Ethyl Acetate Fraction of Aqueous Extract of Lentinula edodes Inhibits Osteoclastogenesis by Suppressing NFATc1 Expression
Abstract
:1. Introduction
2. Results
2.1. LEA Suppresses Osteoclast Differentiation
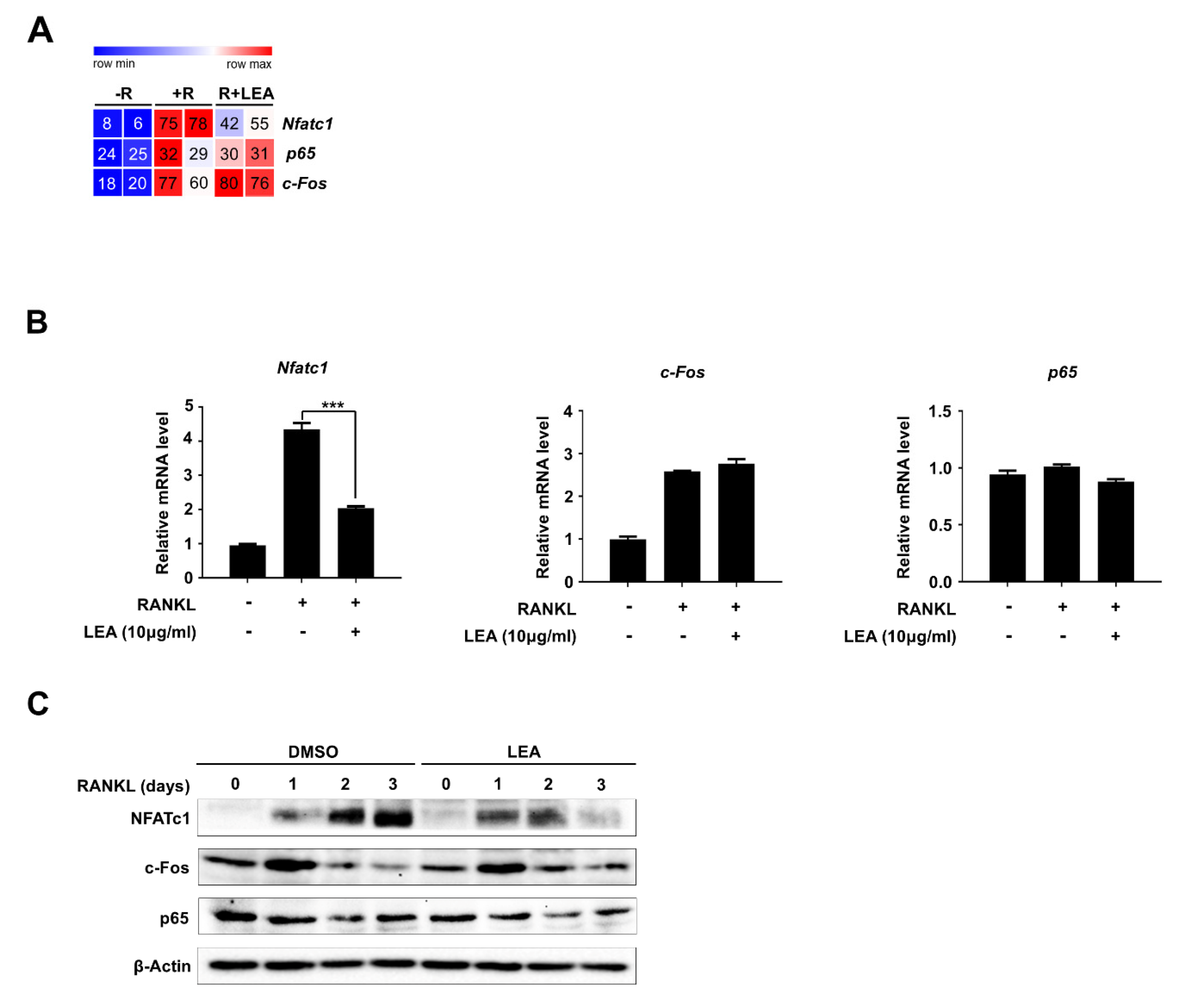
2.2. LEA Modulates a Set of Osteoclast-Related Gene Expression in RANKL-Induced Osteoclastogenesis
2.3. LEA Inhibits NFATc1 Expression during Osteoclastogenesis
2.4. LEA Suppresses NFATc1 Expression by Inhibiting the Transactivities of Both p65 and NFATc1
2.5. LEA Suppresses Prednisolone-Induced Osteoporosis in Zebrafish Larvae
3. Discussion
4. Materials and Methods
4.1. Preparation of Water Extract and Its Fractions Using L. edodes
4.2. Osteoclast Differentiation and TRAP Staining
4.3. Cell Viability Assay
4.4. RNA-seq
4.5. RT-qPCR
4.6. Western Blot Analysis
4.7. Reporter Gene Assay
4.8. Fish Maintenance and Drug Treatment
4.9. Whole-Mount Skeletal Staining
4.10. Statistical Analysis
4.11. Accession Numbers
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| RANKL | Receptor activator of the NF-κB ligand |
| BMM | Bone marrow-derived macrophage |
| GIO | Glucocorticoid-induced osteoporosis |
| TRAP | Tartrate resistant acid phosphatase |
References
- Florencio-Silva, R.; Sasso, G.R.; Sasso-Cerri, E.; Simoes, M.J.; Cerri, P.S. Biology of Bone Tissue: Structure, Function, and Factors That Influence Bone Cells. BioMed Res. Int. 2015, 2015, 421746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crockett, J.C.; Rogers, M.J.; Coxon, F.P.; Hocking, L.J.; Helfrich, M.H. Bone remodelling at a glance. J. Cell Sci. 2011, 124, 991–998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sobacchi, C.; Schulz, A.; Coxon, F.P.; Villa, A.; Helfrich, M.H. Osteopetrosis: Genetics, treatment and new insights into osteoclast function. Nat. Rev. Endocrinol. 2013, 9, 522–536. [Google Scholar] [CrossRef] [PubMed]
- Canalis, E.; Giustina, A.; Bilezikian, J.P. Mechanisms of anabolic therapies for osteoporosis. N. Engl. J. Med. 2007, 357, 905–916. [Google Scholar] [CrossRef] [Green Version]
- Charles, J.F.; Aliprantis, A.O. Osteoclasts: More than ‘bone eaters’. Trends Mol. Med. 2014, 20, 449–459. [Google Scholar] [CrossRef] [Green Version]
- Yi, S.J.; Lee, H.; Lee, J.; Lee, K.; Kim, J.; Kim, Y.; Park, J.I.; Kim, K. Bone Remodeling: Histone Modifications as Fate Determinants of Bone Cell Differentiation. Int. J. Mol. Sci. 2019, 20, 3147. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.H.; Kim, N. Regulation of NFATc1 in Osteoclast Differentiation. J. Bone Metab. 2014, 21, 233–241. [Google Scholar] [CrossRef] [Green Version]
