Abstract
Huanglongbing (HLB), also known as citrus greening, is the most notorious citrus disease worldwide. Candidatus Liberibacter asiaticus (CaLas) is a phloem-restricted bacterium associated with HLB. Because there is no mutant library available, the pathogenesis of CaLas is obscure. In this study, we employed tobacco mosaic virus (TMV) to express two mature secretion proteins CLIBASIA_03915 (m03915) and CLIBASIA_04250 (m04250) in Nicotiana benthamiana (N. benthamiana). Phloem necrosis was observed in the senescent leaves of N. benthamiana that expressed the two low molecular weight proteins, while no phloem necrosis was observed in the plants that expressed the control, green fluorescent protein (GFP). Additionally, no phloem necrosis was observed in the senescent leaves of N. benthamiana that expressed the null mutation of m03915 and frameshifting m04250. The subcellular localizations of m03915 and m04250 were determined by fusion with GFP using confocal microscopy. The subcellular localization of m03915 was found to be as free GFP without a nuclear localization sequence (NLS). However, m04250 did have an NLS. Yeast two-hybrid (Y2H) was carried out to probe the citrus proteins interacting with m03915 and m04250. Six citrus proteins were found to interact with m03915. The identified proteins were involved in the metabolism of compounds, transcription, response to abiotic stress, ubiquitin-mediated protein degradation, etc. The prey of m04250 was involved in the processing of specific pre-mRNAs. Identification of new virulence factors of CaLas will give insight into the pathogenesis of CaLas, and therefore, it will eventually help develop the HLB-resistant citrus.
1. Introduction
Huanglongbing (HLB), also known as citrus greening, is the most notorious citrus disease worldwide. The typical symptoms of HLB include yellowing of the veins, dieback of twigs, decline of roots, fruits unable to develop proper color, and, eventually, the death of the infected trees [1,2]. HLB is associated with a phloem-restricted Gram-negative bacterium, named Candidatus Liberibacter africanus (CaLaf), Ca. Liberibacter asiaticus (CaLas), and Ca. L. americanus (CaLam) based on geographic distribution. CaLas is the most virulent pathogen with global distribution [1]. CaLas caused a 72.2% reduction in orange juice and a 20.5% reduction in the fresh fruit market in the United States from 2007–2008 to 2017–2018, respectively [3]. All commercial citrus cultivars are susceptible to HLB, and there is no cure for HLB [2]. Therefore, effective management of HLB is urgently needed. However, the lack of in-depth knowledge of the virulence mechanism of CaLas prevents the development of effective HLB management.
Though CaLas remains uncultured, the availability of CaLas genomic information [4,5] facilitates research on the virulence mechanism of CaLas. CaLas lacks type III, and type IV secretion systems (TIIISS, TIVSS) to deliver effectors into host cells [4]. The effectors of CaLas mainly rely on Sec secretion machinery to enter host cells. Several reports have used transient expression or viral vectors to identify the effectors of CaLas [6,7,8]. CaL5315 was reported as a lethal effector, causing cell death in infiltrated N. benthamiana leaves by transient expression [6]. CLIBASIA_00460 caused necrosis in the systemic leaves when expressed by Potato virus X (PVX) in N. benthamiana [7]. CLIBASIA_00470 and CLIBASIA_04025 were reported to inhibit N. benthamiana growth when expressed by tobacco mosaic virus (TMV) at two weeks post infiltration (WPI) [8].
Several reports have shown that CaLas encodes proteins to suppress citrus disease resistance as the virulence mechanism [9,10,11]. SC2_gp095 encodes a peroxidase that downregulates RbohB when expressed transiently in plants [10]. CaLas also encodes a salicylic acid (SA) hydroxylase to downregulate the accumulation of SA in plants, therefore, inhibiting citrus disease resistance. CaLas05315, also called Sec-delivered effector 1 (SDE1), was found to interact with citrus papain-like cysteine proteases (PLCPs) and inhibited their activities. PLCPs confer plant disease resistance. SDE1 transgenic citrus exhibited the suppression of PLCPs [11]. The inhibition of defense response is one of the strategies of CaLas to establish colonization in citrus. CaLas caused phloem anatomical changes such as phloem necrosis, phloem plugging, and phloem collapse [12]. The phloem of the flowering plant consists of companion cells and mature sieve elements to transport phloem sap from source to sink [13]. Young sieve elements lose their ribosomes, dictyosomes, nucleus, and cytoskeleton during the transition to mature sieve elements [14]. The mature sieve elements contain smooth endoplasmic reticulum (ER), mitochondria, sieve element plastids, and phloem proteins [15]. It was recently reported that CaLas encodes proteins to interact with the key proteins of lignin production [8], which is the main component of woody cell wall biomass (up to 30%) [16].
We reported two ultra-low molecular weight putative virulence factors CLIBASIA_03915 (m03915) and CLIBASIA_04250 (m04250) that caused phloem necrosis in senescent leaves of N. benthamiana when expressing the mature proteins (m03915 and m04250) by TMV. The two hypothetical proteins were Sec-dependent secretory proteins confirmed by phoA assay. The two putative virulence factors were confirmed by mutation assay to function at the protein level. Green fluorescence protein (GFP) was used to tag m03915 and m04250 to determine their subcellular localizations. It was found that m04250 localized in the nucleolus. Yeast two-hybrid (Y2H) was applied to identify host proteins that interacted with the two effectors. The putative mechanisms of phloem necrosis caused by m03915 and m04250 and the potential application of our findings in citrus improvement are discussed.
