Understanding Mechanisms of Salinity Tolerance in Barley by Proteomic and Biochemical Analysis of Near-Isogenic Lines
Abstract
:1. Introduction
2. Results
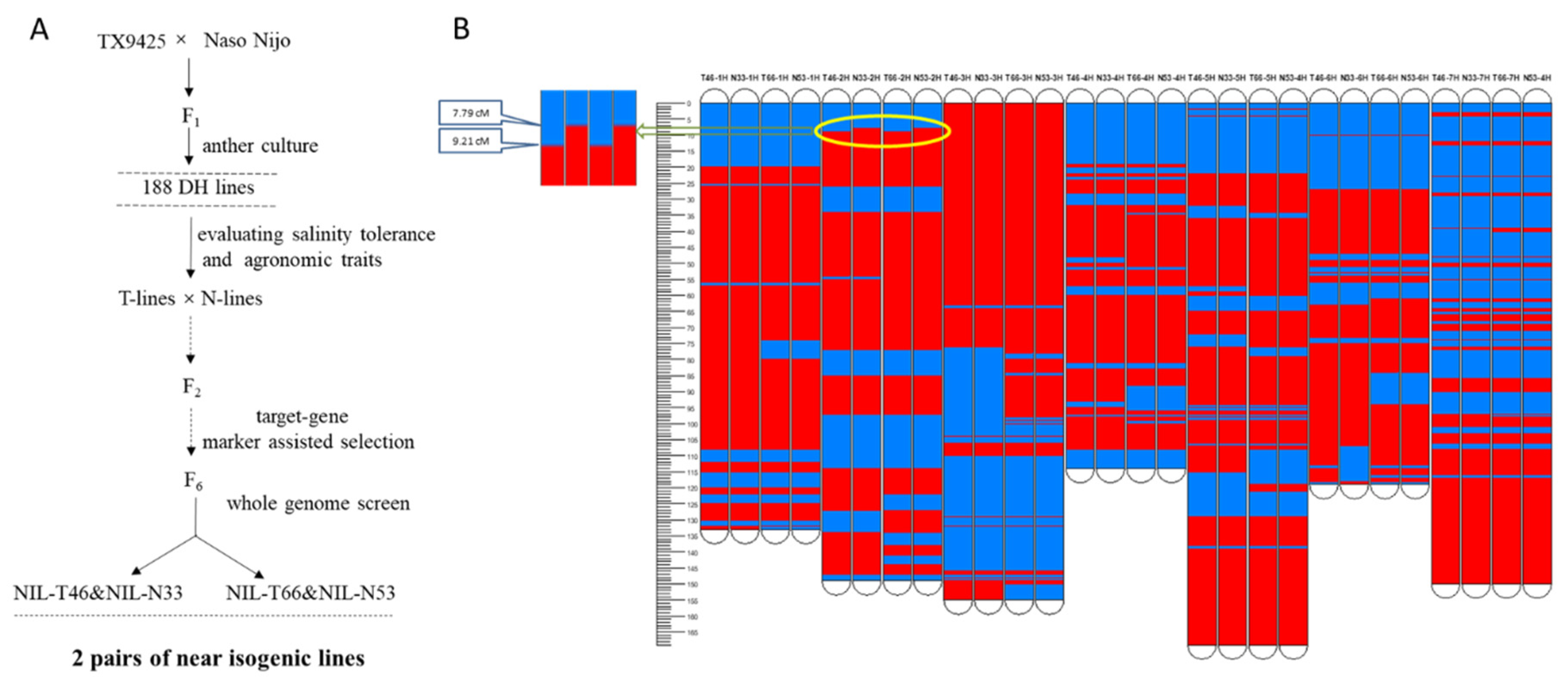
2.1. Near Isogenic Lines Exhibited Contrasting Salinity Tolerance
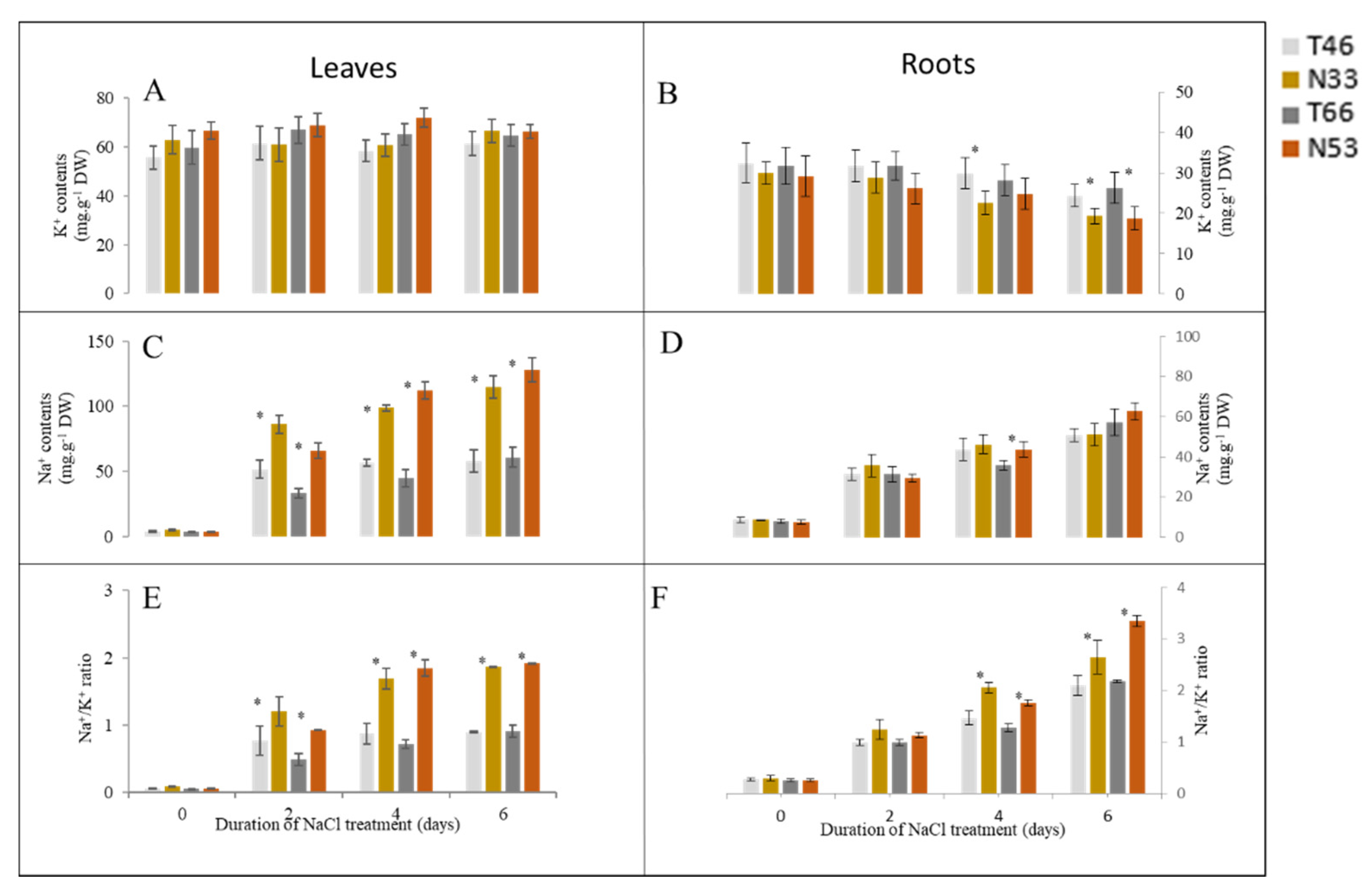
2.2. Effect of Salinity on Na+ and K+ Contents and Na+/ K+ Ratios
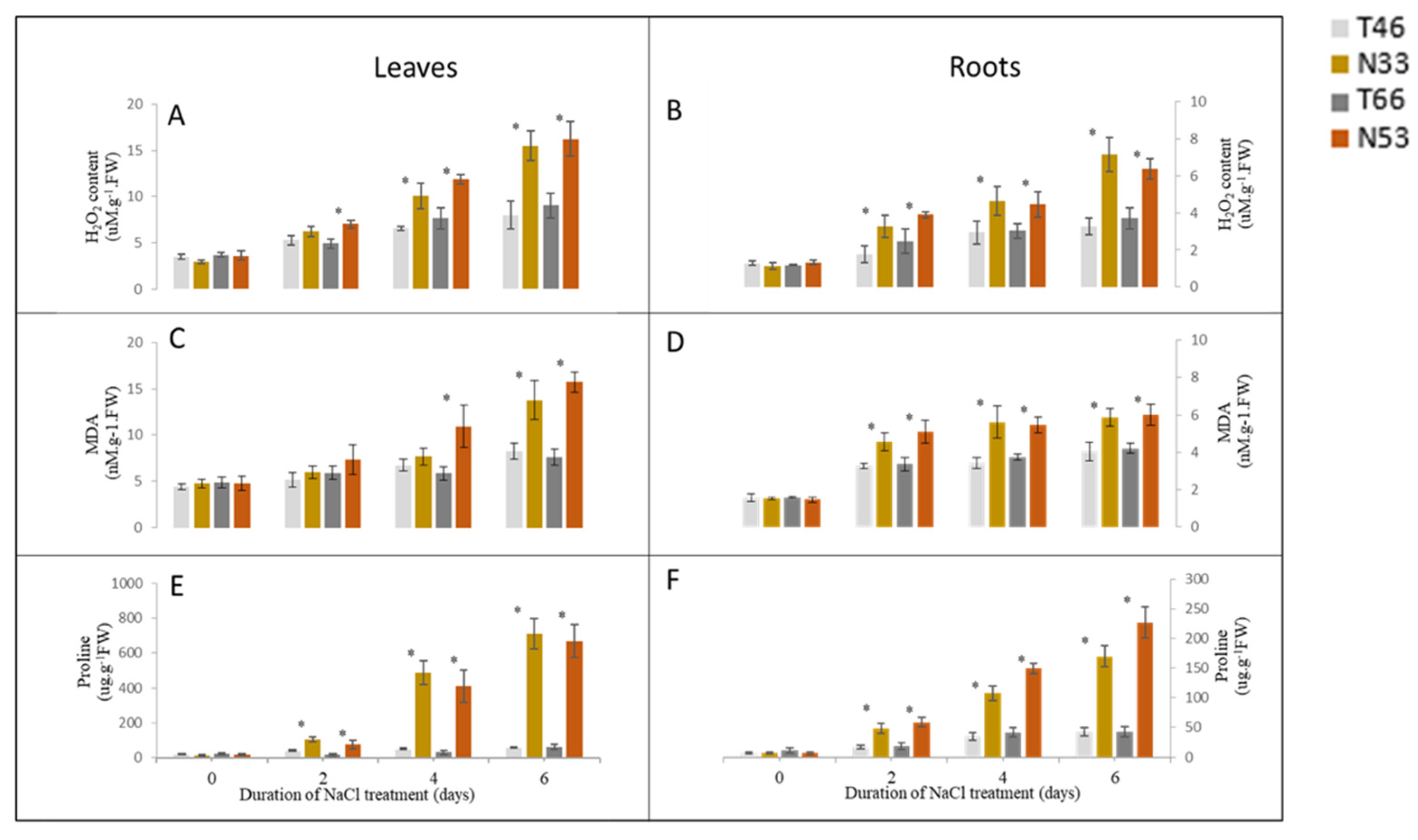
2.3. The Accumulation of H2O2, MDA, and Proline After Salinity Treatment
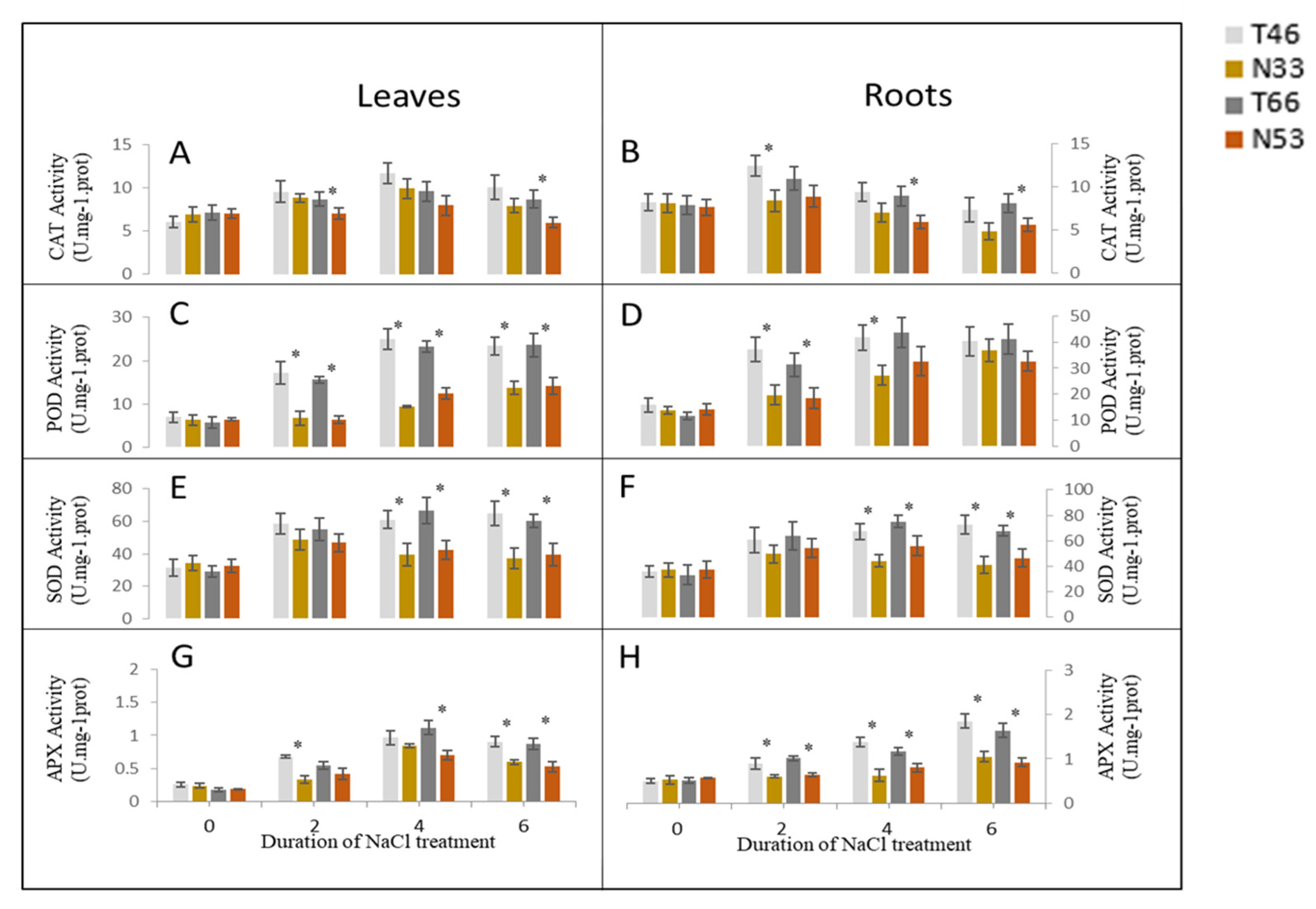
2.4. Changes in Activities of Antioxidant (AO) Enzymes under Salinity Stress
2.5. Identification of Differentially Expressed Proteins
3. Discussion
3.1. Near Isogenic Lines With Contrasting Salinity Tolerance Poses Less Background Noise
3.2. The Effects of QSl.TxNn.2H on Biochemical Characteristics and Plant Ionome
3.3. ROS Scavenging, Photosynthesis, and ATP Synthase Related Proteins Upregulated in Salinity Tolerant Lines and Underly the Tolerance Mechanism
4. Materials and Methods
4.1. DNA Extraction and Genotype Screening
4.2. Development of Near Isogenic Lines (NILs)
4.3. Growth Condition and Materials Collection
4.4. Determination of H2O2, MDA and Proline Measurements Contents
4.5. Antioxidant Enzyme Activity Measurements
4.6. Na+ and K+ Content
4.7. Protein Extraction, 2-DE, and Image Analysis
4.8. Identification of Proteins by Mass Spectrometry (MS)
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Rengasamy, P. World salinization with emphasis on Australia. J. Exp. Bot. 2006, 57, 1017–1023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, J.K. Plant salt tolerance. Trends Plant Sci. 2001, 6, 66–71. [Google Scholar] [CrossRef]
