Muscle Hyperplasia in Japanese Quail by Single Amino Acid Deletion in MSTN Propeptide
Abstract
:1. Introduction
2. Results
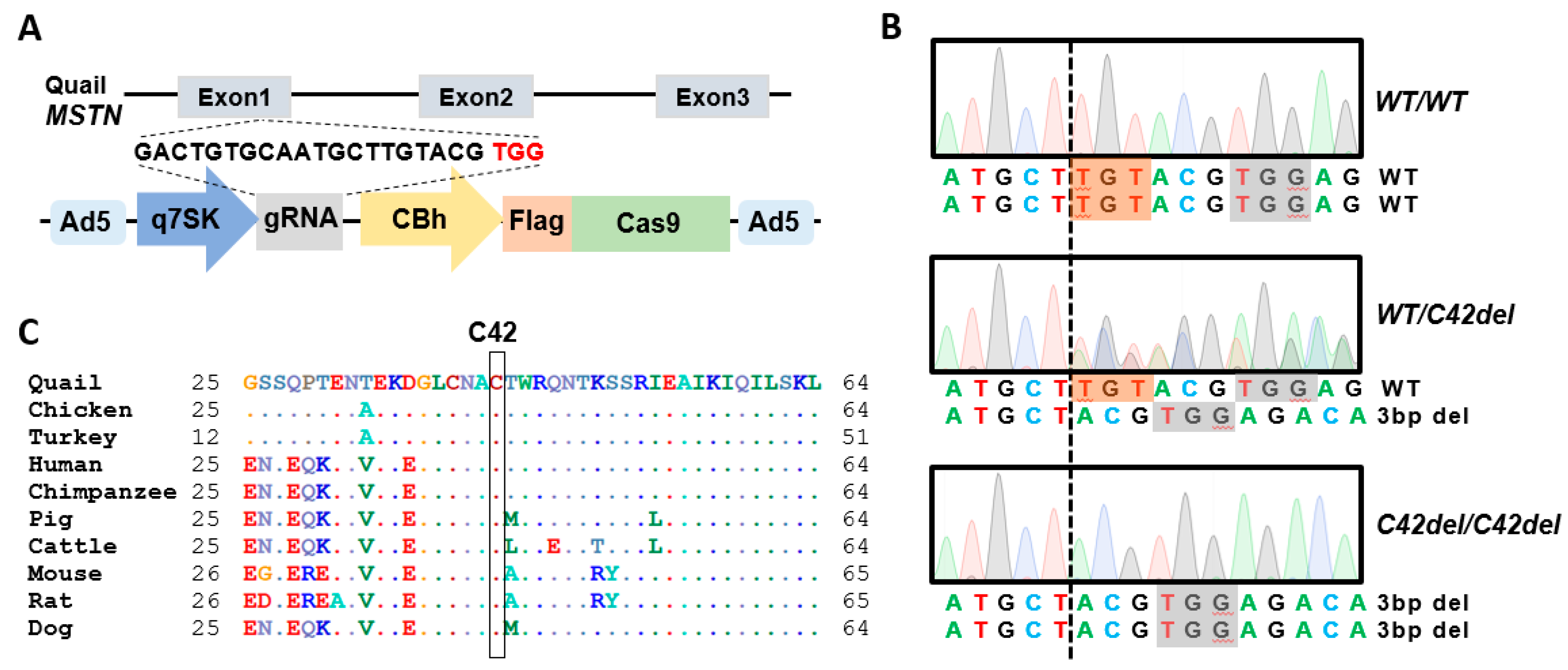
2.1. Generation of MSTN Mutation in Quail Using the Adenovirus-Mediated Method
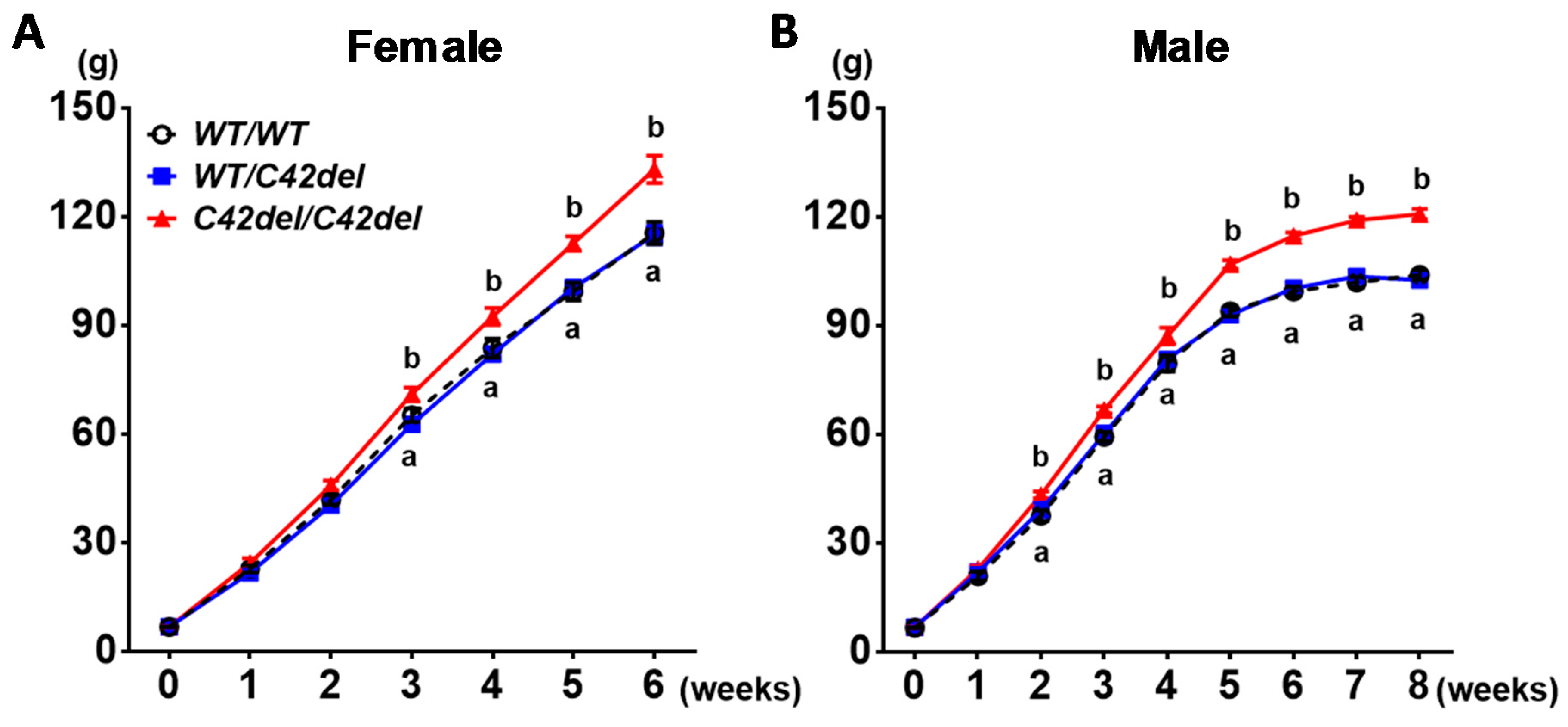
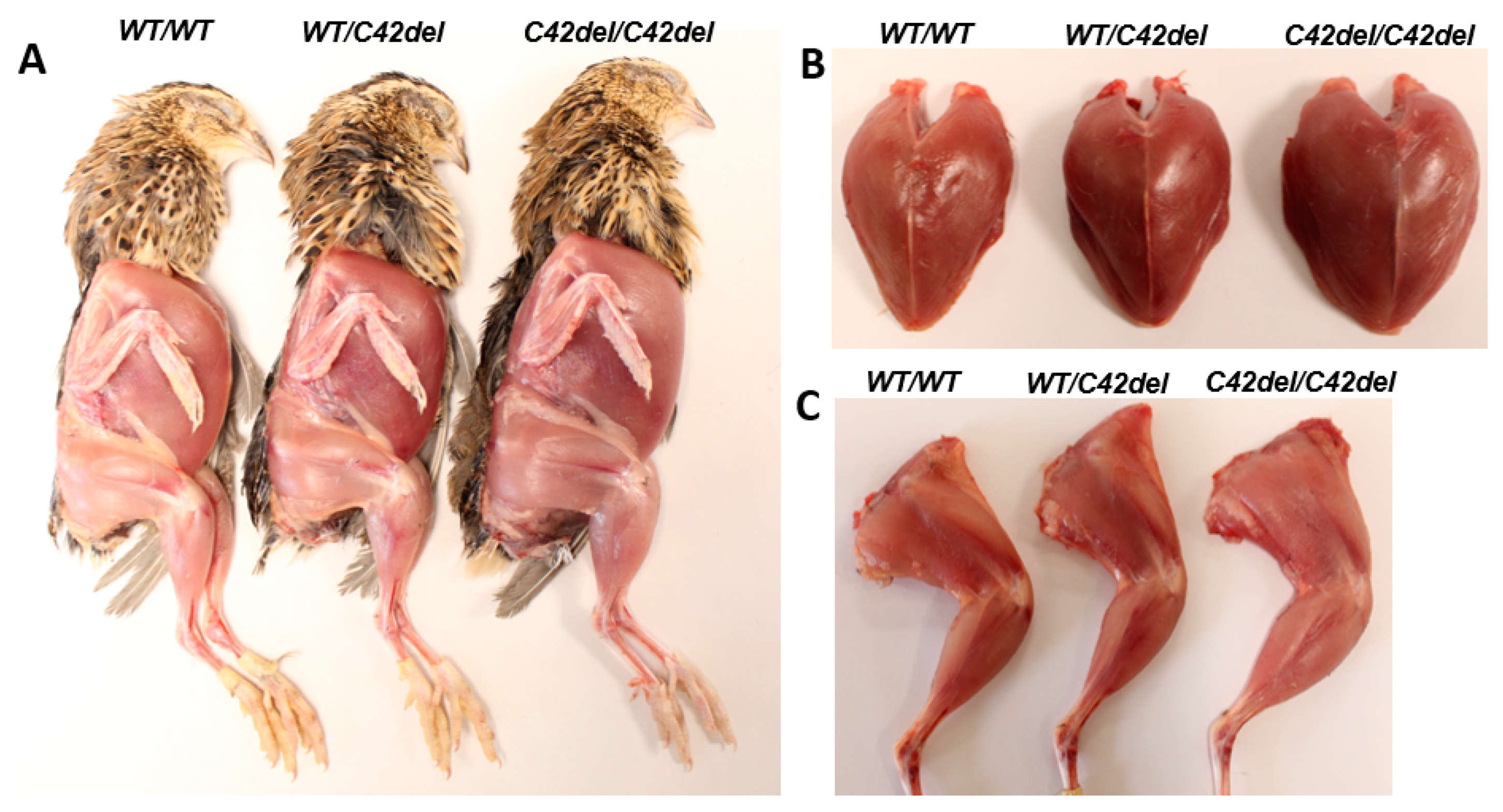
2.2. Positive Effect of MSTN C42del Mutaiton in Quail Muscle Growth
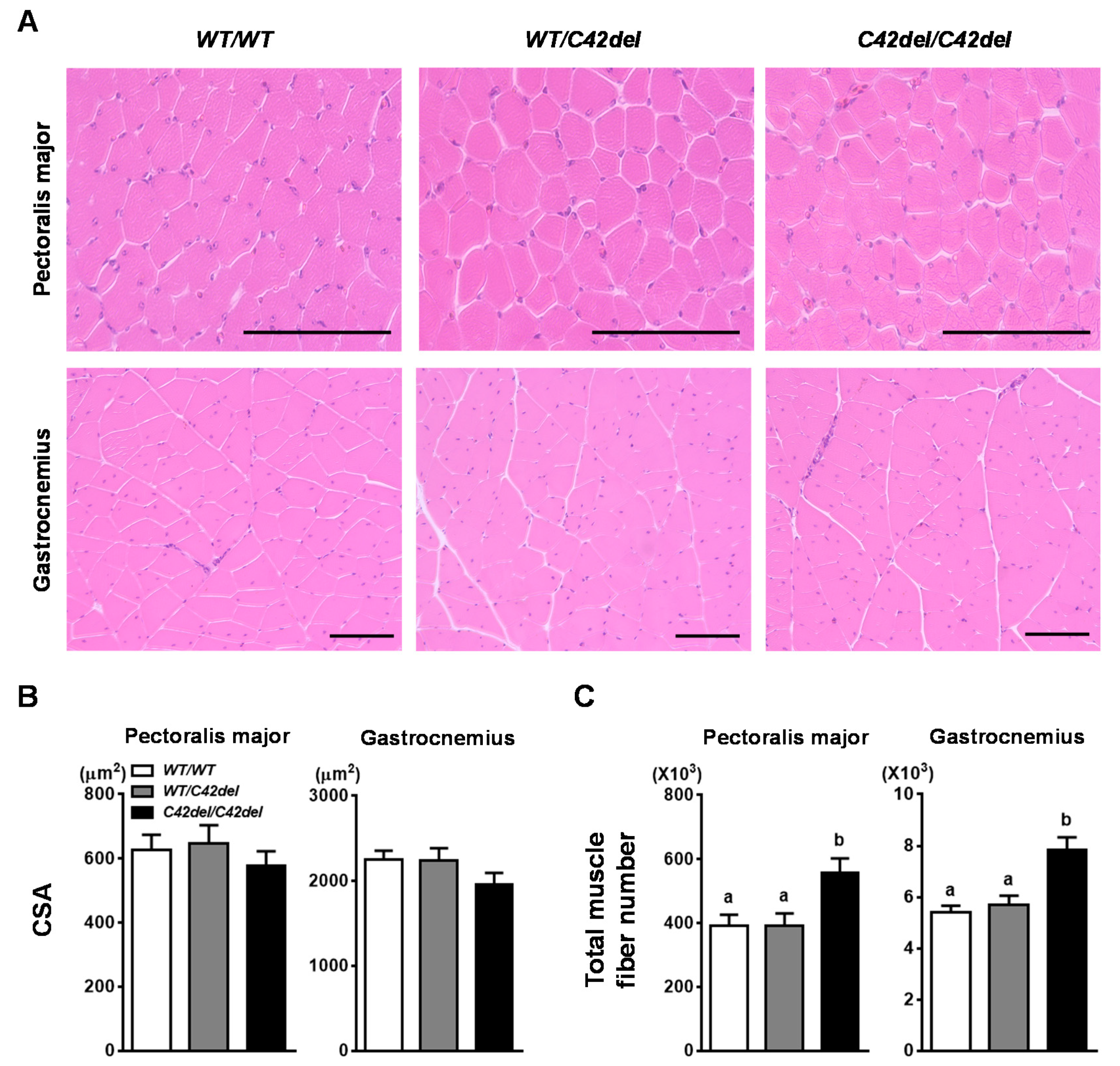
2.3. Muscle Fiber Hyperplasia in C42del/C42del Quail
3. Discussion
4. Materials and Methods
4.1. Animal Care
4.2. Construction and Injection of Adenoviral Vector
4.3. Production of Quail with MSTN Mutation
4.4. Analysis of Off-Target Mutation
4.5. Tissue Sampling
4.6. Histological Processing and Measurement of Muscle Fiber Number and Size
4.7. Statistical Analyses
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ACVR2B | activin receptor type 2B |
| C42del | cysteine 42 deletion |
| CSA | cross-sectional area |
| MSTN | myostatin |
| gRNA | guide RNA |
| PAM | protospacer adjacent motif |
| PCR | polymerase chain reaction |
| PGC | primordial germ cell |
| PM | pectoralis major |
| SEM | standard error of the mean |
| TGF-β | transforming growth factor-β |
| WT | wild-type |
Appendix A
| Quail | Chromosome | Locus | Score | Sequence | PAM | Direction |
|---|---|---|---|---|---|---|
| MSTN | 7 | 4,829,332 | 46.1 | GACTGTGCAATGCTTGTACG | TGG | + |
| Off-target 1 | 1 | 125,916,261 | 30.2 | TCCTAAGAAATGCTTGTACG | TGG | + |
| Off-target 2 | 7 | 7,649,801 | 28.2 | AATGCTGCAATGCTTGGACG | TGG | + |
| Off-target 3 | 5 | 424,027 | 28.2 | GTCTCTTAGATGCTTGTACG | TGG | − |
| Off-target 4 | 1 | 105,890,272 | 28.2 | AATGGACACATGCTTGTACG | GGG | + |
| Off-target 5 | 4 | 22,457,528 | 28.2 | AGTCACTCTATGCTTGTACG | GGG | + |
| Off-target 6 | 1 | 55,771,184 | 28.2 | TGGTATGCAATGCCTGTACG | CGG | − |
| Purpose | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| MSTN | GCATGGACGAGCTGTACAAGTA | CCCTGCTAATGTTAGGTGCTT |
| Off-target 1 | CGCACTATGGAATGGCAAGATTT | TCTCCCTCAATCTTAGTACTGCTT |
| Off-target 2 | AGACCTTCTGCATACTGCCTT | CTTCAGAACTTGCAGGTTTGCTA |
| Off-target 3 | TGTGTTCAACTGCTCAGAAGGAA | GTGGGAAGTTCCAGACAAGTT |
| Off-target 4 | ATGGGAAGAACTGCTACTGGAA | AAGAGGCTTCCTGTGCTTCT |
| Off-target 5 | CACTGAGGAAGTTTGTCTTGGAGTTA | TGGCTGAAAGATCTTATCTTCACTCA |