2. Results
2.1. CLIBASIA_03915 and CLIBASIA_04250 Were Sec-Dependent Proteins
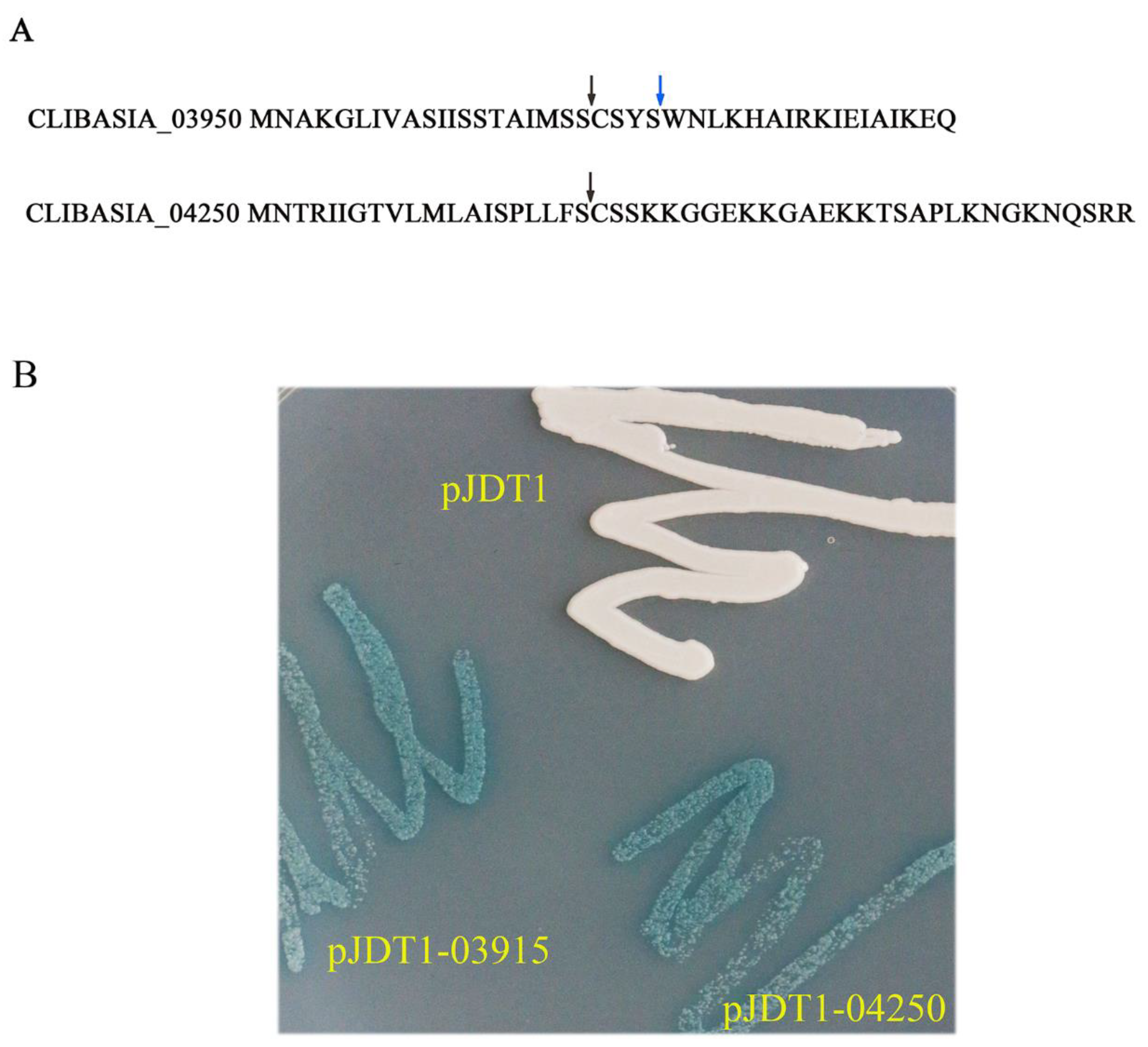
The signal peptides of CLIBASIA_03915 and CLIBASIA_04250 were predicted by SignalP 3.0 and SignalP 5.0. The cleavage site of CLIBASIA_03915 was between 24-25 aa (SignalP 3.0) or 20-21 aa (SignalP 5.0), and the cleavage site of CLIBASIA_04250 was between 20-21 aa (SignalP 3.0 and SignalP 5.0). The full-length DNA fragments of CLIBASIA_03915 and CLIBASIA_04250 with added enzyme sites were introduced into vector pJDT1, which contained the phoA gene (alkaline phosphatase) lacking the signal sequence [17] to have pJDT1-03915 and pJDT1-04250. The Escherichia coli clones harboring the positive recombinant plasmids along with the control pJDT1were streaked on Luria–Bertani (LB) plates amended with 5-bromo-4-chloro-3-indolylphosphate (BCIP) and sodium phosphate. The cells expressing pJDT1-03915 and pJDT1-04250 turned blue while the cells expressing the negative control (pJDT1) remained unchanged in color (Figure 1). This finding indicated that CLIBASIA_03915 and CLIBASIA_04250 were Sec-Dependent proteins.
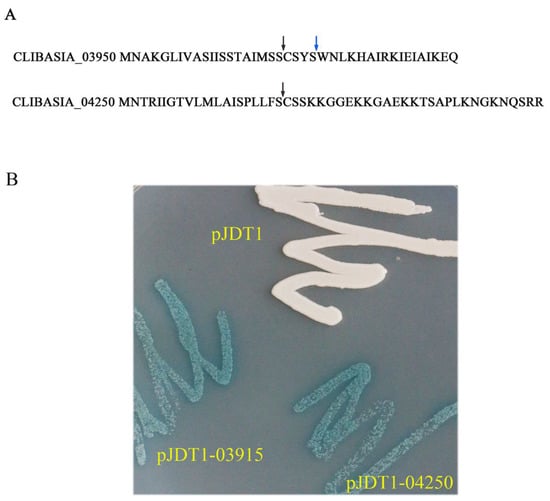
Figure 1.
Prediction and confirmation of signal peptides of CLIBASIA_03915 and CLIBASIA_04250. (A) The signal peptides of CLIBASIA_03915 and CLIBASIA_04250 were predicted by SingalP 3.0 and SingalP 5.0. The cleavage site of CLIBASIA_03915 was between 20–21 aa (SignalP 5.0) or 24-25 aa (SignalP 3.0). SignalP 3.0 and SignalP 5.0 predicted the same cleavage site of CLIBASIA_04250 (20-21 aa). The arrows indicated the cleavage sites. (B) The signal peptides of CLIBASIA_03915 and CLIBASIA_04250 were confirmed by phoA assay. The E. coli cells harboring the recombinant plasmids along with the negative control pJDT1 were streaked on Luria–Bertani (LB) plates amended with 5-bromo-4-chloro-3-indolylphosphate (BCIP) and sodium phosphate. The negative control pJDT1 did not cause the color change. The E. coli cells expressing the recombinant plasmids turned to blue.
2.2. Heterologous Expression of the Mature Proteins of CLIBASIA_03915 (m03915) and CLIBASIA_04250 (m04250) Caused Phloem Necrosis in the Senescent Leaves of N. benthamiana
TMV was used to express virulence factors of CaLas and resulted in identifying two virulence factors that inhibited plant growth at two weeks post infiltration (WPI) [8]. Heterologous expression of m03915 and m04250 in N. benthamiana by TMV showed a similar phenotype to control GFP at 2 WPI. However, phloem necrosis was observed in the senescent leaves of plants that expressed m03915 and m04250 at 5 WPI (Figure 2B,C). No phloem necrosis was observed in the senescent leaves of N. benthamiana that expressed GFP at the same time (Figure 2A).

Figure 2.