- Flowers, T.J. Improving crop salt tolerance. J. Exp. Bot. 2004, 55, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Genc, Y.; Mcdonald, G.K.; Tester, M. Reassessment of tissue Na+ concentration as a criterion for salinity tolerance in bread wheat. Plant Cell Environ. 2007, 30, 1486–1498. [Google Scholar] [CrossRef]
- Zhu, J.K. Genetic analysis of plant salt tolerance using Arabidopsis. Plant Physiol. 2000, 124, 941–948. [Google Scholar] [CrossRef] [Green Version]
- Sudhir, P.; Murthy, S.D.S. Effects of salt stress on basic processes of photosynthesis. Photosynthetica 2004, 42, 481–486. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Shabala, L.; Zhou, M.; Shabala, S. Hydrogen peroxide-induced root Ca2+ and K+ fluxes correlate with salt tolerance in cereals: Towards the cell-based phenotyping. Int. J. Mol. Sci. 2018, 19, 702. [Google Scholar] [CrossRef] [Green Version]
- Demidchik, V.; Cuin, T.A.; Svistunenko, D.; Smith, S.J.; Miller, A.J.; Shabala, S.; Sokolik, A.; Yurin, V. Arabidopsis root K+-efflux conductance activated by hydroxyl radicals: Single-Channel properties, genetic basis and involvement in stress-induced cell death. J. Cell Sci. 2010, 123, 1468–1479. [Google Scholar] [CrossRef] [Green Version]
- Chakraborty, K.; Bose, J.; Shabala, L.; Shabala, S. Difference in root K+ retention ability and reduced sensitivity of K+-permeable channels to reactive oxygen species confer differential salt tolerance in three Brassica species. J. Exp. Bot. 2016, 67, 4611–4625. [Google Scholar] [CrossRef] [Green Version]
- Jacoby, R.P.; Taylor, N.L.; Millar, A.H. The role of mitochondrial respiration in salinity tolerance. Trends Plant Sci. 2011, 16, 614–623. [Google Scholar]