| Off-target 6 | CTGTCTCTGTGTCCAGATCAGAT | AGAGGAGCCTCATGTTGGAA |
References
- Hunton, P. 100 Years of poultry genetics. Worlds Poult. Sci. J. 2006, 62, 417–428. [Google Scholar] [CrossRef]
- McPherron, A.C.; Lawler, A.M.; Lee, S.J. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature 1997, 387, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Schuelke, M.; Wagner, K.R.; Stolz, L.E.; Hubner, C.; Riebel, T.; Komen, W.; Braun, T.; Tobin, J.F.; Lee, S.J. Myostatin mutation associated with gross muscle hypertrophy in a child. N. Engl. J. Med. 2004, 350, 2682–2688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grobet, L.; Martin, L.J.; Poncelet, D.; Pirottin, D.; Brouwers, B.; Riquet, J.; Schoeberlein, A.; Dunner, S.; Ménissier, F.; Massabanda, J.; et al. A deletion in the bovine myostatin gene causes the double-muscled phenotype in cattle. Nat. Genet. 1997, 17, 71–74. [Google Scholar] [CrossRef] [PubMed]
- Kambadur, R.; Sharma, M.; Smith, T.P.; Bass, J.J. Mutations in myostatin (GDF8) in double-muscled Belgian Blue and Piedmontese cattle. Genome Res. 1997, 7, 910–916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mosher, D.S.; Quignon, P.; Bustamante, C.D.; Sutter, N.B.; Mellersh, C.S.; Parker, H.G.; Ostrander, E.A. A mutation in the myostatin gene increases muscle mass and enhances racing performance in heterozygote dogs. PLoS Genet. 2007, 3, e79. [Google Scholar] [CrossRef]
- Bi, Y.; Hua, Z.; Liu, X.; Hua, W.; Ren, H.; Xiao, H.; Zhang, L.; Li, L.; Wang, Z.; Laible, G.; et al. Isozygous and selectable marker-free MSTN knockout cloned pigs generated by the combined use of CRISPR/Cas9 and Cre/LoxP. Sci. Rep. 2016, 6, 31729. [Google Scholar] [CrossRef]
- He, Z.; Zhang, T.; Jiang, L.; Zhou, M.; Wu, D.; Mei, J.; Cheng, Y. Use of CRISPR/Cas9 technology efficiently targetted goat myostatin through zygotes microinjection resulting in double-muscled phenotype in goats. Biosci. Rep. 2018, 38. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Niu, Y.; Zhou, J.; Yu, H.; Kou, Q.; Lei, A.; Zhao, X.; Yan, H.; Cai, B.; Shen, Q.; et al. Multiplex gene editing via CRISPR/Cas9 exhibits desirable muscle hypertrophy without detectable off-target effects in sheep. Sci. Rep. 2016, 6, 32271. [Google Scholar] [CrossRef]
