, 475. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, M.M.; Arya, H.C. Sclerospora graminicola axenic culture. Science 1969, 17, 291–293. [Google Scholar] [CrossRef] [PubMed]
- Hosur, G.P.; Jogaiah, S.; Sreedhara, A.P.; Nagraj, P.G.; Kini, R.K.; Shetty, S.H. Association between accumulation of allene oxide synthase activity and development of resistance against downy mildew disease of pearl millet. Mol. Biol. Rep. 2013, 40, 6821. [Google Scholar] [CrossRef] [Green Version]
- Berens, M.L.; Berry, H.M.; Mine, A.; Argueso, C.T.; Tsuda, K. Evolution of hormone signaling networks in plant defense. Annu. Rev. Phytopathol. 2017, 55, 401–425. [Google Scholar] [CrossRef]
- Swain, S.; Singh, N.; Nandi, A.K. Identification of plant defence regulators through transcriptional profiling of Arabidopsis thaliana cdd1 mutant. J. Biosci. 2015, 40, 137–146. [Google Scholar] [CrossRef]
- Chern, M.; Canlas, P.E.; Ronald, P.C. Strong suppression of systemic acquired resistance in Arabidopsis by NRR is dependent on its ability to interact with NPR1 and its putative repression domain. Mol. Plant 2008, 1, 552–559. [Google Scholar] [CrossRef]
- Wang, F.; Lin, R.; Feng, J.; Chen, W.; Qiu, D.; Xu, S. TaNaNAC1 acts as a negative regulator of stripe rust resistance in wheat, enhances susceptibility to Pscudomonas syringae, and promotes lateral root development in transgenic Arabidopsis thaliana. Front. Plant Sci. 2015, 6, 108. [Google Scholar]
- Tian, G.Z.; Li, Y.F.; Qiu, W.F. Infection between plant hormones and diseases. Plant Physiol. Commun. 1999, 3, 177–184. [Google Scholar]
- Mishra, K.A.; Dhillon, B.S. Levels of endogenous GAs in healthy and malformed panicles of mango (Manifera indica L.). Indian J. Hortic. 1980, 37, 33–34. [Google Scholar]
- Zhu, S.; Gao, F.; Cao, X.; Chen, M.; Ye, G.; Wei, C.; Li, Y. The rice dwarf virus P2 protein interacts with ent-kaurene oxidases in vivo, leading to reduced biosynthesis of GAs and rice dwarf symptoms. Plant Physiol. 2005, 139, 1935–1945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, J.; Liu, M.J.; Dai, L.; Zhou, J.Y. The variations of endogenous hormones in Chinese jujube infected with Witches’ Broom diease. Sci. Agric. Sin. 2006, 39, 2255–2260. [Google Scholar]
- Ghareeb, H.; Becker, A.; Iven, T.; Feussner, I.; Schirawski, J. Sporisorium reilianum infection changes inflorescence and branching architectures of maize. Plant Physiol. 2011, 156, 2037–2052. [Google Scholar] [CrossRef] [Green Version]
- Dernoeden, P.H.; Jackson, N. Infection and mycelia colonization of gramineous hosts by Sclerophthora macrospora. Phytopathology 1980, 70, 1009–1013. [Google Scholar] [CrossRef]
- Dobon, A.; Bunting, D.C.; Cabrera-Quio, L.E.; Uauy, C.; Saunders, D.G. The host-pathogen interaction between wheat and yellow rust induces temporally coordinated waves of gene expression. BMC Genom. 2016, 17, 380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, M.; Zhao, J.; Tzeng, D.T.W.; Liu, Y.; Deng, L.; Yang, T.; Zhai, Q.; Wu, F.; Huang, Z.; Zhou, M.; et al. MYC2 orchestrates a hierarchical transcriptional cascade that regulates jasmonate-mediated plant immunity in tomato. Plant Cell 2017, 29, 1883–1906. [Google Scholar] [CrossRef] [Green Version]
- Jeon, J.E.; Kim, J.G.; Ficher, C.R.; Mehta, N.; Dufour, S.C.; Wemmer, K.; Mudgett, M.B.; Sattely, E. A pathogen-responsive gene cluster for highly modified fatty acids in tomato. Cell 2020, 180, 176–187. [Google Scholar] [CrossRef]
- Sequeira, L. Hormone metabolism in diseased plants. Annu. Rev. Plant Physiol. 1973, 24, 353–380. [Google Scholar] [CrossRef]
- Kubeš, M.; Napier, R. Non-canonical auxin signalling: Fast and curious. J. Exp. Bot. 2019, 70, 2609–2614. [Google Scholar] [CrossRef]
- Austin, M.J.; Muskett, P.; Kahn, K.; Feys, B.J.; Jones, J.D.; Parker, J.E. Regulatory role of SGT1 in early R gene-mediated plant defenses. Science 2002, 295, 2077–2080. [Google Scholar] [CrossRef] [Green Version]