- Munns, R.; Day, D.A.; Fricke, W.; Watt, M.; Arsova, B.; Barkla, B.J.; Bose, J.; Byrt, C.S.; Chen, Z.H.; Foster, K.J.; et al. Energy costs of salt tolerance in crop plants. New Phytol. 2019, 225, 1072–1090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poorter, H.; Remkes, C.; Lambers, H. Carbon and nitrogen economy of 24 wild species differing in relative growth rate. Plant Physiol. 1990, 94, 621–627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, G.; Suzuki, N.; Ciftci-Yilmaz, S.; Mittler, R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010, 33, 453–467. [Google Scholar] [CrossRef]
- Hasegawa, P.M. Sodium (Na+) homeostasis and salt tolerance of plants. Environ. Exp. Bot. 2013, 92, 19–31. [Google Scholar] [CrossRef]
- Zhu, J.K. Regulation of ion homeostasis under salt stress. Curr. Opin. Plant Biol. 2003, 6, 441–445. [Google Scholar] [CrossRef]
- Jin, Y.; Jing, W.; Zhang, Q.; Zhang, W. Cyclic nucleotide gated channel 10 negatively regulates salt tolerance by mediating Na+ transport in Arabidopsis. J. Plant Res. 2015, 128, 211–220. [Google Scholar] [CrossRef]
- Roy, S.J.; Gilliham, M.; Berger, B.; Essah, P.A.; Cheffings, C.; Miller, A.J.; Davenport, R.J.; Liu, L.H.; Skynner, M.J.; Davies, J.M.; et al. Investigating glutamate receptor-like gene co-expression in Arabidopsis thaliana. Plant Cell Environ. 2008, 31, 861–871. [Google Scholar] [CrossRef]
- Demidchik, V.; Maathuis, F.J. Physiological roles of nonselective cation channels in plants: From salt stress to signalling and development. New Phytol. 2007, 175, 387–404. [Google Scholar] [CrossRef]
- Shi, H.; Ishitani, M.; Kim, C.; Zhu, J.K. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. USA 2000, 97, 6896–6901. [Google Scholar] [CrossRef] [Green Version]
- Gaxiola, R.A.; Rao, R.; Sherman, A.; Grisafi, P.; Alper, S.L.; Fink, G.R. The Arabidopsis thaliana proton transporters, AtNhx1 and Avp1, can function in cation detoxification in yeast. Proc. Natl. Acad. Sci. USA 1999, 96, 1480–1485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shabala, S.; Pottosin, I. Regulation of potassium transport in plants under hostile conditions: Implications for abiotic and biotic stress tolerance. Physiol. Plant. 2014, 151, 257–279. [Google Scholar] [CrossRef] [PubMed]