- Lv, Q.; Yuan, L.; Deng, J.; Chen, M.; Wang, Y.; Zeng, J.; Li, Z.; Lai, L. Efficient Generation of Myostatin Gene Mutated Rabbit by CRISPR/Cas9. Sci. Rep. 2016, 6, 25029. [Google Scholar] [CrossRef] [Green Version]
- Chisada, S.; Okamoto, H.; Taniguchi, Y.; Kimori, Y.; Toyoda, A.; Sakaki, Y.; Takeda, S.; Yoshiura, Y. Myostatin-deficient medaka exhibit a double-muscling phenotype with hyperplasia and hypertrophy, which occur sequentially during post-hatch development. Dev. Biol. 2011, 359, 82–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Chen, Y.L.; Bian, W.P.; Xie, S.L.; Qi, G.L.; Liu, L.; Strauss, P.R.; Zou, J.X.; Pei, D.S. Deletion of mstna and mstnb impairs the immune system and affects growth performance in zebrafish. Fish Shellfish Immunol. 2018, 72, 572–580. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Ge, J.; Xu, Z.; Dong, X.; Cao, S.; Pan, J.; Zhao, Q. Generation of myostatin B knockout yellow catfish (Tachysurus fulvidraco) using transcription activator-like effector nucleases. Zebrafish 2014, 11, 265–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McFarlane, C.; Langley, B.; Thomas, M.; Hennebry, A.; Plummer, E.; Nicholas, G.; McMahon, C.; Sharma, M.; Kambadur, R. Proteolytic processing of myostatin is auto-regulated during myogenesis. Dev. Biol. 2005, 283, 58–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.J.; McPherron, A.C. Regulation of myostatin activity and muscle growth. Proc. Natl. Acad. Sci. USA 2001, 98, 9306–9311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolfman, N.M.; McPherron, A.C.; Pappano, W.N.; Davies, M.V.; Song, K.; Tomkinson, K.N.; Wright, J.F.; Zhao, L.; Sebald, S.M.; Greenspan, D.S.; et al. Activation of latent myostatin by the BMP-1/tolloid family of metalloproteinases. Proc. Natl. Acad. Sci. USA 2003, 100, 15842–15846. [Google Scholar] [CrossRef] [Green Version]
- Trendelenburg, A.U.; Meyer, A.; Rohner, D.; Boyle, J.; Hatakeyama, S.; Glass, D.J. Myostatin reduces Akt/TORC1/p70S6K signaling, inhibiting myoblast differentiation and myotube size. Am. J. Physiol. Cell Physiol. 2009, 296, C1258–C1270. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.J.; Reed, L.A.; Davies, M.V.; Girgenrath, S.; Goad, M.E.; Tomkinson, K.N.; Wright, J.F.; Barker, C.; Ehrmantraut, G.; Holmstrom, J.; et al. Regulation of muscle growth by multiple ligands signaling through activin type II receptors. Proc. Natl. Acad. Sci. USA 2005, 102, 18117–18122. [Google Scholar] [CrossRef] [Green Version]
