Rice OsAAA-ATPase1 is Induced during Blast Infection in a Salicylic Acid-Dependent Manner, and Promotes Blast Fungus Resistance
Abstract
:1. Introduction
2. Results
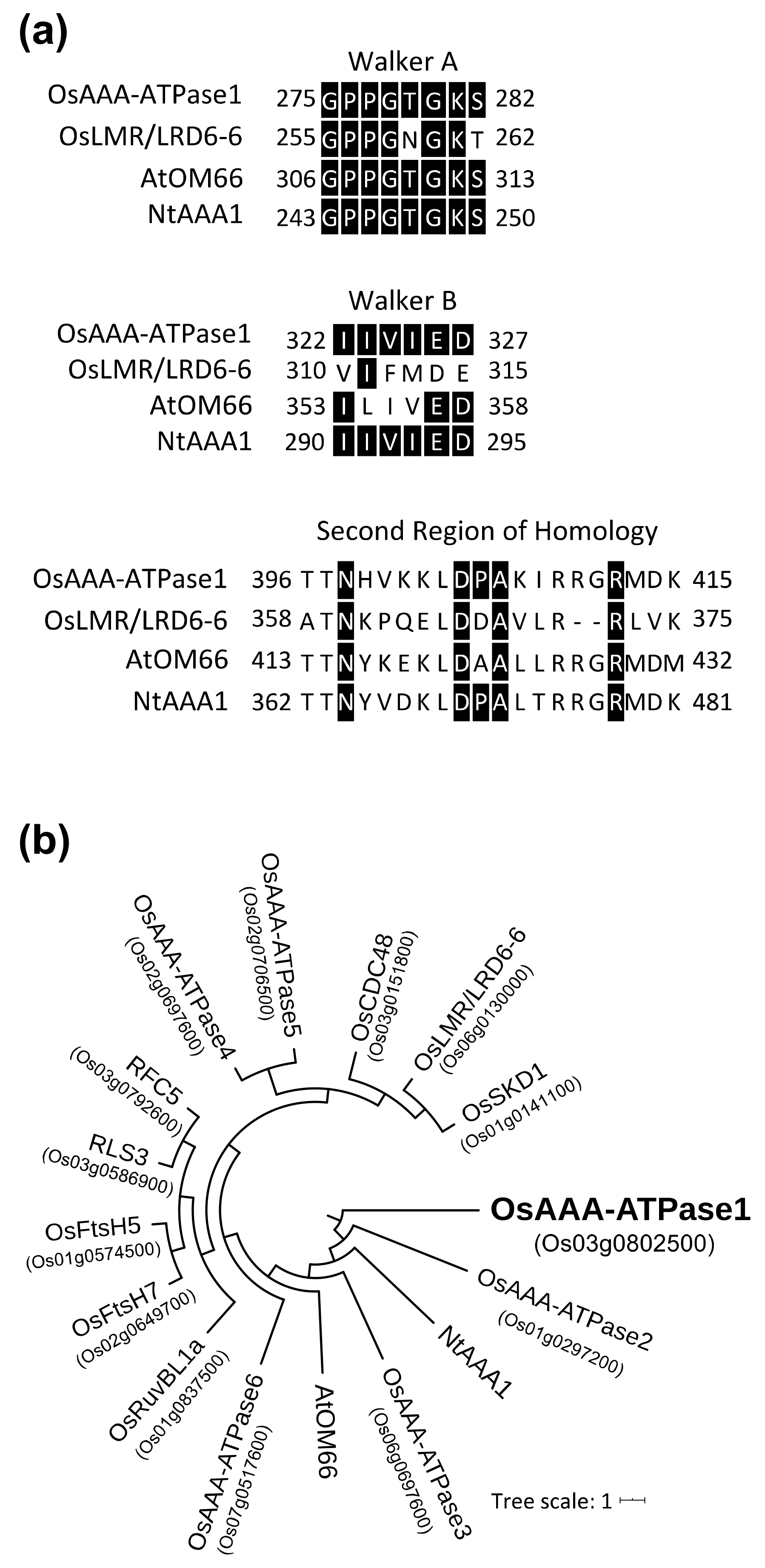
2.1. OsAAA-ATPase1 Encodes an AAA-ATPase Family Protein
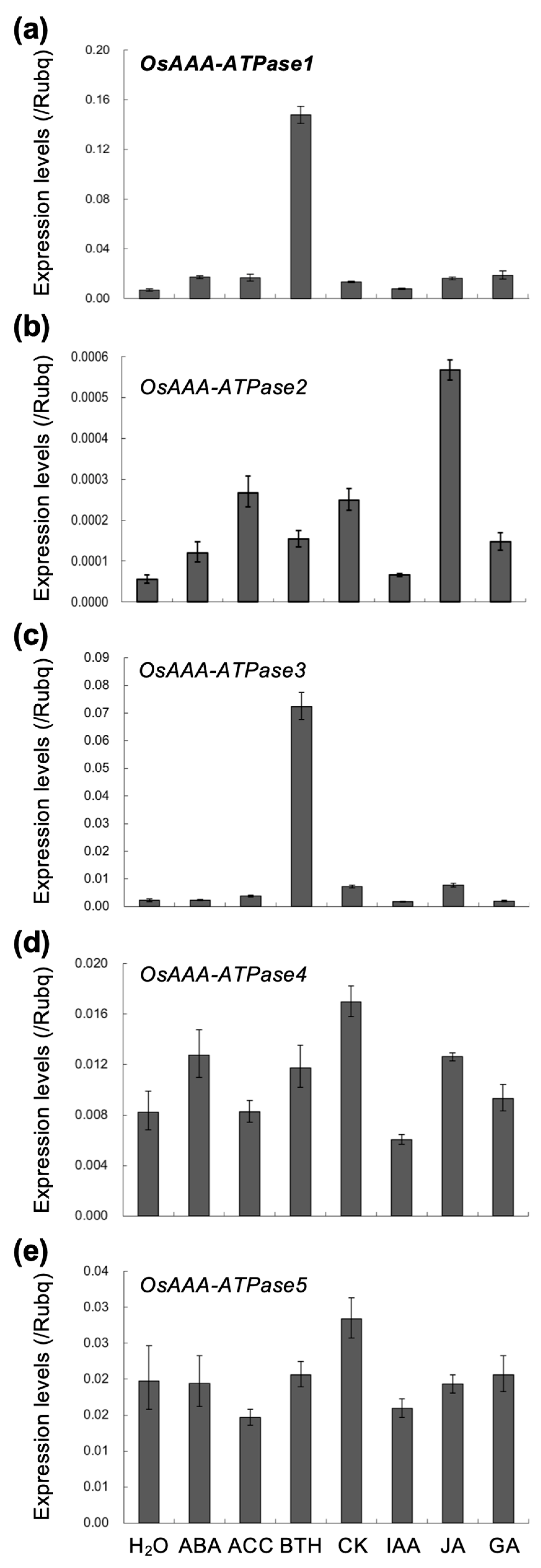
2.2. OsAAA-ATPase1 is Induced by SA Treatment
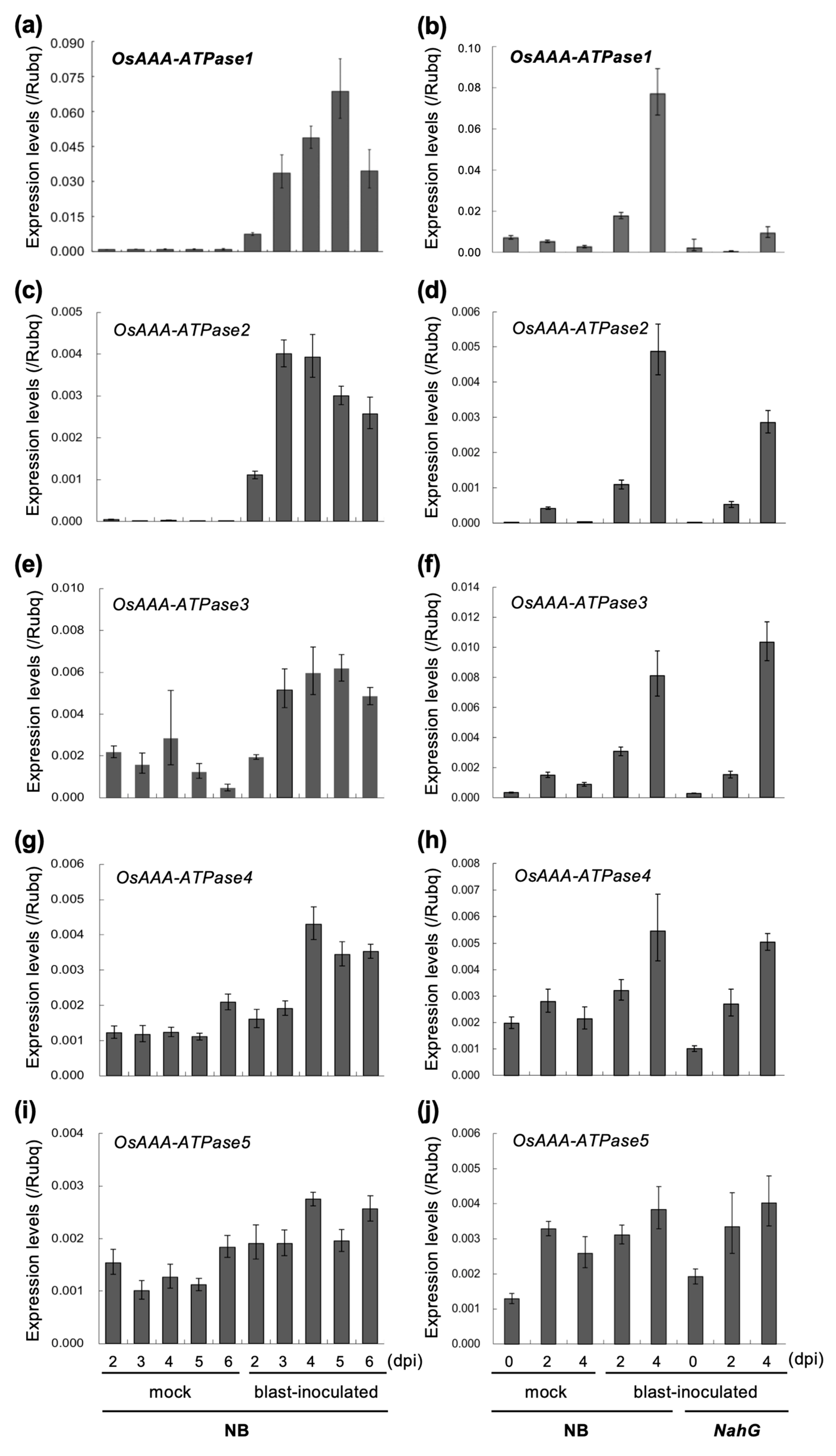
2.3. OsAAA-ATPase1 is Induced in Response to Blast Infection in An SA-Dependent Manner
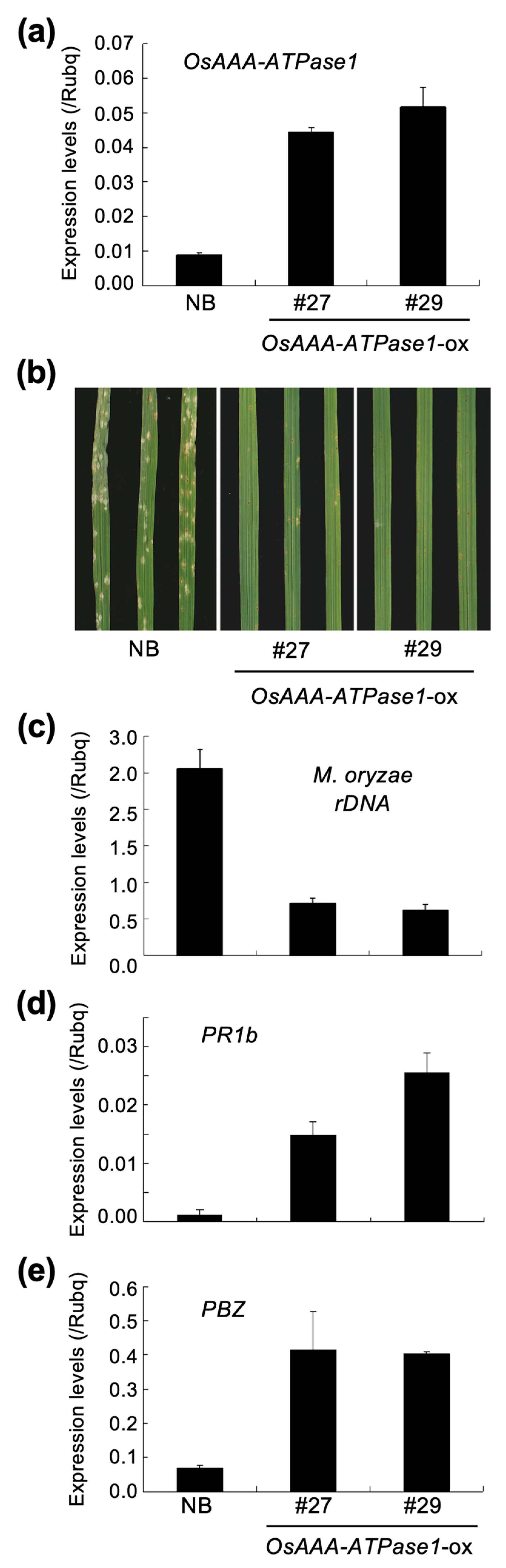
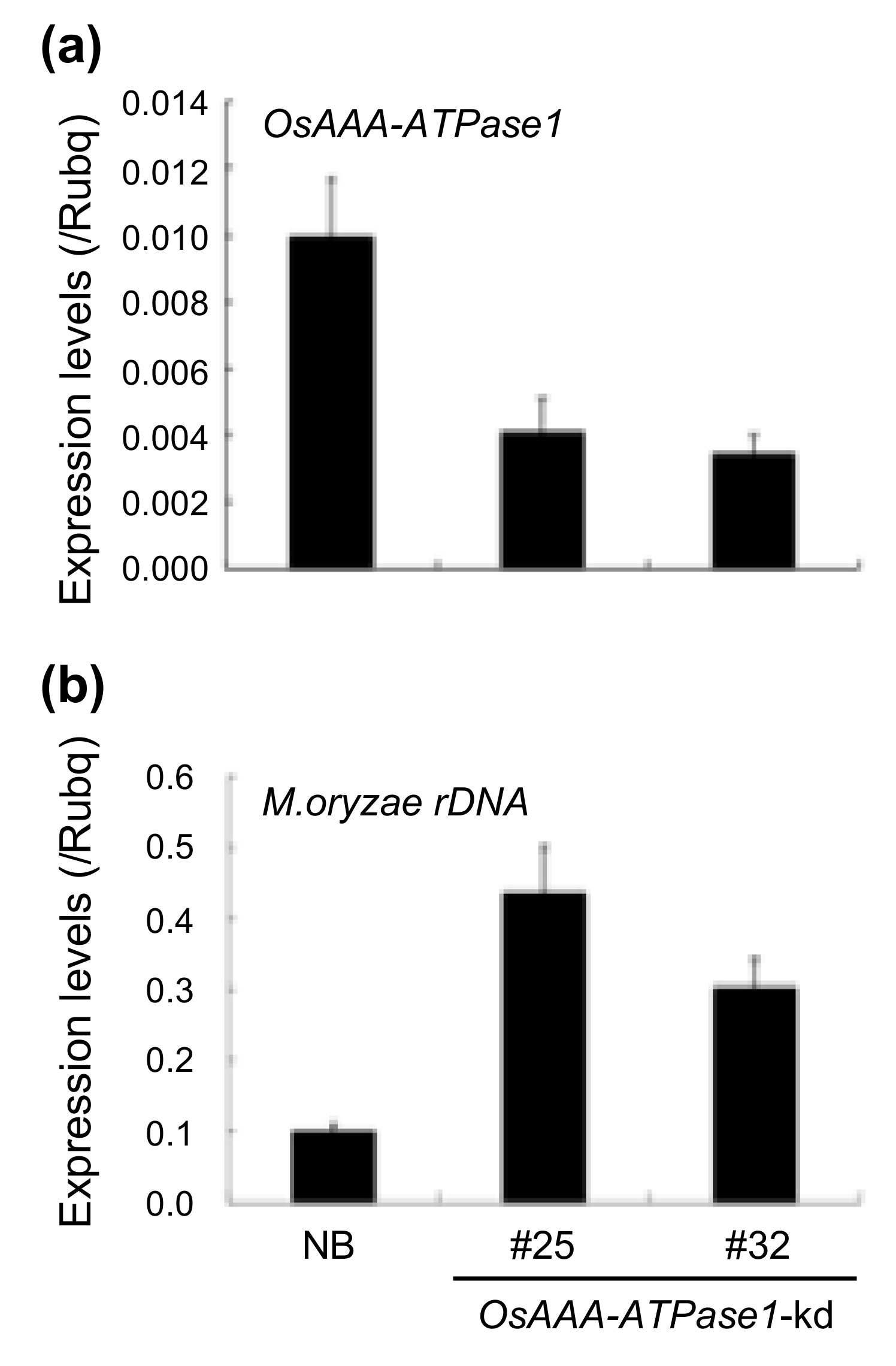
2.4. OsAAA-ATPase1 is Positively Involved in Blast Resistance
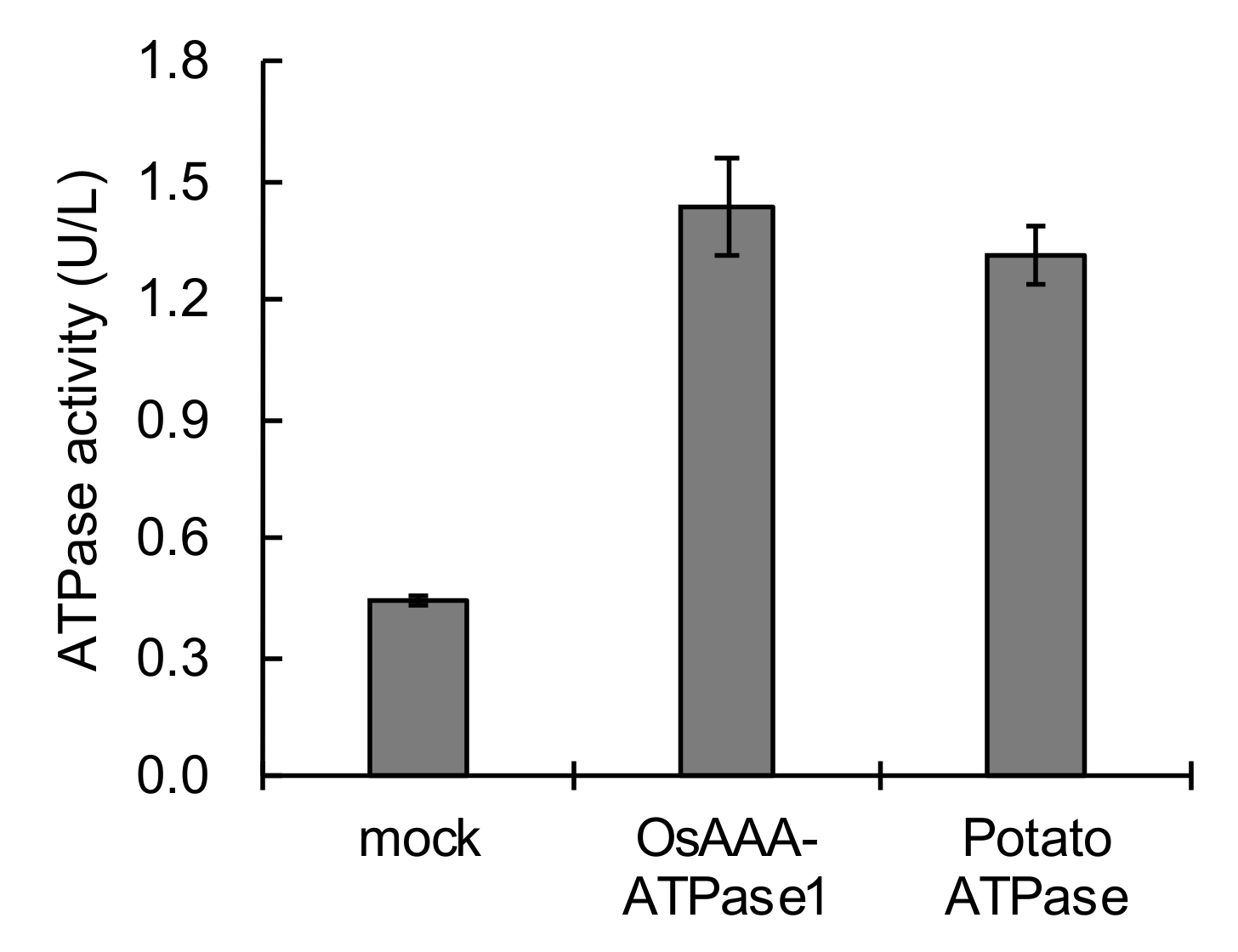
2.5. OsAAA-ATPase1 Has ATPase Activity and Is Localized in the Cytosol
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Plasmid DNA Construction and Rice Transformation
4.3. Chemical Treatments
4.4. Protein Expression, Purification, and ATPase Assay
4.5. Subcellular Localization
4.6. Pathogen Culture and Inoculations
4.7. RNA Analyses
4.8. Amino Acid Sequence Alignment and Phylogenetic Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Klessig, D.F.; Choi, H.W.; Dempsey, D.M.A. Systemic acquired resistance and salicylic acid: Past, present, and future. Mol. Plant Microbe Interact. 2018, 31, 871–888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, X. NPR1, all things considered. Curr. Opin. Plant Biol. 2004, 7, 547–552. [Google Scholar] [CrossRef]
- Wang, D.; Amornsiripanitch, N.; Dong, X. A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. Plos Pathog. 2006, 2, e123. [Google Scholar] [CrossRef] [Green Version]
- Cao, H.; Glazebrook, J.; Clarke, J.D.; Volko, S.; Dong, X. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 1997, 88, 57–63. [Google Scholar] [CrossRef] [Green Version]
- Shimono, M.; Sugano, S.; Nakayama, A.; Jiang, C.J.; Ono, K.; Toki, S.; Takatsuji, H. Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. Plant Cell 2007, 19, 2064–2076. [Google Scholar] [CrossRef] [Green Version]