- Véry, A.A.; Nieves-Cordones, M.; Daly, M.; Khan, I.; Fizames, C.; Sentenac, H. Molecular biology of K+ transport across the plant cell membrane: What do we learn from comparison between plant species? J. Plant Physiol. 2014, 171, 748–769. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Cuin, T.A.; Zhou, M.; Twomey, A.; Naidu, B.P.; Shabala, S. Compatible solute accumulation and stress-mitigating effects in barley genotypes contrasting in their salt tolerance. J. Exp. Bot. 2007, 58, 4245–4255. [Google Scholar] [CrossRef] [Green Version]
- Bose, J.; Rodrigo-Moreno, A.; Shabala, S. ROS homeostasis in halophytes in the context of salinity stress tolerance. J. Exp. Bot. 2014, 65, 1241–1257. [Google Scholar] [CrossRef]
- Xu, R.; Wang, J.; Li, C.; Johnson, P.; Lu, C.; Zhou, M. A single locus is responsible for salinity tolerance in a Chinese landrace barley (Hordeum vulgare L.). PLoS ONE 2012, 7, e43079. [Google Scholar] [CrossRef]
- Fatehi, F.; Hosseinzadeh, A.; Alizadeh, H.; Brimavandi, T.; Struik, P.C. The proteome response of salt-resistant and salt-sensitive barley genotypes to long-term salinity stress. Mol. Biol. Rep. 2012, 39, 6387–6397. [Google Scholar] [CrossRef]
- Rasoulnia, A.; Bihamta, M.R.; Peyghambari, S.A.; Alizadeh, H.; Rahnama, A. Proteomic response of barley leaves to salinity. Mol. Biol. Rep. 2011, 38, 5055–5063. [Google Scholar] [CrossRef]