- Kocsis, T.; Trencsenyi, G.; Szabo, K.; Baan, J.A.; Muller, G.; Mendler, L.; Garai, I.; Reinauer, H.; Deak, F.; Dux, L.; et al. Myostatin propeptide mutation of the hypermuscular Compact mice decreases the formation of myostatin and improves insulin sensitivity. Am. J. Physiol. Endocrinol. Metab. 2017, 312, E150–E160. [Google Scholar] [CrossRef] [Green Version]
- Shin, S.; Song, Y.; Ahn, J.; Kim, E.; Chen, P.; Yang, S.; Suh, Y.; Lee, K. A novel mechanism of myostatin regulation by its alternative splicing variant during myogenesis in avian species. Am. J. Physiol. Cell Physiol. 2015, 309, C650–C659. [Google Scholar] [CrossRef]
- Chen, P.R.; Suh, Y.; Shin, S.; Woodfint, R.M.; Hwang, S.; Lee, K. Exogenous Expression of an Alternative Splicing Variant of Myostatin Prompts Leg Muscle Fiber Hyperplasia in Japanese Quail. Int. J. Mol. Sci. 2019, 20, 4617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhattacharya, T.K.; Shukla, R.; Chatterjee, R.N.; Bhanja, S.K. Comparative analysis of silencing expression of myostatin (MSTN) and its two receptors (ACVR2A and ACVR2B) genes affecting growth traits in knock down chicken. Sci. Rep. 2019, 9, 7789. [Google Scholar] [CrossRef] [PubMed]
- Park, T.S.; Lee, H.J.; Kim, K.H.; Kim, J.S.; Han, J.Y. Targeted gene knockout in chickens mediated by TALENs. Proc. Natl. Acad. Sci. USA 2014, 111, 12716–12721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oishi, I.; Yoshii, K.; Miyahara, D.; Kagami, H.; Tagami, T. Targeted mutagenesis in chicken using CRISPR/Cas9 system. Sci. Rep. 2016, 6, 23980. [Google Scholar] [CrossRef] [Green Version]
- Park, T.S.; Park, J.; Lee, J.H.; Park, J.W.; Park, B.C. Disruption of G0/G1 switch gene 2 (G0S2) reduced abdominal fat deposition and altered fatty acid composition in chicken. FASEB J. 2019, 33, 1188–1198. [Google Scholar] [CrossRef]
- Dimitrov, L.; Pedersen, D.; Ching, K.H.; Yi, H.; Collarini, E.J.; Izquierdo, S.; van de Lavoir, M.C.; Leighton, P.A. Germline Gene Editing in Chickens by Efficient CRISPR-Mediated Homologous Recombination in Primordial Germ Cells. PLoS ONE 2016, 11, e0154303. [Google Scholar] [CrossRef]