- Sugano, S.; Jiang, C.J.; Miyazawa, S.; Masumoto, C.; Yazawa, K.; Hayashi, N.; Shimono, M.; Nakayama, A.; Miyao, M.; Takatsuji, H. Role of OsNPR1 in rice defense program as revealed by genome-wide expression analysis. Plant Mol. Biol. 2010, 74, 549–562. [Google Scholar] [CrossRef]
- Kachroo, P.; Shanklin, J.; Shah, J.; Whittle, E.J.; Klessig, D.F. A fatty acid desaturase modulates the activation of defense signaling pathways in plants. Proc. Natl. Acad. Sci. USA 2001, 98, 9448–9453. [Google Scholar] [CrossRef] [Green Version]
- Shah, J.; Kachroo, P.; Nandi, A.; Klessig, D.F. A recessive mutation in the Arabidopsis SSI2 gene confers SA- and NPR1-independent expression of PR genes and resistance against bacterial and oomycete pathogens. Plant J. 2001, 25, 563–574. [Google Scholar] [CrossRef] [PubMed]
- Kachroo, P.; Kachroo, A.; Lapchyk, L.; Hildebrand, D.; Klessig, D.F. Restoration of defective cross talk in ssi2 mutants: Role of salicylic acid, jasmonic acid, and fatty acids in SSI2-mediated signaling. Mol. Plant Microbe Interact. 2003, 16, 1022–1029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sekine, K.T.; Nandi, A.; Ishihara, T.; Hase, S.; Ikegami, M.; Shah, J.; Takahashi, H. Enhanced resistance to Cucumber mosaic virus in the Arabidopsis thaliana ssi2 mutant is mediated via an SA-independent mechanism. Mol. Plant Microbe Interact. 2004, 17, 623–632. [Google Scholar] [CrossRef]
- Kachroo, A.; Fu, D.Q.; Havens, W.; Navarre, D.; Kachroo, P.; Ghabrial, S.A. An oleic acid-mediated pathway induces constitutive defense signaling and enhanced resistance to multiple pathogens in soybean. Mol. Plant Microbe Interact. 2008, 21, 564–575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, C.J.; Shimono, M.; Maeda, S.; Inoue, H.; Mori, M.; Hasegawa, M.; Sugano, S.; Takatsuji, H. Suppression of the rice fatty-acid desaturase gene OsSSI2 enhances resistance to blast and leaf blight diseases in rice. Mol. Plant Microbe Interact. 