Mature proteins of CLIBASIA_03915 (m03915) and CLIBASIA_04250 (m04250) caused the phloem necrosis in the senescent leaves of Nicotiana benthamiana. Plants expressing control green fluorescent protein (GFP) did not show phloem necrosis (A). On the contrary, expression of m03915 (B) and m04250 (C) by tobacco mosaic virus in N. benthamiana plants caused phloem necrosis in senescent leaves. The pictures were taken at 5 weeks post infiltration (WPI).
2.3. The Mutated m03915 and m04250 Could Not Cause Phloem Necrosis in the Senescent Leaves
To exclude the possibility that phloem necrosis was caused by virus-induced gene silencing (VIGS), mutations were introduced into m03915 and m04250. The null mutation was introduced into m03915 by replacing the start codon ATG with the stop codon TAA to generate the mutant m03915Nu. The frameshifting mutation was introduced into m04250. The frameshifting mutation of m04250FS led to early termination of translation (after two amino acids FF). At 5 WPI, m03915 caused phloem necrosis in the senescent leaves, while no phloem necrosis was observed in the senescent leaves of N. benthamiana expressing m03915Nu (Figure 3A). Plants expressing m04250FS did not show the phenotype of phloem necrosis as native m04250 (Figure 3B). The results indicated that m03915 and m04250 functioned at the protein level rather than at the RNA level to cause phloem necrosis.

Figure 3.
The mutated m03915 (m03915Nu, on right) and m04250 (m04250FS, on right) could not cause phloem necrosis, while m03915 (on left) and m04250 (on left) caused phloem necrosis at 5 WPI. (A) The mutated m03915 (m03915Nu, on right) could not cause phloem necrosis, while m03915 (on left) caused phloem necrosis at 5 WPI. (B) The mutated m04250 (m04250FS, on right) did not cause phloem necrosis, while m04250 (on left) caused phloem necrosis at 5 WPI.
2.4. Determination of the Subcellular Localizations of m03915 and m04250
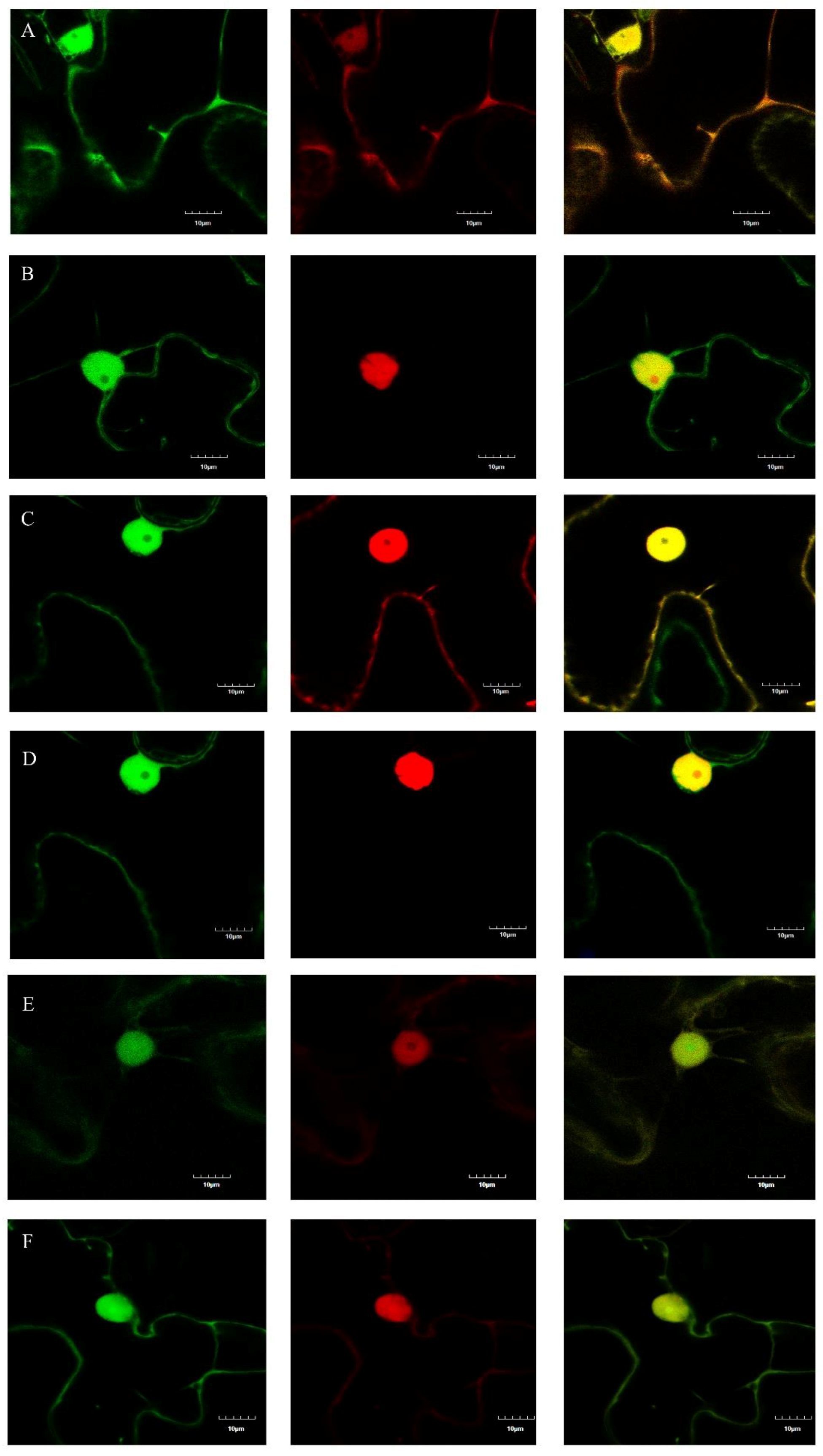
GFP was used to detect the subcellular localizations of m03915 and m04250. The patterns of distribution of free GFP, m03915-GFP, and m04250-GFP were similar in N. benthamiana cells (Figure S1). Red fluorescent protein (RFP) and RFP-nuclear localization signal (NLS) were co-expressed with GFP/m03915-GFP/m04250-GFP in N. benthamiana to determine whether an NLS was in m03915-GFP or m04250-GFP. Free GFP and free RFP were found to be distributed in the cytosol and nuclear lumen (Figure 4A). RFP was observed in the cell nucleus when containing an NLS. Free GFP could not enter the nucleolus (Figure 4B). The subcellular localization of m03915 was the same as free GFP, no NLS was present (Figure 4C,D). However, m04250-GFP showed stronger fluorescence in the nucleolus (Figure 4E,F). This indicated that m04250 localized in multiple cellular compartments, including the nucleolus.