- Witzel, K.; Weidner, A.; Surabhi, G.K.; Börner, A.; Mock, H.P. Salt stress-induced alterations in the root proteome of barley genotypes with contrasting response towards salinity. J. Exp. Bot. 2009, 60, 3545–3557. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, I.M.; Dai, H.; Zheng, W.; Cao, F.; Zhang, G.; Sun, D.; Wu, F. Genotypic differences in physiological characteristics in the tolerance to drought and salinity combined stress between Tibetan wild and cultivated barley. Plant Physiol. Biochem. 2013, 63, 49–60. [Google Scholar] [CrossRef]
- Shabala, S.; Shabala, S.; Cuin, T.A.; Pang, J.; Percey, W.; Chen, Z.; Conn, S.; Eing, C.; Wegner, L.H. Xylem ionic relations and salinity tolerance in barley. Plant J. 2010, 61, 839–853. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.D.; Li, H.B.; Zheng, Z.; Wei, Y.M.; Zheng, Y.L.; McIntyre, C.L.; Zhou, M.X.; Liu, C.J. Characterization of a QTL affecting spike morphology on the long arm of chromosome 3H in barley (Hordeum vulgare L.) based on near isogenic lines and a NIL-derived population. Theor. Appl. Genet. 2012, 125, 1385–1392. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Zhu, M.; Shabala, S.; Li, C.D.; Johnson, P.; Zhou, M.X. Antioxidant sctivity in salt-stressed barley leaves: Evaluating time-and age-dependence and suitability for the use as a biochemical marker in breeding programs. J. Agron. Crop. Sci. 2014, 200, 261–272. [Google Scholar] [CrossRef]
- Horie, T.; Hauser, F.; Schroeder, J.I. HKT transporter-mediated salinity resistance mechanisms in Arabidopsis and monocot crop plants. Trends Plant Sci. 2009, 14, 660–668. [Google Scholar] [CrossRef] [Green Version]
- Shabala, S. Learning from halophytes: Physiological basis and strategies to improve abiotic stress tolerance in crops. Ann. Bot. 2013, 112, 1209–1221. [Google Scholar] [CrossRef]
- Ishikawa, T.; Shabala, S. Control of xylem Na+ loading and transport to the shoot in rice and barley as a determinant of differential salinity stress tolerance. Physiol. Plant. 2019, 165, 619–631. [Google Scholar] [CrossRef] [Green Version]