2009, 22, 820–829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- HU, L.-q.; MU, J.-j.; SU, P.-s.; WU, H.-y.; YU, G.-h.; WANG, G.-p.; Liang, W.; Xin, M.; LI, A.-f.; WANG, H.-w. Multi-functional roles of TaSSI2 involved in Fusarium head blight and powdery mildew resistance and drought tolerance. J. Integr. Agr. 2018, 17, 368–380. [Google Scholar] [CrossRef]
- Song, N.; Hu, Z.; Li, Y.; Li, C.; Peng, F.; Yao, Y.; Peng, H.; Ni, Z.; Xie, C.; Sun, Q. Overexpression of a wheat stearoyl-ACP desaturase (SACPD) gene TaSSI2 in Arabidopsis ssi2 mutant compromise its resistance to powdery mildew. Gene 2013, 524, 220–227. [Google Scholar] [CrossRef] [PubMed]
- Vale, R.D. AAA proteins. Lords of the ring. J. Cell Biol. 2000, 150, F13–F19. [Google Scholar] [CrossRef] [PubMed]
- Yedidi, R.S.; Wendler, P.; Enenkel, C. AAA-ATPases in protein degradation. Front. Mol. Biosci. 2017, 4, 42. [Google Scholar] [CrossRef] [Green Version]
- Snider, J.; Houry, W.A. AAA+ proteins: Diversity in function, similarity in structure. Biochem. Soc. Trans. 2008, 36, 72–77. [Google Scholar] [CrossRef]
- Sugimoto, M.; Yamaguchi, Y.; Nakamura, K.; Tatsumi, Y.; Sano, H. A hypersensitive response-induced ATPase associated with various cellular activities (AAA) protein from tobacco plants. Plant Mol. Biol. 2004, 56, 973–985. [Google Scholar] [CrossRef]
- Lee, M.H.; Sano, H. Attenuation of the hypersensitive response by an ATPase associated with various cellular activities (AAA) protein through suppression of a small GTPase, ADP ribosylation factor, in tobacco plants. Plant J. 2007, 51, 127–139. [Google Scholar] [CrossRef]