- Oishi, I.; Yoshii, K.; Miyahara, D.; Tagami, T. Efficient production of human interferon beta in the white of eggs from ovalbumin gene-targeted hens. Sci. Rep. 2018, 8, 10203. [Google Scholar] [CrossRef]
- Lee, H.J.; Yoon, J.W.; Jung, K.M.; Kim, Y.M.; Park, J.S.; Lee, K.Y.; Park, K.J.; Hwang, Y.S.; Park, Y.H.; Rengaraj, D.; et al. Targeted gene insertion into Z chromosome of chicken primordial germ cells for avian sexing model development. FASEB J. 2019, 33, 8519–8529. [Google Scholar] [CrossRef]
- Lee, J.; Ma, J.; Lee, K. Direct delivery of adenoviral CRISPR/Cas9 vector into the blastoderm for generation of targeted gene knockout in quail. Proc. Natl. Acad. Sci. USA 2019, 116, 13288–13292. [Google Scholar] [CrossRef] [Green Version]
- Ahn, J.; Lee, J.; Park, J.Y.; Oh, K.B.; Hwang, S.; Lee, C.W.; Lee, K. Targeted genome editing in a quail cell line using a customized CRISPR/Cas9 system. Poult. Sci. 2017, 96, 1445–1450. [Google Scholar] [CrossRef]
- Yang, S.; Suh, Y.; Choi, Y.M.; Shin, S.; Han, J.Y.; Lee, K. Loss of fat with increased adipose triglyceride lipase-mediated lipolysis in adipose tissue during laying stages in quail. Lipids 2013, 48, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Li, E.J.; Wall, R.J.; Yang, J. Coordinated patterns of gene expressions for adult muscle build-up in transgenic mice expressing myostatin propeptide. BMC Genomics 2009, 10, 305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.J. Quadrupling muscle mass in mice by targeting TGF-beta signaling pathways. PLoS ONE 2007, 2, e789. [Google Scholar] [CrossRef] [PubMed]
- Oldham, J.M.; Martyn, J.A.; Sharma, M.; Jeanplong, F.; Kambadur, R.; Bass, J.J. Molecular expression of myostatin and MyoD is greater in double-muscled than normal-muscled cattle fetuses. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2001, 280, R1488–R1493. [Google Scholar] [CrossRef] [PubMed]
- Hennebry, A.; Berry, C.; Siriett, V.; O′Callaghan, P.; Chau, L.; Watson, T.; Sharma, M.; Kambadur, R. Myostatin regulates fiber-type composition of skeletal muscle by regulating MEF2 and MyoD gene expression. Am. J. Physiol. Cell Physiol. 2009, 296, C525–C534. [Google Scholar] [CrossRef] [PubMed]
- Fakhfakh, R.; Michaud, A.; Tremblay, J.P. Blocking the myostatin signal with a dominant negative receptor improves the success of human myoblast transplantation in dystrophic mice. Mol. Ther. 2011, 19, 204–210. [Google Scholar] [CrossRef]