- Shi, H.; Quintero, F.J.; Pardo, J.M.; Zhu, J.K. The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 2002, 14, 465–477. [Google Scholar] [CrossRef] [Green Version]
- Colmenero-Flores, J.M.; Martínez, G.; Gamba, G.; Vázquez, N.; Iglesias, D.J.; Brumós, J.; Talón, M. Identification and functional characterization of cation-chloride cotransporters in plants. Plant J. 2007, 50, 278–292. [Google Scholar] [CrossRef]
- Maggio, A.; Miyazaki, S.; Veronese, P.; Fujita, T.; Ibeas, J.I.; Damsz, B.; Narasimhan, M.L.; Hasegawa, P.M.; Joly, R.J.; Bressan, R.A. Does proline accumulation play an active role in stress-induced growth reduction? Plant J. 2002, 31, 699–712. [Google Scholar] [CrossRef] [Green Version]
- Smirnoff, N.; Cumbes, Q.J. Hydroxyl radical scavenging activity of compatible solutes. Phytochemistry 1989, 28, 1057–1060. [Google Scholar] [CrossRef]
- Jin, X.; Huang, Y.; Zeng, F.; Zhou, M.; Zhang, G. Genotypic difference in response of peroxidase and superoxide dismutase isozymes and activities to salt stress in barley. Acta Physiol. Plant. 2009, 31, 1103. [Google Scholar] [CrossRef]
- Moradi, F.; Ismail, A.M. Responses of photosynthesis, chlorophyll fluorescence and ROS-scavenging systems to salt stress during seedling and reproductive stages in rice. Ann. Bot. 2007, 99, 1161–1173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meloni, D.A.; Oliva, M.A.; Martinez, C.A.; Cambraia, J. Photosynthesis and activity of superoxide dismutase, peroxidase and glutathione reductase in cotton under salt stress. Environ. Exp. Bot. 2003, 49, 69–76. [Google Scholar] [CrossRef]
- Mansour, M.M.F.; Ali, E.F. Evaluation of proline functions in saline conditions. Phytochemistry 2017, 140, 52–68. [Google Scholar] [CrossRef] [PubMed]