- Zhang, B.; Van Aken, O.; Thatcher, L.; De Clercq, I.; Duncan, O.; Law, S.R.; Murcha, M.W.; Van der Merwe, M.; Seifi, H.S.; Carrie, C. The mitochondrial outer membrane AAA ATPase AtOM66 affects cell death and pathogen resistance in Arabidopsis thaliana. Plant J. 2014, 80, 709–727. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.; Yin, J.; Liang, S.; Liang, R.; Zhou, X.; Chen, Z.; Zhao, W.; Wang, J.; Li, W.; He, M. The multivesicular bodies (MVBs)-localized AAA ATPase LRD6-6 inhibits immunity and cell death likely through regulating MVBs-mediated vesicular trafficking in rice. Plos Genet. 2016, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fekih, R.; Tamiru, M.; Kanzaki, H.; Abe, A.; Yoshida, K.; Kanzaki, E.; Saitoh, H.; Takagi, H.; Natsume, S.; Undan, J.R. The rice (Oryza sativa L.) LESION MIMIC RESEMBLING, which encodes an AAA-type ATPase, is implicated in defense response. Mol. Genet. Genom. 2015, 290, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, G.K.; Rakwal, R.; Jwa, N.S. Rice (Oryza sativa L.) OsPR1b gene is phytohormonally regulated in close interaction with light signals. Biochem. Biophys. Res. Commun. 2000, 278, 290–298. [Google Scholar] [CrossRef] [PubMed]
- Nakashita, H.; Yoshioka, K.; Takayama, M.; Kuga, R.; Midoh, N.; Usami, R.; Horikoshi, K.; Yoneyama, K.; Yamaguchi, I. Characterization of PBZ1, a probenazole-inducible gene, in suspension-cultured rice cells. Biosci. Biotechnol. Biochem. 2001, 65, 205–208. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Jiang, J.; Oard, J.H. Structure, expression and promoter activity of two polyubiquitin genes from rice (Oryza sativa L.). Plant Sci. 2000, 156, 201–211. [Google Scholar] [CrossRef]
- Qi, M.; Yang, Y. Quantification of Magnaporthe grisea during infection of rice plants using real-time polymerase chain reaction and northern blot/phosphoimaging analyses. Phytopathology 2002, 92, 870–876. [Google Scholar] [CrossRef] [Green Version]
- Huang, Q.N.; Shi, Y.F.; Zhang, X.B.; Song, L.X.; Feng, B.H.; Wang, H.M.; Xu, X.; Li, X.H.; Guo, D.; Wu, J.L. Single base substitution in OsCDC48 is responsible for premature senescence and death phenotype in rice. J. Integ. Plant Biol. 2016, 58, 12–28. [Google Scholar] [CrossRef] [Green Version]