- Lee, N.K.; Choi, Y.G.; Baik, J.Y.; Han, S.Y.; Jeong, D.W.; Bae, Y.S.; Kim, N.; Lee, S.Y. A crucial role for reactive oxygen species in RANKL-induced osteoclast differentiation. Blood 2005, 106, 852–859. [Google Scholar] [CrossRef] [Green Version]
- Boyce, B.F.; Yamashita, T.; Yao, Z.; Zhang, Q.; Li, F.; Xing, L. Roles for NF-kappaB and c-Fos in osteoclasts. J. Bone Miner. Metab. 2005, 23, 11–15. [Google Scholar] [CrossRef]
- Cherian, K.E.; Kapoor, N.; Paul, T.V. Glucocorticoid-induced Osteoporosis. Indian J. Endocrinol. Metab. 2017, 21, 652–654. [Google Scholar] [CrossRef]
- Rodan, G.A.; Fleisch, H.A. Bisphosphonates: Mechanisms of action. J. Clin. Investig. 1996, 97, 2692–2696. [Google Scholar] [CrossRef] [PubMed]
- Cheon, Y.H.; Baek, J.M.; Park, S.H.; Ahn, S.J.; Lee, M.S.; Oh, J.; Kim, J.Y. Stauntonia hexaphylla (Lardizabalaceae) leaf methanol extract inhibits osteoclastogenesis and bone resorption activity via proteasome-mediated degradation of c-Fos protein and suppression of NFATc1 expression. BMC Complement. Altern. Med. 2015, 15, 280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, Y.H.; Jang, S.A.; Kim, T.; Ha, H. Forsythia suspensa Protects against Bone Loss in Ovariectomized Mice. Nutrients 2019, 11, 1831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bisen, P.S.; Baghel, R.K.; Sanodiya, B.S.; Thakur, G.S.; Prasad, G.B. Lentinus edodes: A macrofungus with pharmacological activities. Curr. Med. Chem. 2010, 17, 2419–2430. [Google Scholar] [CrossRef] [PubMed]
- Fang, N.; Li, Q.; Yu, S.; Zhang, J.; He, L.; Ronis, M.J.; Badger, T.M. Inhibition of growth and induction of apoptosis in human cancer cell lines by an ethyl acetate fraction from shiitake mushrooms. J. Altern. Complement. Med. 2006, 12, 125–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finimundy, T.C.; Gambato, G.; Fontana, R.; Camassola, M.; Salvador, M.; Moura, S.; Hess, J.; Henriques, J.A.; Dillon, A.J.; Roesch-Ely, M. Aqueous extracts of Lentinula edodes and Pleurotus sajor-caju exhibit high antioxidant capability and promising in vitro antitumor activity. Nutr. Res. 2013, 33, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.H.; Belury, M.A. Selective induction of apoptosis in murine skin carcinoma cells (CH72) by an ethanol extract of Lentinula edodes. Cancer Lett. 2005, 220, 21–28. [Google Scholar] [CrossRef]
- Hirasawa, M.; Shouji, N.; Neta, T.; Fukushima, K.; Takada, K. Three kinds of antibacterial substances from Lentinus edodes (Berk.) Sing. (Shiitake, an edible mushroom). Int. J. Antimicrob. Agents 1999, 11, 151–157. [Google Scholar] [CrossRef]