- Ding, X.; Cao, Y.; Huang, L.; Zhao, J.; Xu, C.; Li, X.; Wang, S. Activation of the indole-3-acetic acid-amido synthetase GH3-8 suppresses expansin expression and promotes salicylate- and jasmonate-independent basal immunity in rice. Plant Cell 2008, 20, 228–240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hauvermale, A.L.; Ariizumi, T.; Steber, C.M. Gibberellin signaling: A theme and variations on DELLA repression. Plant Physiol. 2012, 160, 83–92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ueguchi-Tanaka, M.; Nakajima, M.; Katoh, E.; Ohmiya, H.; Asano, K.; Saji, S.; Honqyu, X.; Ashikari, M.; Kitano, H.; Yamaquchi, I.; et al. Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1, and gibberellin. Plant Cell 2007, 19, 2140–2155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murase, K.; Hirano, Y.; Hakoshima, T. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 2008, 27, 459–463. [Google Scholar] [CrossRef] [PubMed]
- Chahtane, H.; Nogueira, F.T.; Allard, P.M.; Marcourt, L.; Ferreira-queiroz, E.; Shanmuqabalaji, V.; Wolfender, J.L.; Lopez-Molina, L. The plant pathogen Pseudomonas aeruginosa triggers a DELLA-dependent seed germination arrest in Arabidopsis. Elife 2018, 7, 37082. [Google Scholar] [CrossRef] [PubMed]
- Miao, L.; Miao, X.; Wang, J.; Liu, Z.; Zhang, B.; Li, W.; Chang, X.; Reynolds, M.; Wang, Z.; Jing, R. Elite haplotypes of a protein kinase gene TaSnRK2.3 associated with important agronomic traits in common wheat. Front. Plant Sci. 2017, 8, 368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prasad, K.; Parameswaran, S.; Vijayraqhavan, U. OsMADS1, a rice MADS-box factor, controls differentiation of specific cell types in the lemma and palea and is an early-acting regulator of inner floral organs. Plant J. 2005, 43, 915–928. [Google Scholar] [CrossRef]
- Rio, D.C.; Ares, M.; Hannon, G.J.; Nilsen, T.W. Purification of RNA using TRIzol (TRI reagent). Cold Spring Harb. Protoc. 2010, 6. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acid Res. 1999, 20, 29–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, L.; E-Futschik, M. Mfuzz: A software package for soft clustering of microarray data. Bioinformation 2007, 2, 5–7. [Google Scholar] [CrossRef]
- Kolde, R. Pheatmap: Pretty Heatmaps [EB/OL]. Available online: http://CRAN.R-project.org/package=pheatmap (accessed on 19 November 2019).
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]











© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, R.; Han, Y.; Zhang, Q.; Chang, G.; Han, Y.; Li, X.; Zhang, B. Transcriptome Profiling Analysis Reveals Co-Regulation of Hormone Pathways in Foxtail Millet during Sclerospora graminicola Infection. Int. J. Mol. Sci. 2020, 21, 1226. https://doi.org/10.3390/ijms21041226
Li R, Han Y, Zhang Q, Chang G, Han Y, Li X, Zhang B. Transcriptome Profiling Analysis Reveals Co-Regulation of Hormone Pathways in Foxtail Millet during Sclerospora graminicola Infection. International Journal of Molecular Sciences. 2020; 21(4):1226. https://doi.org/10.3390/ijms21041226
Chicago/Turabian StyleLi, Renjian, Yanqing Han, Qi Zhang, Guorong Chang, Yuanhuai Han, Xukai Li, and Baojun Zhang. 2020. "Transcriptome Profiling Analysis Reveals Co-Regulation of Hormone Pathways in Foxtail Millet during Sclerospora graminicola Infection" International Journal of Molecular Sciences 21, no. 4: 1226. https://doi.org/10.3390/ijms21041226
APA StyleLi, R., Han, Y., Zhang, Q., Chang, G., Han, Y., Li, X., & Zhang, B. (2020). Transcriptome Profiling Analysis Reveals Co-Regulation of Hormone Pathways in Foxtail Millet during Sclerospora graminicola Infection. International Journal of Molecular Sciences, 21(4), 1226. https://doi.org/10.3390/ijms21041226