- Saifi, S.K.; Passricha, N.; Tuteja, R.; Tuteja, N. Stress-induced Oryza sativa RuvBL1a is DNA-independent ATPase and unwinds DNA duplex in 3′ to 5′ direction. Protoplasma 2018, 255, 669–684. [Google Scholar] [CrossRef]
- Lin, Y.; Tan, L.; Zhao, L.; Sun, X.; Sun, C. RLS3, a protein with AAA+ domain localized in chloroplast, sustains leaf longevity in rice. J. Integr. Plant Biol. 2016, 58, 971–982. [Google Scholar] [CrossRef]
- Xia, Z.; Wei, Y.; Sun, K.; Wu, J.; Wang, Y.; Wu, K. The maize AAA-type protein SKD1 confers enhanced salt and drought stress tolerance in transgenic tobacco by interacting with Lyst-interacting protein 5. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Wang, F.; Liu, J.; Chen, M.; Zhou, L.; Li, Z.; Zhao, Q.; Pan, G.; Zaidi, S.-H.-R.; Cheng, F. Involvement of abscisic acid in PSII photodamage and D1 protein turnover for light-induced premature senescence of rice flag leaves. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, T.; Ishibashi, T.; Kimura, S.; Tanaka, H.; Hashimoto, J.; Sakaguchi, K. Characterization of all the subunits of replication factor C from a higher plant, rice (Oryza sativa L.), and their relation to development. Plant Mol. Biol. 2003, 53, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Santos, L. Molecular mechanisms of the AAA proteins in plants. Adv. Agril. Food Biotechnol. 2006, 37, 1–15. [Google Scholar]
- Zhang, P.; Zhang, Y.; Sun, L.; Sinumporn, S.; Yang, Z.; Sun, B.; Xuan, D.; Li, Z.; Yu, P.; Wu, W. The rice AAA-ATPase OsFIGNL1 is essential for male meiosis. Front. Plant Sci. 2017, 8, 1639. [Google Scholar] [CrossRef]
- Shahriari, M.; Keshavaiah, C.; Scheuring, D.; Sabovljevic, A.; Pimpl, P.; Häusler, R.E.; Hülskamp, M.; Schellmann, S. The AAA-type ATPase AtSKD1 contributes to vacuolar maintenance of Arabidopsis thaliana. Plant J. 2010, 64, 71–85. [Google Scholar] [CrossRef]
