A High Resolution Mass Spectrometry Study Reveals the Potential of Disulfide Formation in Human Mitochondrial Voltage-Dependent Anion Selective Channel Isoforms (hVDACs)
Abstract
:1. Introduction
2. Results
2.1. Mass Spectrometry Analysis of Human VDAC1
2.2. Mass Spectrometry Analysis of Human VDAC2
2.3. Mass Spectrometry Analysis of Human VDAC3
3. Discussion
3.1. Oxidation States of Methionines in Human and Rat VDAC Isoforms
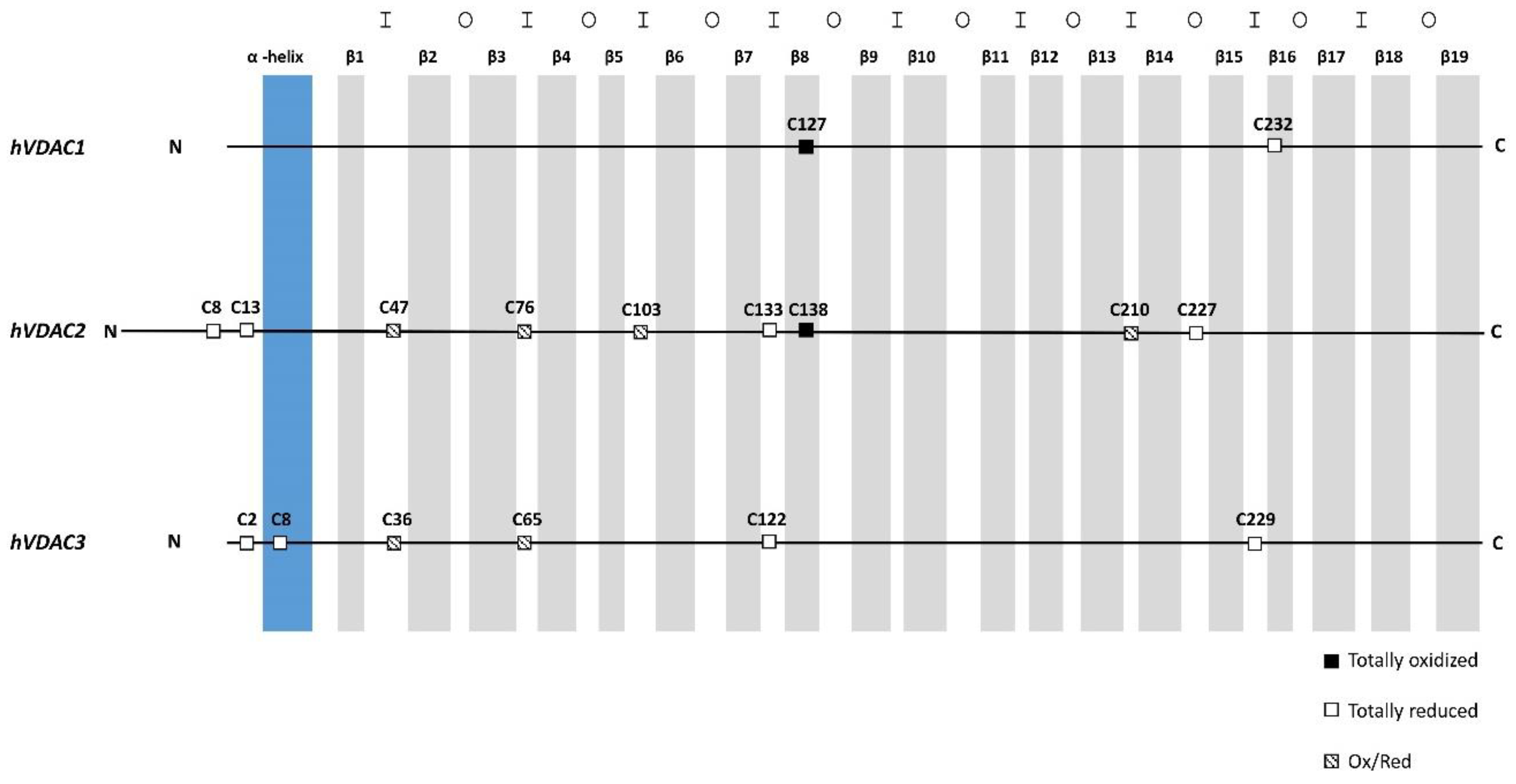
3.2. Oxidation States of Cysteines in Human and Rat VDAC Isoforms
3.3. Sulfur Oxidation State of VDAC Isoforms is Conserved Between Rat and Human, and Between Tissues and Cultured Cells
4. Materials and Methods
4.1. Chemicals
4.2. Extraction of Proteins from Human HAP1 Cell Culture Mitochondria Under Reducing Condition
4.3. Liquid Chromatography and Tandem Mass Spectrometry (LC–MS/MS) Analysis
4.4. Database Search
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| PBS | phosphate buffered saline |
| EGTA | ethylene glycol tetraacetic acid |
| EDTA | ethylenediaminetetraacetic acid |
| FA | formic acid |
| HEPES | 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid |
| DTT | dithiothreitol |
| IAA | iodoacetamide |
| FBS | fetal bovine serum |
| IMDM | Iscove’s Modified Dulbecco’s Medium |
References
- Reina, S.; De Pinto, V. Anti-cancer compounds targeted to VDAC: Potential and perspectives. Curr. Med. Chem. 2017, 24, 4447–4469. [Google Scholar] [CrossRef] [PubMed]
- Magrì, A.; Belfiore, R.; Reina, S.; Tomasello, M.F.; Di Rosa, M.C.; Guarino, F.; Leggio, L.; De Pinto, V.; Messina, A. Hexokinase I N-terminal based peptide prevents the VDAC1-SOD1 G93A interaction and re-establishes ALS cell viability. Sci. Rep. 2016, 6, 1–14. [Google Scholar] [CrossRef]
- Messina, A.; Reina, S.; Guarino, F.; De Pinto, V. VDAC isoforms in mammals. Biochim. Biophys. Acta 2012, 1818, 1466–1476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blachly-Dyson, E.; Zambronicz, E.B.; Yu, W.H.; Adams, V.; McCabe, E.R.; Adelman, J.; Colombini, M.; Forte, M. Cloning and functional expression in yeast of two human isoforms of the outer mitochondrial membrane channel, the voltage-dependent anion channel. J. Biol. Chem. 1993, 268, 1835–1841. [Google Scholar] [PubMed]