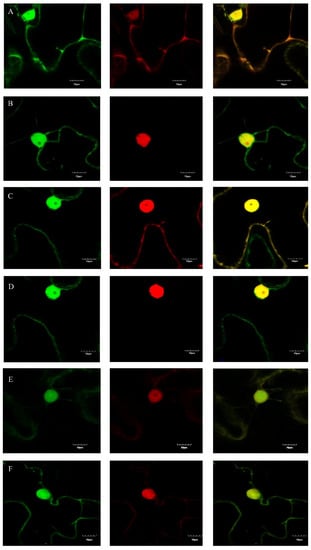
Figure 4.
Determination of subcellular localizations of m03915 and m04250. GFP was used to determine the subcellular localizations of m03915 and m04250 by fusion at C-terminal of m03915 and m04250 using confocal microscopy. Free GFP was used as a control. Six true leaf-stage N. benthamiana were infiltrated with Agrobacterium EHA 105 harboring the desired plasmid. Red fluorescent protein and red fluorescent protein with nuclear localization signal (NLS) (RFP-NLS) were used for coinfiltration to determine whether m03915 and m04250 had an NLS. The samples were collected at two days post infiltration and observed under a confocal microscope. (A) free GFP coinfiltrated with RFP; (B) free GFP coinfiltrated with RFP-NLS; (C) m03915-GFP coinfiltrated with RFP; (D) m03915-GFP coinfiltrated with RFP-NLS; (E) m04250-GFP coinfiltrated with RFP; (F) m04250-GFP infiltrated with RFP-NLS. The left images showed the GFP channel, the middle images showed the RFP channel; the right images showed the overlay of GFP and RFP. GFP and m03915-GFP did not have an NLS. On the contrary, m04250-GFP localized in the nucleus.
2.5. Identification of the Host Proteins Interacting with m03915 and m04250
Yeast two-hybrid (Y2H) was applied to identify citrus proteins interacting with m03915 and m04250. The second Glutamic acid of m03915 was changed to Alanine (m03915E20A) to eliminate the autoactivation activity. Six genes (seven isolates) were identified from the library that interacted with m03915E20A. XM_006465843 was annotated as a cytochrome P450 71A1-like protein. XM_006467727.3 encoded the transcription factor bHLH16. The annotation of XM_006478039.3 was a glutamate decarboxylase 5-like protein. XM_006452415 encoded a cold and drought-regulated protein. Interestingly, XM_006481033 was annotated as an ubiquitin-activating enzyme E1 1-like protein. Two isolates were annotated as an EG45-like domain-containing protein (accession number XM_006474049). One isolate which interacted with m04250 was annotated as a suppressor of mec-8 and unc-52 protein homolog 2 protein (access number XM_006474512) (Table 1).

Table 1.
The host proteins interacted with mature proteins of CLIBASIA_03915 and CLIBASIA_04250.
3. Discussion
CaLas is a phloem restricted uncultured Gram-negative bacterium. It is directly delivered into citrus phloem by the vector of Asian citrus psyllid (ACP) under natural conditions [18]. The phloem sap of pineapple sweet orange contains 20 amino acids, 8 sugars, and 7 organic acids as nutrition for CaLas and psyllid [19]. ACP delivers CaLas directly into citrus phloem to avoid triggering citrus immunity responses. Moreover, CaLas suppresses plant disease resistance by the degradation of defense signals [9,10], such as SA and H2O2, which participate in local hypersensitivity reactions as well as systemic acquired resistance [20,21]. Efforts have been made to understand the HLB-tolerance mechanism of citrus [22,23,24,25,26,27] and how HLB symptoms develop in citrus [28,29,30]. However, it is still unclear how citrus develops HLB symptoms upon CaLas infection, including phloem necrosis.
Based on the genomic information of CaLas, several reports focused on the virulence mechanisms of CaLas [7,8,9,10]. The identified effectors could cause cell death of infiltrated leaves [6,8], systemic leaves necrosis [7], and inhibition of plant growth [8]. Mature proteins of CLIBASIA_03915 (m03915) and CLIBASIA_04250 (m04250) caused phloem necrosis in the senescent leaves of N. benthamiana when expressed by TMV (Figure 2) and functioned at the protein level (Figure 3). Six proteins were identified to interact with m03915E20A (Table 1). Cytochrome P450s play critical roles in the synthesis and metabolism of lignin, sterols, isoflavonoids, terpenes, flavonoids, furanocoumarins, and other secondary plant products [31]. The expression of cytochrome P450 was upregulated in the HLB tolerant citrus under biotic and abiotic stress [25]. Genes involved in cytochrome P450-related reactions were upregulated in HLB tolerant citrus [32]. Basic helix–loop–helix (bHLH) transcription factors are the second largest group of transcription factors in plants that are also involved in plant development, phytohormone signaling, and stress responses [33]. It was reported that AtbHLH162 was involved in the response to fungal pathogen [34]. Glutamate decarboxylase 5 catalyzes the production of γ-aminobutyric acid (GABA) [35]. The concentration of GABA was higher in CaLas infected ACP than in the control, and therefore, GABA might be from HLB infected citrus [36]. XM_006452415 encoded a cold and drought-regulated protein by annotation. However, the proteins that hit XM_006452415 can be grouped into cold and drought-related protein, phase-change related protein, glycine-rich protein, glycine-rich cell wall structural protein-like, etc. The exact function of XM_006452415 needs to be determined. Interestingly, a protein annotated as a ubiquitin-activating enzyme E1 1-like protein (UBA1) was found to interact with m03915E20A. The ubiquitin–proteasome system regulates plant development, flowering, responses to abiotic stresses, and biotic stresses [37]. UBA1 was required for disease resistance in Arabidopsis [38]. Plant pathogens target the key enzymes in the ubiquitin-conjugating cascade to suppress plant disease resistance [39]. It was reported that CLIBASIA_00470 inhibited plant growth and interacted with polyubiquitin-like proteins and a putative E3 protein [8]. It is possible that m03915 downregulated genes in cell wall synthesis to cause phloem necrosis via ubiquitin–mediated protein degradation system. CaLas may hitchhike the host ubiquitin–proteasome system to suppress citrus disease resistance and interfere with citrus development. EG45-like domain-containing protein may play a role in water and solute homeostasis [40]. Only one prey was identified to interact with m04250. It was annotated as a suppressor of mec-8 and unc-52 protein homolog 2 protein, also known as SMU2. SMU2 regulates plant pre-mRNA splicing cooperatively with SMU1 to regulate plant development [41]. The alternative splicing of genes varied significantly between HLB tolerant citrus cultivar and HLB-susceptible citrus cultivar when infected with HLB [25]. The mature protein m04250 may disturb the normal pre-mRNA splicing to cause phloem necrosis.