- Kaplan, C.P.; Thomas, J.E.; Charlton, W.L.; Baker, A. Identification and characterisation of PEX6 orthologues from plants. Bba-Mol. Cell Res. 2001, 1539, 173–180. [Google Scholar] [CrossRef] [Green Version]
- Chung, K.; Tasaka, M. RPT2a, a 26S proteasome AAA-ATPase, is directly involved in Arabidopsis CC-NBS-LRR protein uni-1D-induced signaling pathways. Plant Cell Physiol. 2011, 52, 1657–1664. [Google Scholar] [CrossRef] [Green Version]
- Su, Y.; Hu, S.; Zhang, B.; Ye, W.; Niu, Y.; Guo, L.; Qian, Q. Characterization and fine mapping of a new early leaf senescence mutant es3 (t) in rice. Plant Growth Regul. 2017, 81, 419–431. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Ji, J.; Xu, Q.; Qi, X.; Weng, Y.; Chen, X. The major-effect quantitative trait locus Cs ARN 6.1 encodes an AAA ATP ase domain-containing protein that is associated with waterlogging stress tolerance by promoting adventitious root formation. Plant J. 2018, 93, 917–930. [Google Scholar] [CrossRef] [Green Version]
- Siebers, M.; Brands, M.; Wewer, V.; Duan, Y.; Holzl, G.; Dormann, P. Lipids in plant-microbe interactions. Bba-Mol. Cell Biol. L. 2016, 1861, 1379–1395. [Google Scholar] [CrossRef]
- Lim, G.H.; Singhal, R.; Kachroo, A.; Kachroo, P. Fatty Acid- and Lipid-Mediated Signaling in Plant Defense. Annu. Rev. Phytopathol. 2017, 55, 505–536. [Google Scholar] [CrossRef] [PubMed]
- Kachroo, A.; Venugopal, S.C.; Lapchyk, L.; Falcone, D.; Hildebrand, D.; Kachroo, P. Oleic acid levels regulated by glycerolipid metabolism modulate defense gene expression in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 5152–5157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delteil, A.; Zhang, J.; Lessard, P.; Morel, J.-B. Potential candidate genes for improving rice disease resistance. Rice 2010, 3, 56. [Google Scholar] [CrossRef]