- Li, H.; Wang, G.; Hao, Z.; Zhang, G.; Qing, Y.; Liu, S.; Qing, L.; Pan, W.; Chen, L.; Liu, G.; et al. Generation of biallelic knock-out sheep via gene-editing and somatic cell nuclear transfer. Sci. Rep. 2016, 22, 33675. [Google Scholar] [CrossRef] [Green Version]
- Du, W.; Zhang, Y.; Yang, J.Z.; Li, H.B.; Xia, J.; Li, N.; Zhang, J.S.; Yan, X.M.; Zhou, Z.Y. Effect of MSTN propeptide protein on the growth and development of Altay lamb muscle. Genet. Mol. Res. 2016, 15. [Google Scholar] [CrossRef]
- Kumar, R.; Singh, S.P.; Mitra, A. Short-hairpin Mediated Myostatin Knockdown Resulted in Altered Expression of Myogenic Regulatory Factors with Enhanced Myoblast Proliferation in Fetal Myoblast Cells of Goats. Anim. Biotechnol. 2018, 29, 59–67. [Google Scholar] [CrossRef]
- Li, R.; Zeng, W.; Ma, M.; Wei, Z.; Liu, H.; Liu, X.; Wang, M.; Shi, X.; Zeng, J.; Yang, L.; et al. Precise editing of myostatin signal peptide by CRISPR/Cas9 increases the muscle mass of Liang Guang Small Spotted pigs. Transgenic Res. 2020, 29, 149–163. [Google Scholar] [CrossRef]
- Rehfeldt, C.; Fiedler, I.; Dietl, G.; Ender, K. Myogenesis and postnatal skeletal muscle cell growth as influenced by selection. Livest. Prod. Sci. 2000, 66, 177–188. [Google Scholar] [CrossRef]
- Picard, B.; Lefaucheur, L.; Berri, C.; Duclos, M.J. Muscle fiber ontogenesis in farm animal species. Reprod. Nutr. Dev. 2002, 42, 415–431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castelhano-Barbosa, E.C.; Gabriel, J.E.; Alvares, L.E.; Monteiro-Vitorello, C.B.; Coutinho, L.L. Temporal and spatial expression of the myostatin gene during chicken embryo development. Growth Dev. Aging 2005, 69, 3–12. [Google Scholar]
- Wang, X.; Niu, Y.; Zhou, J.; Zhu, H.; Ma, B.; Yu, H.; Yan, H.; Hua, J.; Huang, X.; Qu, L.; et al. CRISPR/Cas9-mediated MSTN disruption and heritable mutagenesis in goats causes increased body mass. Anim. Genet. 2018, 49, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Kambadur, R.; Matthews, K.G.; Somers, W.G.; Devlin, G.P.; Conaglen, J.V.; Fowke, P.J.; Bass, J.J. Myostatin, a transforming growth factor-beta superfamily member, is expressed in heart muscle and is upregulated in cardiomyocytes after infarct. J. Cell Physiol. 1999, 180, 1–9. [Google Scholar] [CrossRef]