Since conventional breeding cannot create HLB-tolerance/resistance citrus, the transgenic approach has been used to enhance citrus HLB tolerance. The approach of overexpression has been applied to improve citrus HLB resistance [42,43,44]. However, clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated proteins (CRISPR/Cas) cannot be used in citrus HLB improvement without known HLB susceptible gene [45] in spite of the advantages [46,47] and successful generation of plants that were resistant to bacterial pathogens and fungal pathogens [48] including citrus canker [49,50] by targeting LOB1 which is the susceptibility gene of citrus canker [51]. Meanwhile, CRISPR is a powerful weapon to combat viruses [52]. However, CRISPR is only a promising tool when the virulence mechanism of CaLas is still vague [45,53]. Based on our results and previous reports, we propose that E3 enzymes differential expressed in HLB tolerant and HLB susceptible citrus upon CaLas infection [25] may be considered as the target of CRISPR based on the possibility of CaLas interference with citrus development and disease resistance via targeting the specific proteins of the ubiquitin–proteasome system. Overexpression of the key genes in cell wall synthesis may be another way to enhance citrus HLB tolerance.
4. Materials and methods
4.1. Plant Growth Conditions
N. benthamiana was grown in a greenhouse at 25 °C with 16 h light and 8 h dark.
4.2. Prediction and Confirmation of CLIBASIA_03915 and CLIBASIA_04250 Signal Peptides
Signal peptides of CLIBASIA_03915 and CLIBASIA_04250 were predicted by combination of SignalP 3.0 [54], SignalP 5.0 [55], Phobius [56], and TMHMM [57]. To confirm the signal peptides of CLIBASIA_03915 and CLIBASIA_04250, phoA assay was employed. Vector pJDT1 [17] without the signal peptide sequence of phoA was used. The DNA fragments of CLIBASIA_03915 and CLIBASIA_04250 containing signal peptides were amplified with primers CLIBASIA_03915FF/CLIBASIA_03915FR, CLIBASIA_04250FF/CLIBASIA_04250FR, and respectively cloned into pGEM-T Easy vector (Promega, Madison, WI, USA, Cat. No. A1360). The correct plasmids were digested with SacI and MfeI, and the recovered DNA fragment was inserted into vector pJDT1. The plasmids were confirmed by sequencing, then were transformed into Escherichia coli (E. coli) strain BL21 DE3 (NEB, cat. C2527I). The E. coli single clone harboring CLIBASIA_03915/ CLIBASIA_04250 was streaked along with negative control pJDT1 on Luria–Bertani (LB) plates amended with 5-bromo-4-chloro-3-indolylphosphate (BCIP) (90 mg/L) and sodium phosphate (75 mM, pH 7.2).
4.3. Heterologous Expression of the Mature Proteins of CLIBASIA_03915 (m03915) and CLIBASIA_04250 (m04250) in N. benthamiana
N. benthamiana was used to express m03915 and m04250 by TMV via agroinfiltration, as described previously [8]. Primers m03915 EF/m03915 ER and m04250 EF/m04250 ER were used to amplify the DNA fragments from HLB-infected citrus without the predicted SP (SP of 03915 1-24 aa, SP of 04250 1-20 aa). Primers m03915 NF/ m03915 ER and m04250 NF/ m04250 ER were used to introduce mutations into m03915 (m03915Nu) and m04250 (m04250FS). Agrobacterium strain EHA105 was used to express m03915, m04250, m03915Nu, and m04250FS in N. benthamiana by TMV. The six-leaf stage N. benthamiana was used for agroinfiltration. The agrobacterium culture was adjusted to optical density (OD) value 0.6 by solution with 10 mM MgCl2, 10 mM MES pH 5.6, and 10 µM acetosyringone [58].
4.4. Determination of the Subcellular Localizations of m03915 and m04250
Transient expression was employed to determine the subcellular localizations of m03915 and m04250. Primers m03915 GF/m03915 GR and m04250 GF/m04250 GR were used to amplify DNA fragments to fuse with the green fluorescent protein (GFP). XbaI and XmaI sites were added at the 5′ and 3′ end of the DNA fragments, respectively. The GFP DNA fragment was amplified from plasmid p30B-GFPC3 [59] with primers GFP FF/GFP FR to introduce enzyme sites of XmaI and SacI. The amplified DNA fragments were cloned into the pGEM-T Easy vector. The plasmids were digested with XbaI and SacI, then the recovered DNA fragments were inserted into pBI121 [60] to get the final vectors for determination of the subcellular localizations of m03915 and m04250. The control GFP DNA fragment was amplified with primers GFP F/GFP R and inserted into pBI121. Agrobacterium EHA105 carrying free GFP, m03915-GFP, and m04250-GFP were cultured and OD was adjusted to 0.8 for agroinfiltration. To determine whether m03915-GFP and m04250-GFP had an NLS, red fluorescent protein (RFP) and RFP with a nuclear localization signal (NLS) were used. Plasmid pCAMBIA1380 was used as the backbone to express RFP and RFP-NLS. The DNA fragment from pBI121 between HindIII and EcoRI with a CaMV 35S promoter-GUS-NOS terminator was inserted into pCAMBIA1380 to get pCAMBIA1380-GUS. To eliminate the leaking expression of RFP, an intron was added at 5′ of the RFP and RFP-NLS. The amplified intron was from pBI121-GUS-INTRON with primers HAint F/ HAint R. The DNA fragment containing the intron was inserted into pCAMBIA1380-GUS between XbaI and SacI sites to get pCAMBIA1380-INT. RFP was amplified from plasmid pSAT6-RFP-C1 [61] with primers RFP F/RFP R. The DNA fragment was recovered and inserted into pCAMBIA1380-INT between BamHI and SacI sites. RFP-NLS was amplified from pSAT6-mRFP-VirD2NLS [62] with primers RFP-NLS F/RFP-NLS R. The DNA fragment with RFP-NLS was inserted into pCAMBIA1380-INT between BamHI and SacI sites. Agrobacterium EHA105 carrying RFP or RFP-NLS was coinfiltrated with m03915-GFP or m04250-GFP to determine whether there was any NLS in the candidates, while GFP was control. The N. benthamiana leaves were collected at two days post infiltration (DPI) and were observed under an Olympus FV1000 MPE multiphoton laser scanning microscope (Tokyo, Japan).