- Kleene, R.; Pfanner, N.; Pfaller, R.; Link, T.A.; Sebald, W.; Neupert, W.; Tropschug, M. Mitochondrial porin of Neurospora crassa: cDNA cloning, in vitro expression and import into mitochondria. Embo J. 1987, 6, 2627–2633. [Google Scholar] [CrossRef]
- Messina, A.; Neri, M.; Perosa, F.; Caggese, C.; Marino, M.; Caizzi, R.; De Pinto, V. Cloning and chromosomal localization of a cDNA encoding a mitochondrial porin from Drosophila melanogaster. FEBS Lett. 1996, 384, 9–13. [Google Scholar] [CrossRef] [Green Version]
- Troll, H.; Malchow, D.; Muller-Taubenberger, A.; Humbel, B.; Lottspeich, F.; Ecke, M.; Gerisch, G.; Schmid, A.; Benz, R. Purification, functional characterization, and cDNA sequencing of mitochondrial porin from Dictyostelium discoideum. J. Biol. Chem. 1992, 267, 21072–21079. [Google Scholar]
- Menzel, V.A.; Cassarà, M.C.; Benz, R.; De Pinto, V.; Messina, A.; Cunsolo, V.; Saletti, R.; Hinsch, K.D.; Hinsch, E. Molecular and functional characterization of VDAC2 purified from mammal spermatozoa. Biosci. Rep. 2009, 29, 351–362. [Google Scholar] [CrossRef] [Green Version]
- Reina, S.; Checchetto, V.; Saletti, R.; Gupta, A.; Chaturvedi, D.; Guardiani, C.; Guarino, F.; Scorciapino, M.A.; Magrì, A.; Foti, S.; et al. VDAC3 as a sensor of oxidative state of the intermembrane space of mitochondria: The putative role of cysteine residue modifications. Oncotarget 2016, 7, 2249–2268. [Google Scholar] [CrossRef] [Green Version]
- Saletti, R.; Reina, S.; Pittalà, M.G.G.; Belfiore, R.; Cunsolo, V.; Messina, A.; De Pinto, V.; Foti, S. High resolution mass spectrometry characterization of the oxidation pattern of methionine and cysteine residues in rat liver mitochondria Voltage-Dependent Anion selective Channel 3 (VDAC3). Biochim. Biophys. Acta Biomembr. 2017, 1859, 301–311. [Google Scholar] [CrossRef]