- McPherron, A.C.; Lee, S.J. Suppression of body fat accumulation in myostatin-deficient mice. J. Clin. Investig. 2002, 109, 595–601. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Arnold, H.B.; Della-Fera, M.A.; Azain, M.J.; Hartzell, D.L.; Baile, C.A. Myostatin knockout in mice increases myogenesis and decreases adipogenesis. Biochem. Biophys. Res. Commun. 2002, 291, 701–706. [Google Scholar] [CrossRef]
- Shahin, K.A.; Berg, R.T. Growth and distribution of individual muscles in Double Muscled and normal cattle. Can. J. Anim. Sci. 1985, 65, 279–293. [Google Scholar]
- Fiems, L.O. Double Muscling in Cattle: Genes, Husbandry, Carcasses and Meat. Animals (Basel) 2012, 2, 472–506. [Google Scholar] [CrossRef]
- Cai, C.; Qian, L.; Jiang, S.; Sun, Y.; Wang, Q.; Ma, D.; Xiao, G.; Li, B.; Xie, S.; Gao, T.; et al. Loss-of-function myostatin mutation increases insulin sensitivity and browning of white fat in Meishan pigs. Oncotarget 2017, 8, 34911–34922. [Google Scholar] [CrossRef] [Green Version]
- Jackson, M.F.; Luong, D.; Vang, D.D.; Garikipati, D.K.; Stanton, J.B.; Nelson, O.L.; Rodgers, B.D. The aging myostatin null phenotype: Reduced adiposity, cardiac hypertrophy, enhanced cardiac stress response, and sexual dimorphism. J. Endocrinol. 2012, 213, 263–275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, X.J. Gender modulates cardiac phenotype development in genetically modified mice. Cardiovasc. Res. 2004, 63, 510–519. [Google Scholar] [CrossRef] [PubMed]
- Romvári, R.; Petrási, Z.; Süto, Z.; Szabó, A.; Andrássy, G.; Garamvölgyi, R.; Horn, P. Noninvasive characterization of the turkey heart performance and its relationship to skeletal muscle volume. Poult. Sci. 2004, 83, 696–700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, S.; Choi, Y.M.; Han, J.Y.; Lee, K. Inhibition of lipolysis in the novel transgenic quail model overexpressing G0/G1 switch gene 2 in the adipose tissue during feed restriction. PLoS ONE 2014, 9, e100905. [Google Scholar] [CrossRef] [Green Version]
- Choi, Y.M.; Suh, Y.; Shin, S.; Lee, K. Skeletal muscle characterization of Japanese quail line selectively bred for lower body weight as an avian model of delayed muscle growth with hypoplasia. PLoS ONE 2014, 9, e95932. [Google Scholar] [CrossRef]