4.5. Identification of host Proteins Interacting with m03915 and m04250 by Yeast Two-Hybrid (Y2H)
The construction of the library used for Y2H screening was described previously [8]. The Saccharomyces cerevisiae strain Y187 and pGADT7-Rec (SmaI-linearized) were used to construct the library of HLB infected sweet orange ‘Valencia’. The library construction was as the manual of the “Mate & Plate” Library System (Clontech, Cat no. 630490). Vector pGBKT7 DNA-BD (Clontech, Cat. No. 630443) was used to construct the bait plasmids. Primers m03915 YF/m03915 YMR (eliminate the autoactivation activity of m03915) and m04250 YF/m04250 YR were used to amplify the DNA fragments for construction bait plasmids pGBKT7-m03915E20A and pGBKT7-m04250. Saccharomyces cerevisiae Y2HGold was transformed with the two bait plasmids. The library screening was performed according to Matchmaker Gold Yeast Two-Hybrid (Clontech, Cat. No. 630489). The annotation and accession number of the identified positive prey were retrieved from the National Center for Biotechnology Information (NCBI).
Supplementary Materials
The following are available online at https://www.mdpi.com/1422-0067/21/4/1414/s1, Figure S1: Distribution pattern of free GFP, m03915-GFP, and m04250-GFP in the epidermic cells of N. benthamiana; Table S1: The primers used in this research.
Author Contributions
D.L., H.L. and X.Y. conceived the study and designed the research; H.L., X.Y., L.S., X.X., F.F.X. and S.W. performed research; J.Z., H.J., and H.Y. analyzed the data; X.Y. and H.L. wrote the manuscript; The manuscript was edited by B.R., N.K., and D.L.; Funding acquisition, D.L. All authors have read and agreed to the published version of the manuscript.
Acknowledgments
This research was supported by grants from the Natural Science Foundation of China (31471753), the Science and Technology Assistance Project to Developing Countries (KY201904016), Chongqing Technology Innovation and Application Development Project (cstc2019jscx-msxmX0398) and the Talent Introduction Program of Southwest University (SWU019008).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bové, J.M. Huanglongbing: A destructive, newly-emerging, century-old disease of citrus. J. Plant Pathol. 2006, 88, 7–37. [Google Scholar]
- Gottwald, T.R. Current epidemiological understanding of citrus Huanglongbing. Annu. Rev. Phytopathol. 2010, 48, 119–139. [Google Scholar] [CrossRef]
- Dala-Paula, B.M.; Plotto, A.; Bai, J.; Manthey, J.A.; Baldwin, E.A.; Ferrarezi, R.S.; Gloria, M.B.A. Effect of Huanglongbing or Greening Disease on Orange Juice Quality, a Review. Front. Plant Sci. 2018, 9, 1976. [Google Scholar] [CrossRef]
- Duan, Y.; Zhou, L.; Hall, D.G.; Li, W.; Doddapaneni, H.; Lin, H.; Liu, L.; Vahling, C.M.; Gabriel, D.W.; Williams, K.P.; et al. Complete genome sequence of citrus huanglongbing bacterium, ‘Candidatus Liberibacter asiaticus’ obtained through metagenomics. Mol. Plant Microb. Interact. 2009, 22, 1011–1020. [Google Scholar] [CrossRef]
- Tyler, H.L.; Roesch, L.F.; Gowda, S.; Dawson, W.O.; Triplett, E.W. Confirmation of the sequence of ‘Candidatus Liberibacter asiaticus’ and assessment of microbial diversity in Huanglongbing-infected citrus phloem using a metagenomic approach. Mol. Plant Microb. Interact. 2009, 22, 1624–1634. [Google Scholar] [CrossRef]
- Pitino, M.; Armstrong, C.M.; Cano, L.M.; Duan, Y.P. Transient Expression of Candidatus Liberibacter Asiaticus Effector Induces Cell Death in Nicotiana benthamiana. Front. Plant Sci. 2016, 7, 982. [Google Scholar] [CrossRef]
- Liu, X.; Fan, Y.; Zhang, C.; Dai, M.; Wang, X.; Li, W. Nuclear Import of a Secreted “Candidatus Liberibacter asiaticus” Protein is Temperature Dependent and Contributes to Pathogenicity in Nicotiana benthamiana. Front. Microbiol. 2019, 10, 1684. [Google Scholar] [CrossRef]
- Ying, X.; Wan, M.; Hu, L.; Zhang, J.; Li, H.; Lv, D. Identification of the Virulence Factors of Candidatus Liberibacter asiaticus via Heterologous Expression in Nicotiana benthamiana using Tobacco Mosaic Virus. Int. J. Mol. Sci. 2019, 20, 5575. [Google Scholar] [CrossRef]
- Li, J.Y.; Pang, Z.Q.; Trivedi, P.; Zhou, X.F.; Ying, X.B.; Jia, H.G.; Wang, N.A. ‘Candidatus Liberibacter asiaticus’ Encodes a Functional Salicylic Acid (SA) Hydroxylase That Degrades SA to Suppress Plant Defenses. Mol. Plant Microb. Interact. 2017, 30, 620–630. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Fleites, L.A.; Gabriel, D.W. Prophage-Encoded Peroxidase in ‘Candidatus Liberibacter asiaticus’ Is a Secreted Effector That Suppresses Plant Defenses. Mol. Plant Microb. Interact. 2015, 28, 1330–1337. [Google Scholar] [CrossRef] [PubMed]
- Clark, K.; Franco, J.Y.; Schwizer, S.; Pang, Z.; Hawara, E.; Liebrand, T.W.H.; Pagliaccia, D.; Zeng, L.; Gurung, F.B.; Wang, P.; et al. An effector from the Huanglongbing-associated pathogen targets citrus proteases. Nat. Commun. 2018, 9, 1718. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.H.; Achor, D.; Exteberria, E.; Yu, Q.B.; Du, D.L.; Stanton, D.; Liang, G.; Gmitter, F.G. Phloem Regeneration Is a Mechanism for Huanglongbing-Tolerance of “Bearss” Lemon and “LB8-9” Sugar Belle (R) Mandarin. Front. Plant Sci. 2019, 10, 277. [Google Scholar] [CrossRef] [PubMed]
- De Schepper, V.; De Swaef, T.; Bauweraerts, I.; Steppe, K. Phloem transport: A review of mechanisms and controls. J. Exp. Bot. 2013, 64, 4839–4850. [Google Scholar] [CrossRef] [PubMed]