- Saletti, R.; Reina, S.; Pittalà, M.G.G.; Magrì, A.; Cunsolo, V.; Foti, S.; De Pinto, V. Post-translational modifications of VDAC1 and VDAC2 cysteines from rat liver mitochondria. Biochim. Biophys. Acta Bioenerg. 2018, 1859, 806–816. [Google Scholar] [CrossRef] [PubMed]
- Guan, Z.Q.; Yates, N.A.; Bakhtiar, R. Detection and characterization of methionine oxidation in peptides by collision-induced dissociation and electron capture dissociation. J. Am. Soc. Mass Spectrom. 2003, 14, 605–613. [Google Scholar] [CrossRef] [Green Version]
- Gupta, A.; Mahalakshmi, R. Helix–strand interaction regulates stability and aggregation of the human mitochondrial membrane protein channel VDAC3. J. Gen. Physiol. 2019, 151, 489–504. [Google Scholar] [CrossRef] [PubMed]
- Stadtman, E.R.; Moskovitz, J.; Levine, R.L. Oxidation of methionine residues of proteins: Biological consequences. Antioxid. Redox Signal. 2003, 5, 577–582. [Google Scholar] [CrossRef] [PubMed]
- Stadtman, E.R. Protein oxidation and aging. Free Radic. Res. 2006, 40, 1250–1258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ugarte, N.; Petropoulos, I.; Friguet, B. Oxidized mitochondrial protein degradation and repair in aging and oxidative stress. Antioxid. Redox Signal. 2010, 13, 539–549. [Google Scholar] [CrossRef]
- Levine, R.L.; Mosoni, L.; Berlett, B.S.; Stadtman, E.R. Methionine residues as endogenous antioxidants in proteins. Proc. Natl. Acad. Sci. USA 1996, 93, 15036–15040. [Google Scholar] [CrossRef] [Green Version]
- Stadtman, E.R.; Moskovitz, J.; Berlett, B.S.; Levine, R.L. Cyclic oxidation and reduction of protein methionine residues is an important antioxidant mechanism. Mol. Cell Biochem. 2002, 234, 3–9. [Google Scholar] [CrossRef]
- Bachi, A.; Dalle-Donne, I.; Scaloni, A. Redox proteomics: Chemical principles, methodological approaches and biological/biomedical promises. Chem. Rev. 2013, 113, 596–698. [Google Scholar] [CrossRef]
- Zaid, H.; Abu-Hamad, S.; Israelson, A.; Nathan, I.; Shoshan-Barmatz, V. The voltage-dependent anion channel-1 modulates apoptotic cell death. Cell Death Differ. 2005, 12, 751–760. [Google Scholar] [CrossRef]