- Haque, E.; Taniguchi, H.; Hassan, M.; Bhowmik, P.; Karim, M.R.; Śmiech, M.; Zhao, K.; Rahman, M.; Islam, T. Application of CRISPR/Cas9 genome editing technology for the improvement of crops cultivated in tropical climates: Recent progress, prospects, and challenges. Front. Plant Sci. 2018, 9, 617. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Dong, R.; Liu, L.; Hu, Z.; Li, J.; Wang, Y.; Ding, X.; Chu, Z. A novel mutant allele of SSI2 confers a better balance between disease resistance and plant growth inhibition on Arabidopsis thaliana. Bmc Plant Biol. 2016, 16, 208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mandal, M.K.; Chandra-Shekara, A.; Jeong, R.-D.; Yu, K.; Zhu, S.; Chanda, B.; Navarre, D.; Kachroo, A.; Kachroo, P. Oleic acid–dependent modulation of NITRIC OXIDE ASSOCIATED1 protein levels regulates nitric oxide–mediated defense signaling in Arabidopsis. Plant Cell 2012, 24, 1654–1674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreau, M.; Lee, G.I.; Wang, Y.; Crane, B.R.; Klessig, D.F. AtNOS/AtNOA1 is a functional Arabidopsis thaliana cGTPase and not a nitric-oxide synthase. J. Biol. Chem. 2008, 283, 32957–32967. [Google Scholar] [CrossRef] [Green Version]
- Miki, D.; Itoh, R.; Shimamoto, K. RNA silencing of single and multiple members in a gene family of rice. Plant Physiol. 2005, 138, 1903–1913. [Google Scholar] [CrossRef] [Green Version]
- Miki, D.; Shimamoto, K. Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol. 2004, 45, 490–495. [Google Scholar] [CrossRef]
- Toki, S.; Hara, N.; Ono, K.; Onodera, H.; Tagiri, A.; Oka, S.; Tanaka, H. Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J. 2006, 47, 969–976. [Google Scholar] [CrossRef]
- Yang, Y.; Qi, M.; Mei, C. Endogenous salicylic acid protects rice plants from oxidative damage caused by aging as well as biotic and abiotic stress. Plant J. 2004, 40, 909–919. [Google Scholar] [CrossRef] [PubMed]
- Inoue, H.; Hayashi, N.; Matsushita, A.; Liu, X.; Nakayama, A.; Sugano, S.; Jiang, C.-J.; Takatsuji, H. Blast resistance of CC-NB-LRR protein Pb1 is mediated by WRKY45 through protein–protein interaction. Proc. Natl. Acad. Sci. USA 2013, 110, 9577–9582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Gene Name (RAP+DB ID) | Primer Sequences (5’→3’) | References |