- Knoblauch, M.; Peters, W.S. Münch, morphology, microfluidics—Our structural problem with the phloem. Plant Cell Environ. 2010, 33, 1439–1452. [Google Scholar] [CrossRef] [PubMed]
- Froelich, D.R.; Mullendore, D.L.; Jensen, K.H.; Ross-Elliott, T.J.; Anstead, J.A.; Thompson, G.A.; Pelissier, H.C.; Knoblauch, M. Phloem ultrastructure and pressure flow: Sieve-Element-Occlusion-Related agglomerations do not affect translocation. Plant Cell 2011, 23, 4428–4445. [Google Scholar] [CrossRef] [PubMed]
- Campbell, M.M.; Sederoff, R.R. Variation in Lignin Content and Composition (Mechanisms of Control and Implications for the Genetic Improvement of Plants). Plant Physiol. 1996, 110, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Mdluli, K.E.; Treit, J.D.; Kerr, V.J.; Nano, F.E. New vectors for the in vitro generation of alkaline phosphatase fusions to proteins encoded by G+C-rich DNA. Gene 1995, 155, 133–134. [Google Scholar] [CrossRef]
- Grafton-Cardwell, E.E.; Stelinski, L.L.; Stansly, P.A. Biology and management of Asian citrus psyllid, vector of the huanglongbing pathogens. Annu. Rev. Entomol. 2013, 58, 413–432. [Google Scholar] [CrossRef]
- Hijaz, F.; Killiny, N. Collection and chemical composition of phloem sap from Citrus sinensis L. Osbeck (sweet orange). PLoS ONE 2014, 9, e101830. [Google Scholar] [CrossRef]
- Baxter, A.; Mittler, R.; Suzuki, N. ROS as key players in plant stress signalling. J. Exp. Bot. 2014, 65, 1229–1240. [Google Scholar] [CrossRef]
- Delaney, T.P.; Uknes, S.; Vernooij, B.; Friedrich, L.; Weymann, K.; Negrotto, D.; Gaffney, T.; Gut-Rella, M.; Kessmann, H.; Ward, E.; et al. A central role of salicylic Acid in plant disease resistance. Science 1994, 266, 1247–1250. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zhong, X.; Liu, X.L.; Lou, B.H.; Zhou, C.Y.; Wang, X.F. Comparative transcriptome analysis unveils the tolerance mechanisms of Citrus hystrix in response to ‘Candidatus Liberibacter asiaticus’ infection. PLoS ONE 2017, 12, e0189229. [Google Scholar] [CrossRef] [PubMed]
- Hijaz, F.; Nehela, Y.; Killiny, N. Possible role of plant volatiles in tolerance against huanglongbing in citrus. Plant Signal. Behav. 2016, 11, e1138193. [Google Scholar] [CrossRef]
- Martinelli, F.; Reagan, R.L.; Dolan, D.; Fileccia, V.; Dandekar, A.M. Proteomic analysis highlights the role of detoxification pathways in increased tolerance to Huanglongbing disease. BMC Plant Biol. 2016, 16, 167. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhou, L.; Yu, X.; Stover, E.; Luo, F.; Duan, Y. Transcriptome Profiling of Huanglongbing (HLB) Tolerant and Susceptible Citrus Plants Reveals the Role of Basal Resistance in HLB Tolerance. Front. Plant Sci. 2016, 7, 933. [Google Scholar] [CrossRef] [PubMed]
- Albrecht, U.; Bowman, K.D. Transcriptional response of susceptible and tolerant citrus to infection with Candidatus Liberibacter asiaticus. Plant Sci. 2012, 185–186, 118–130. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Chen, C.; Yu, Q.; Khalaf, A.; Achor, D.S.; Brlansky, R.H.; Moore, G.A.; Li, Z.G.; Gmitter, F.G., Jr. Comparative transcriptional and anatomical analyses of tolerant rough lemon and susceptible sweet orange in response to ‘Candidatus Liberibacter asiaticus’ infection. Mol. Plant Microb. Interact. 2012, 25, 1396–1407. [Google Scholar] [CrossRef] [PubMed]
- Nwugo, C.C.; Lin, H.; Duan, Y.P.; Civerolo, E.L. The effect of ‘Candidatus Liberibacter asiaticus’ infection on the proteomic profiles and nutritional status of pre-symptomatic and symptomatic grapefruit (Citrus paradisi) plants. BMC Plant Biol. 2013, 13, 59. [Google Scholar] [CrossRef]
- Martinelli, F.; Uratsu, S.L.; Albrecht, U.; Reagan, R.L.; Phu, M.L.; Britton, M.; Buffalo, V.; Fass, J.; Leicht, E.; Zhao, W.; et al. Transcriptome profiling of citrus fruit response to huanglongbing disease. PLoS ONE 2012, 7, e38039. [Google Scholar] [CrossRef]
- Martinelli, F.; Reagan, R.L.; Uratsu, S.L.; Phu, M.L.; Albrecht, U.; Zhao, W.; Davis, C.E.; Bowman, K.D.; Dandekar, A.M. Gene regulatory networks elucidating huanglongbing disease mechanisms. PLoS ONE 2013, 8, e74256. [Google Scholar] [CrossRef]
- Schuler, M.A. Plant Cytochrome P450 Monooxygenases. Crit. Rev. Plant Sci. 1996, 15, 235–284. [Google Scholar] [CrossRef]
- Balan, B.; Ibanez, A.M.; Dandekar, A.M.; Caruso, T.; Martinelli, F. Identifying Host Molecular Features Strongly Linked With Responses to Huanglongbing Disease in Citrus Leaves. Front. Plant Sci. 2018, 9, 277. [Google Scholar] [CrossRef]
- Feller, A.; Machemer, K.; Braun, E.L.; Grotewold, E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011, 66, 94–116. [Google Scholar] [CrossRef] [PubMed]
- Huibers, R.P.; de Jong, M.; Dekter, R.W.; Van den Ackerveken, G. Disease-specific expression of host genes during downy mildew infection of Arabidopsis. Mol. Plant Microb. Interact. 2009, 22, 1104–1115. [Google Scholar] [CrossRef] [PubMed]
- Scholz, S.S.; Reichelt, M.; Mekonnen, D.W.; Ludewig, F.; Mithofer, A. Insect Herbivory-Elicited GABA Accumulation in Plants is a Wound-Induced, Direct, Systemic, and Jasmonate-Independent Defense Response. Front. Plant Sci. 2015, 6, 1128. [Google Scholar] [CrossRef]
- Killiny, N.; Nehela, Y.; Hijaz, F.; Vincent, C.I. A plant pathogenic bacterium exploits the tricarboxylic acid cycle metabolic pathway of its insect vector. Virulence 2018, 9, 99–109. [Google Scholar] [CrossRef]