- Abu-Hamad, S.; Zaid, H.; Israelson, A.; Nahon, E.; Shoshan-Barmatz, V. Hexokinase-I protection against apoptotic cell death is mediated via interaction with the voltage-dependent anion channel-1: Mapping the site of binding. J. Biol. Chem. 2008, 283, 13482–13490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bayrhuber, M.; Meins, T.; Habeck, M.; Becker, S.; Giller, K.; Villinger, S.; Vonrhein, C.; Griesinger, C.; Zweckstetter, M.; Zeth, K. Structure of the human voltage-dependent anion channel. Proc. Natl. Acad. Sci. USA 2008, 105, 15370–15375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mader, A.; Abu-Hamad, S.; Arbel, N.; Gutiérrez-Aguilar, M.; Shoshan-Barmatz, V. Dominant-negative VDAC1 mutants reveal oligomeric VDAC1 to be the active unit in mitochondria-mediated apoptosis. Biochem. J. 2010, 429, 147–155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hiller, S.; Garces, R.G.; Malia, T.J.; Orekhov, V.Y.; Colombini, M.; Wagner, G. Solution structure of the integral human membrane protein VDAC-1 in detergent micelles. Science 2008, 321, 1206–1210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ujwal, R.; Cascio, D.; Colletier, J.P.; Faham, S.; Zhang, J.; Toro, L.; Ping, P.; Abramson, J. The crystal structure of mouse VDAC1 at 2.3 angstrom resolution reveals mechanistic insights into metabolite gating. Proc. Natl. Acad. Sci. USA 2008, 105, 17742–17747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schindler, A.; Foley, E. Hexokinase 1 blocks apoptotic signals at the mitochondria. Cell. Signal. 2013, 25, 2685–2692. [Google Scholar] [CrossRef]
- De Pinto, V.; Benz, R.; Caggese, C.; Palmieri, F. Characterization of the mitochondrial porin from Drosophila melanogaster. Biochim. Biophys. Acta 1989, 905, 499–502. [Google Scholar] [CrossRef]
- Bizcaino, J.A.; Cote, R.G.; Csordas, A.; Dianes, J.A.; Fabregat, A.; Foster, J.M.; Griss, J.; Alpi, E.; Birim, M.; Contell, J.; et al. The Proteomics Identifications (PRIDE) database and associated tools: Status in 2013. Nucleic Acids Res. 2013, 41, D1063–D1069. [Google Scholar] [CrossRef]