|---|---|---|
| OsAAA-ATPase1 Os03g0802500 | AGTGGTTGCTAGCTTCTCGT ACAACATGTGGTCAAATTATTCCA | [12] |
| OsAAA-ATPase2 Os01g0297200 | CTAGGTTCTGCGATGGACAC CTCCTTTGCAATTGTTCCAC | [12] |
| OsAAA-ATPase3 Os06g0697600 | GTTGTGATCGTGTCATGGTTGCG CAGAAAGCCACACACCATTGC | [12] |
| OsAAA-ATPase4 Os02g0697600 | TTGCCTGAACGGCCAGGTGAT CCCATGTAAGGGTAAGGATTGC | [12] |
| OsAAA-ATPase5 Os02g0706500 | GTTCCATCTCTTTGCCTGTAGC CATGCGCATCTCAGTCTTACC | [12] |
| OsAAA-ATPase6 Os07g0517600 | TCAGTGGCCTCGTCGAGTTC CTACTTGCCTGCTTCACACAT | [12] |
| OsPR1b Os01g0382000 | ACGGGCGTACGTACTGGCTA CTCGGTATGGACCGTGAAG | [23] |
| PBZ1 Os12g0555000 | GCGTTTGAGTCCGTGAGAGT TCACCCATTGATGAAGCAAA | [24] |
| Rubq1 Os06g0681400 | GGAGCTGCTGCTGTTCTAGG TTCAGACACCATCAAACCAGA | [25] |
| M. oryzae 28S rDNA | ACGAGAGGAACCGCTCATTCAGATAATT TCAGCAGATCGTAACGATAAAGCTACTC | [26] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Inoue, H.; Tang, X.; Tan, Y.; Xu, X.; Wang, C.; Jiang, C.-J. Rice OsAAA-ATPase1 is Induced during Blast Infection in a Salicylic Acid-Dependent Manner, and Promotes Blast Fungus Resistance. Int. J. Mol. Sci. 2020, 21, 1443. https://doi.org/10.3390/ijms21041443
Liu X, Inoue H, Tang X, Tan Y, Xu X, Wang C, Jiang C-J. Rice OsAAA-ATPase1 is Induced during Blast Infection in a Salicylic Acid-Dependent Manner, and Promotes Blast Fungus Resistance. International Journal of Molecular Sciences. 2020; 21(4):1443. https://doi.org/10.3390/ijms21041443
Chicago/Turabian StyleLiu, Xinqiong, Haruhiko Inoue, Xianying Tang, Yanping Tan, Xin Xu, Chuntai Wang, and Chang-Jie Jiang. 2020. "Rice OsAAA-ATPase1 is Induced during Blast Infection in a Salicylic Acid-Dependent Manner, and Promotes Blast Fungus Resistance" International Journal of Molecular Sciences 21, no. 4: 1443. https://doi.org/10.3390/ijms21041443