- Huang, W.; Kim, J.S.; Chung, H.Y. Antioxidant activity and total phenolic content in shiitake mycelial exudates. Nat. Prod. Commun. 2011, 6, 845–850. [Google Scholar] [CrossRef] [Green Version]
- Lull, C.; Wichers, H.J.; Savelkoul, H.F. Antiinflammatory and immunomodulating properties of fungal metabolites. Mediat. Inflamm. 2005, 2005, 63–80. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, K.; Matsui, Y.; Ishikawa, S.; Kawanishi, T.; Harada, M. Oral ingestion of Lentinula edodes mycelia extract can restore the antitumor T cell response of mice inoculated with colon-26 cells into the subserosal space of the cecum. Oncol. Rep. 2012, 27, 325–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saif, A.; Wende, K.; Lindequist, U. In vitro bone inducing effects of Lentinula edodes (shiitake) water extract on human osteoblastic cell cultures. Nat. Prod. Bioprospecting 2013, 3, 282–287. [Google Scholar] [CrossRef] [Green Version]
- Negishi-Koga, T.; Takayanagi, H. Ca2+-NFATc1 signaling is an essential axis of osteoclast differentiation. Immunol. Rev. 2009, 231, 241–256. [Google Scholar] [CrossRef] [PubMed]
- Ross, F.P. M-CSF, c-Fms, and signaling in osteoclasts and their precursors. Ann. N. Y. Acad. Sci. 2006, 1068, 110–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wada, T.; Nakashima, T.; Hiroshi, N.; Penninger, J.M. RANKL-RANK signaling in osteoclastogenesis and bone disease. Trends Mol. Med. 2006, 12, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Barrett, R.; Chappell, C.; Quick, M.; Fleming, A. A rapid, high content, in vivo model of glucocorticoid-induced osteoporosis. Biotechnol. J. 2006, 1, 651–655. [Google Scholar] [CrossRef]
- He, H.; Wang, C.; Tang, Q.; Yang, F.; Xu, Y. Possible mechanisms of prednisolone-induced osteoporosis in zebrafish larva. Biomed. Pharmacother. Biomed. Pharmacother. 2018, 101, 981–987. [Google Scholar] [CrossRef]
- Erjavec, I.; Brkljacic, J.; Vukicevic, S.; Jakopovic, B.; Jakopovich, I. Mushroom Extracts Decrease Bone Resorption and Improve Bone Formation. Int. J. Med. Mushrooms 2016, 18, 559–569. [Google Scholar] [CrossRef]
- Jones, D.H.; Kong, Y.Y.; Penninger, J.M. Role of RANKL and RANK in bone loss and arthritis. Ann. Rheum. Dis. 2002, 61, ii32–ii39. [Google Scholar] [CrossRef] [Green Version]
- Takayanagi, H.; Kim, S.; Koga, T.; Nishina, H.; Isshiki, M.; Yoshida, H.; Saiura, A.; Isobe, M.; Yokochi, T.; Inoue, J.; et al. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev. Cell 2002, 3, 889–901. [Google Scholar] [CrossRef] [Green Version]