- Sadanandom, A.; Bailey, M.; Ewan, R.; Lee, J.; Nelis, S. The ubiquitin-proteasome system: Central modifier of plant signalling. New Phytol. 2012, 196, 13–28. [Google Scholar] [CrossRef]
- Goritschnig, S.; Zhang, Y.; Li, X. The ubiquitin pathway is required for innate immunity in Arabidopsis. Plant J. 2007, 49, 540–551. [Google Scholar] [CrossRef]
- Zeng, L.R.; Vega-Sanchez, M.E.; Zhu, T.; Wang, G.L. Ubiquitination-mediated protein degradation and modification: An emerging theme in plant-microbe interactions. Cell Res. 2006, 16, 413–426. [Google Scholar] [CrossRef]
- Ludidi, N.N.; Heazlewood, J.L.; Seoighe, C.; Irving, H.R.; Gehring, C.A. Expansin-like molecules: Novel functions derived from common domains. J. Mol. Evol. 2002, 54, 587–594. [Google Scholar] [CrossRef]
- Chung, T.; Wang, D.; Kim, C.S.; Yadegari, R.; Larkins, B.A. Plant SMU-1 and SMU-2 homologues regulate pre-mRNA splicing and multiple aspects of development. Plant Physiol 2009, 151, 1498–1512. [Google Scholar] [CrossRef] [PubMed]
- Dutt, M.; Barthe, G.; Irey, M.; Grosser, J. Transgenic Citrus Expressing an Arabidopsis NPR1 Gene Exhibit Enhanced Resistance against Huanglongbing (HLB.; Citrus Greening). PLoS ONE 2015, 10, e0137134. [Google Scholar] [CrossRef] [PubMed]
- Hao, G.; Stover, E.; Gupta, G. Overexpression of a Modified Plant Thionin Enhances Disease Resistance to Citrus Canker and Huanglongbing (HLB). Front. Plant Sci. 2016, 7, 1078. [Google Scholar] [CrossRef] [PubMed]
- Zou, X.; Jiang, X.; Xu, L.; Lei, T.; Peng, A.; He, Y.; Yao, L.; Chen, S. Transgenic citrus expressing synthesized cecropin B genes in the phloem exhibits decreased susceptibility to Huanglongbing. Plant Mol. Biol. 2017, 93, 341–353. [Google Scholar] [CrossRef]
- Song, X.; Bhattarai, K.; Lv, D.; Gao, F.; Ying, X. Can CRISPR Win the Battle against Huanglongbing? J. Plant Pathol. Microbiol. 2017, 8, 2. [Google Scholar]
- Noman, A.; Aqeel, M.; He, S. CRISPR-Cas9: Tool for Qualitative and Quantitative Plant Genome Editing. Front. Plant Sci. 2016, 7, 1740. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, H.Y.; Zhu, H.L. CRISPR technology is revolutionizing the improvement of tomato and other fruit crops. Hortic. Res. Engl. 2019, 6, 77. [Google Scholar] [CrossRef]
- Borrelli, V.M.G.; Brambilla, V.; Rogowsky, P.; Marocco, A.; Lanubile, A. The Enhancement of Plant Disease Resistance Using CRISPR/Cas9 Technology. Front. Plant Sci. 2018, 9, 1245. [Google Scholar] [CrossRef]
- Jia, H.; Zhang, Y.; Orbovic, V.; Xu, J.; White, F.F.; Jones, J.B.; Wang, N. Genome editing of the disease susceptibility gene CsLOB1 in citrus confers resistance to citrus canker. Plant Biotechn. J. 2017, 15, 817–823. [Google Scholar] [CrossRef]
- Peng, A.; Chen, S.; Lei, T.; Xu, L.; He, Y.; Wu, L.; Yao, L.; Zou, X. Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol. J. 2017, 15, 1509–1519. [Google Scholar] [CrossRef]
- Hu, Y.; Zhang, J.L.; Jia, H.G.; Sosso, D.; Li, T.; Frommer, W.B.; Yang, B.; White, F.F.; Wang, N.A.; Jones, J.B. Lateral organ boundaries 1 is a disease susceptibility gene for citrus bacterial canker disease. Proc. Natl. Acad. Sci. USA 2014, 111, E521–E529. [Google Scholar] [CrossRef] [PubMed]
- Ali, Z.; Abulfaraj, A.; Idris, A.; Ali, S.; Tashkandi, M.; Mahfouz, M.M. CRISPR/Cas9-mediated viral interference in plants. Genome Biol. 2015, 16, 238. [Google Scholar] [CrossRef] [PubMed]
- Sun, L..; Nie, Z.; Wang, P.; Xu, J. Citrus Genetic Engineering for Disease Resistance: Past, Present and Future. Int. J. Mol. Sci. 2019, 20, 5256. [Google Scholar] [CrossRef] [PubMed]
- Bendtsen, J.D.; Nielsen, H.; von Heijne, G.; Brunak, S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004, 340, 783–795. [Google Scholar] [CrossRef] [PubMed]
- Armenteros, J.J.A.; Tsirigos, K.D.; Sonderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420. [Google Scholar] [CrossRef] [PubMed]
- Käll, L.; Krogh, A.; Sonnhammer, E.L. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 2004, 338, 1027–1036. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Ying, X.B.; Dong, L.; Zhu, H.; Duan, C.G.; Du, Q.S.; Lv, D.Q.; Fang, Y.Y.; Garcia, J.A.; Fang, R.X.; Guo, H.S. RNA-Dependent RNA Polymerase 1 from Nicotiana tabacum Suppresses RNA Silencing and Enhances Viral Infection in Nicotiana benthamiana. Plant Cell 2010, 22, 1358–1372. [Google Scholar] [CrossRef]
- Shivprasad, S.; Pogue, G.P.; Lewandowski, D.J.; Hidalgo, J.; Donson, J.; Grill, L.K.; Dawson, W.O. Heterologous sequences greatly affect foreign gene expression in tobacco mosaic virus-based vectors. Virology 1999, 255, 312–323. [Google Scholar] [CrossRef]
- Chen, P.-Y.; Wang, C.-K.; Soong, S.-C.; To, K.-Y. Complete sequence of the binary vector pBI121 and its application in cloning T-DNA insertion from transgenic plants. Mol. Breed. 2003, 11, 287–293. [Google Scholar] [CrossRef]
- Tzfira, T.; Tian, G.W.; Lacroix, B.; Vyas, S.; Li, J.; Leitner-Dagan, Y.; Krichevsky, A.; Taylor, T.; Vainstein, A.; Citovsky, V. pSAT vectors: A modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol. Biol. 2005, 57, 503–516. [Google Scholar] [CrossRef] [PubMed]
- Citovsky, V.; Lee, L.Y.; Vyas, S.; Glick, E.; Chen, M.H.; Vainstein, A.; Gafni, Y.; Gelvin, S.B.; Tzfira, T. Subcellular localization of interacting proteins by bimolecular fluorescence complementation in planta. J. Mol. Biol. 2006, 362, 1120–1131. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).