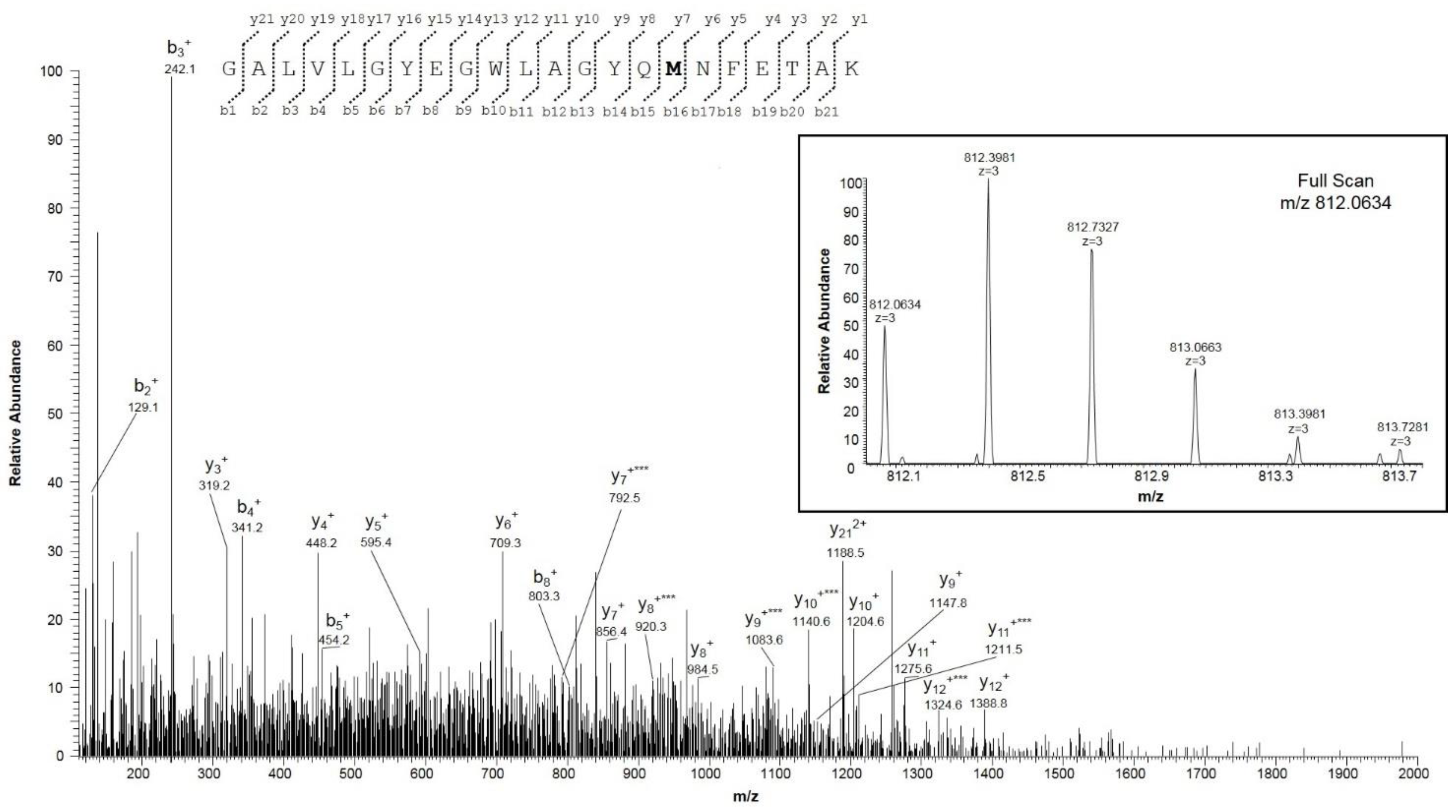


| Peptide | Position in the Sequence | Measured Monoisotopic m/z | Absolute Intensity | Ratio Ox/Red |
|---|---|---|---|---|
| EHINLGCDM129DFDIAGPSIR | 121–139 | 1083.9756 (+2) | 1.6 × 105 | 1.5 |
| EHINLGCDMDFDIAGPSIR | 1075.9784 (+2) | 1.08 × 105 | ||
| GALVLGYEGWLAGYQM155NFETAK | 140–161 | 812.0634 (+3) | 6.4 × 105 | 0.8 |
| GALVLGYEGWLAGYQMNFETAK | 1209.5944 (+2) | 8.5 × 105 |
| Peptide | Position in the Sequence | Measured Monoisotopic m/z | Absolute Intensity | Ratio Ox/Red |
|---|---|---|---|---|
| PMCIPPSYADLGK | 11–23 | 732.8476 (+2) | 8.79 × 105 | 9.8 |
| PMCIPPSYADLGK | 724.8497 (+2) | 8.99 × 104 | ||
| SCSGVEFSTSGSSNTDTGK | 46–64 | 949.8815 (+2) | 1.80 × 104 | 0.01 |
| SCSGVEFSTSGSSNTDTGK | 954.4005 (+2) | 1.59 × 106 | ||
| YKWCEYGLTFTEK | 73–85 | 858.3821 (+2) | 1.53 × 104 | 0.1 |
| YKWCEYGLTFTEK | 862.9037 (+2) | 1.38 × 105 | ||
| WNTDNTLGTEIAIEDQICQGLK | 86–107 | 837.3958 (+3) | 1.09 × 105 | 0.04 |
| WNTDNTLGTEIAIEDQICQGLK | 1260.1086 (+2) | 2.98 × 106 | ||
| ECINLGCDVDFDFAGPAIHGSAVFGYEGW LAGYQMTFDSAKa | 132–172 | 1508.3225 (+3) | 2.94 × 104 | 1.4 |
| ECINLGCDVDFDFAGPAIHGSAVFGYEGW LAGYQMTFDSAKa | 1502.9921 (+3) | 2.12 × 104 | ||
| VCEDLDTSVNLAWTSGTNCTR | 209–229 | 1195.5161 (+2) | 4.40 × 104 | 0.1 |
| VCEDLDTSVNLAWTSGTNCTR | 800.3594 (+3) | 4.40 × 105 |
| Peptide | Position in the Sequence | Measured Monoisotopic m/z | Absolute Intensity | Ratio Ox/Red |
|---|---|---|---|---|
| GYGFGMVK | 21–28 | 437.7098 (+2) | 1.46 × 104 | 0.06 |
| GYGFGMVK | 429.7118 (+2) | 2.51 × 105 | ||
| SCSGVEFSTSGHAYTDTGK | 35–53 | 991.4069 (+2) | 1.81 × 104 | 0.2 |
| SCSGVEFSTSGHAYTDTGK | 995.9267 (+2) | 8.80 × 104 | ||
| YKVCNYGLTFTQK | 62–74 | 806.8889 (+2) | 1.01 × 104 | 0.05 |
| YKVCNYGLTFTQK | 811.4081 (+2) | 1.97 × 105 | ||
| DCFSVGSNVDIDFSGPTIYGWAVLAFE GWLAGYQMSFDTAK | 121–161 | 1513.3688 (+3) | 1.74 × 104 | 1.1 |
| DCFSVGSNVDIDFSGPTIYGWAVLAFE GWLAGYQMSFDTAK | 1508.0270 (+3) | 1.59 × 104 | ||