- Asagiri, M.; Sato, K.; Usami, T.; Ochi, S.; Nishina, H.; Yoshida, H.; Morita, I.; Wagner, E.F.; Mak, T.W.; Serfling, E.; et al. Autoamplification of NFATc1 expression determines its essential role in bone homeostasis. J. Exp. Med. 2005, 202, 1261–1269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ha, H.; Shim, K.S.; Ma, J.Y. Water extract of Uncaria sinensis suppresses RANKL-induced bone loss by attenuating osteoclast differentiation and bone resorption. Integr. Med. Res. 2017, 6, 434–442. [Google Scholar] [CrossRef] [PubMed]
- Geidam, Y.A.; Ambali, A.G.; Onyeyili, P.A. Phytochemical Screening and Antibacterial Properties of Organic Solvent Fractions of Psidium guajava Aqueous Leaf Extracts. Int. J. Pharmacol. 2007, 3, 68–73. [Google Scholar] [CrossRef]
- An, D.; Kim, K.; Lu, W. Defective entry into mitosis 1 (Dim1) negatively regulates osteoclastogenesis by inhibiting the expression of nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 (NFATc1). J. Biol. Chem. 2014, 289, 24366–24373. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.; Kim, J.; Lee, H.; Shin, W.R.; Lee, S.; Lee, J.; Park, J.I.; Jhun, B.H.; Kim, Y.H.; Yi, S.J.; et al. Tetracycline Analogs Inhibit Osteoclast Differentiation by Suppressing MMP-9-Mediated Histone H3 Cleavage. Int. J. Mol. Sci. 2019, 20, 4038. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, H.; Lee, K.; Lee, S.; Lee, J.; Jeong, W.T.; Lim, H.B.; Hyun, T.K.; Yi, S.-J.; Kim, K. Ethyl Acetate Fraction of Aqueous Extract of Lentinula edodes Inhibits Osteoclastogenesis by Suppressing NFATc1 Expression. Int. J. Mol. Sci. 2020, 21, 1347. https://doi.org/10.3390/ijms21041347
Lee H, Lee K, Lee S, Lee J, Jeong WT, Lim HB, Hyun TK, Yi S-J, Kim K. Ethyl Acetate Fraction of Aqueous Extract of Lentinula edodes Inhibits Osteoclastogenesis by Suppressing NFATc1 Expression. International Journal of Molecular Sciences. 2020; 21(4):1347. https://doi.org/10.3390/ijms21041347
Chicago/Turabian StyleLee, Hyerim, Kyubin Lee, Sheunghun Lee, Jisu Lee, Won Tae Jeong, Heung Bin Lim, Tae Kyung Hyun, Sun-Ju Yi, and Kyunghwan Kim. 2020. "Ethyl Acetate Fraction of Aqueous Extract of Lentinula edodes Inhibits Osteoclastogenesis by Suppressing NFATc1 Expression" International Journal of Molecular Sciences 21, no. 4: 1347. https://doi.org/10.3390/ijms21041347
APA StyleLee, H., Lee, K., Lee, S., Lee, J., Jeong, W. T., Lim, H. B., Hyun, T. K., Yi, S.-J., & Kim, K. (2020). Ethyl Acetate Fraction of Aqueous Extract of Lentinula edodes Inhibits Osteoclastogenesis by Suppressing NFATc1 Expression. International Journal of Molecular Sciences, 21(4), 1347. https://doi.org/10.3390/ijms21041347