| Tissue | WT/WT | WT/C42del | C42del/C42del |
|---|---|---|---|
| Pectoralis Major (g) | 16.19 ± 0.26 a | 16.15 ± 0.500 a | 20.12 ± 0.71 b |
| Pectoralis Minor (g) | 5.56 ± 0.10 a | 5.61 ± 0.16 a | 6.92 ± 0.20 b |
| Biceps Femoris (g) | 2.14 ± 0.06 a | 2.20 ± 0.04 a | 2.68 ± 0.08 b |
| Semitendinosus (g) | 0.95 ± 0.02 a | 0.98 ± 0.03 a | 1.17 ± 0.04 b |
| Gastrocnemius (g) | 0.80 ± 0.03 a | 0.79 ± 0.02 a | 0.10 ± 0.04 b |
| Tricep Brachii (g) | 0.56 ± 0.02 a | 0.54 ± 0.03 a | 0.69 ± 0.02 b |
| Leg Fat (g) | 0.34 ± 0.03 a | 0.25 ± 0.02 ab | 0.24 ± 0.02 b |
| Abdominal Fat (g) | 0.23 ± 0.04 a | 0.19 ± 0.02 ab | 0.16 ± 0.02 b |
| Heart (g) | 0.87 ± 0.03 NS | 0.87 ± 0.03 NS | 0.87 ± 0.02 NS |
| Tissue | WT/WT | WT/C42del | C42del/C42del |
|---|---|---|---|
| Pectoralis Major (g) | 14.00 ± 0.52 a | 13.95 ± 0.32 a | 17.96 ± 0.26 b |
| Pectoralis Minor (g) | 4.93 ± 0.17 a | 4.59 ± 0.32 a | 6.35 ± 0.09 b |
| Biceps Femoris (g) | 2.02 ± 0.06 a | 2.06 ± 0.05 a | 2.58 ± 0.05 b |
| Semitendinosus (g) | 0.89 ± 0.03 a | 0.92 ± 0.02 a | 1.14 ± 0.03 b |
| Gastrocnemius (g) | 0.67 ± 0.02 a | 0.70 ± 0.02 a | 0.91 ± 0.03 b |
| Tricep Brachii (g) | 0.48 ± 0.01 a | 0.50 ± 0.01 a | 0.61 ± 0.02 b |
| Heart (g) | 0.82 ± 0.03 a | 0.92 ± 0.03 b | 0.95 ± 0.04 b |
| Leg Fat (g) | 0.41 ± 0.06 NS | 0.30 ± 0.032 NS | 0.32 ± 0.04 NS |
| Abdominal Fat (g) | 0.26 ± 0.04 NS | 0.17 ± 0.02 NS | 0.21 ± 0.03 NS |
| Leg Fat (g, 12 weeks) | 0.70 ± 0.13 NS | 0.65 ± 0.05 NS | 0.58 ± 0.09 NS |
| Abdominal Fat (g, 12 weeks) | 0.44 ± 0.08 NS | 0.39 ± 0.03 NS | 0.37 ± 0.07 NS |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, J.; Kim, D.-H.; Lee, K. Muscle Hyperplasia in Japanese Quail by Single Amino Acid Deletion in MSTN Propeptide. Int. J. Mol. Sci. 2020, 21, 1504. https://doi.org/10.3390/ijms21041504
Lee J, Kim D-H, Lee K. Muscle Hyperplasia in Japanese Quail by Single Amino Acid Deletion in MSTN Propeptide. International Journal of Molecular Sciences. 2020; 21(4):1504. https://doi.org/10.3390/ijms21041504
Chicago/Turabian StyleLee, Joonbum, Dong-Hwan Kim, and Kichoon Lee. 2020. "Muscle Hyperplasia in Japanese Quail by Single Amino Acid Deletion in MSTN Propeptide" International Journal of Molecular Sciences 21, no. 4: 1504. https://doi.org/10.3390/ijms21041504