| YMLDCR | 225–230 | 437.1832 (+2) | 5.88 × 105 | 3.2 |
| YMLDCR | 429.1861 (+2) | 1.86 × 105 |
| Cysteine Position | Oxidative State | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| VDAC1 | VDAC2 | VDAC3 | VDAC1 | VDAC2 | VDAC3 | ||||||
| rat | hu | rat | hu | rat | hu | rat | hu | rat | hu | rat | hu |
| 4 | t.r. | ||||||||||
| 5 | t.r. | ||||||||||
| 9 | 8 | 2 | 2 | - | t.r. | t.r. | t.r. | t.r. | |||
| 14 | 13 | 8 | 8 | - | t.r. | t.r. | t.r. | t.r. | |||
| 48 | 47 | 36 | 36 | - | p.o. | p.o. | p.o. | p.o. | |||
| 77 | 76 | 65 | 65 | - | p.o. | p.o. | p.o. | p.o. | |||
| 104 | 103 | - | - | p.o. | p.o. | - | |||||
| 134 | 133 | 122 | 122 | - | N.D. | t.r. | N.D. | t.r. | |||
| 127 | 127 | 139 | 138 | - | p.o. | t.o. | N.D. | t.o. | - | ||
| 165 | p.o. | ||||||||||
| 211 | 210 | - | - | p.o. | p.o. | - | |||||
| 228 | 227 | - | - | p.o. | t.r. | - | |||||
| 229 | 229 | - | - | t.o. | t.r. | ||||||
| 232 | 232 | - | p.o. | t.r. | - | - | |||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pittalà, M.G.G.; Saletti, R.; Reina, S.; Cunsolo, V.; De Pinto, V.; Foti, S. A High Resolution Mass Spectrometry Study Reveals the Potential of Disulfide Formation in Human Mitochondrial Voltage-Dependent Anion Selective Channel Isoforms (hVDACs). Int. J. Mol. Sci. 2020, 21, 1468. https://doi.org/10.3390/ijms21041468
Pittalà MGG, Saletti R, Reina S, Cunsolo V, De Pinto V, Foti S. A High Resolution Mass Spectrometry Study Reveals the Potential of Disulfide Formation in Human Mitochondrial Voltage-Dependent Anion Selective Channel Isoforms (hVDACs). International Journal of Molecular Sciences. 2020; 21(4):1468. https://doi.org/10.3390/ijms21041468
Chicago/Turabian StylePittalà, Maria G. G., Rosaria Saletti, Simona Reina, Vincenzo Cunsolo, Vito De Pinto, and Salvatore Foti. 2020. "A High Resolution Mass Spectrometry Study Reveals the Potential of Disulfide Formation in Human Mitochondrial Voltage-Dependent Anion Selective Channel Isoforms (hVDACs)" International Journal of Molecular Sciences 21, no. 4: 1468. https://doi.org/10.3390/ijms21041468