- Maksimović, J.D.; Zhang, J.; Zeng, F.; Živanović, B.D.; Shabala, L.; Zhou, M.; Shabala, S. Linking oxidative and salinity stress tolerance in barley: Can root antioxidant enzyme activity be used as a measure of stress tolerance? Plant Soil 2013, 365, 141–155. [Google Scholar] [CrossRef]
- Mittler, R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef]
- Chen, S.; Gollop, N.; Heuer, B. Proteomic analysis of salt-stressed tomato (Solanum lycopersicum) seedlings: Effect of genotype and exogenous application of glycinebetaine. J. Exp. Bot. 2009, 60, 2005–2019. [Google Scholar] [CrossRef] [Green Version]
- Kav, N.N.; Srivastava, S.; Goonewardene, L.; Blade, S.F. Proteome-Level changes in the roots of Pisum sativum in response to salinity. Ann. Appl. Biol. 2004, 145, 217–230. [Google Scholar] [CrossRef]
- Sugimoto, M.; Takeda, K. Proteomic analysis of specific proteins in the root of salt-tolerant barley. Biosci. Biotechnol. Biochem. 2009, 73, 2762–2765. [Google Scholar] [CrossRef]
- Ashraf, M.; Harris, P.J.C. Photosynthesis under stressful environments: An overview. Photosynthetica 2013, 51, 163–190. [Google Scholar] [CrossRef]
- Ifuku, K. The PsbP and PsbQ family proteins in the photosynthetic machinery of chloroplasts. Plant Physiol. Biochem. 2014, 81, 108–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yi, X.; Hargett, S.R.; Liu, H.; Frankel, L.K.; Bricker, T.M. The PsbP protein is required for photosystem II complex assembly/stability and photoautotrophy in Arabidopsis thaliana. J. Biol. Chem. 2007, 282, 24833–24841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simkin, A.J.; McAusland, L.; Lawson, T.; Raines, C.A. Overexpression of the RieskeFeS protein increases electron transport rates and biomass yield. Plant Physiol. 2017, 175, 134–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, C.; Sibicky, T.; Huang, B. Protein profile analysis of salt-responsive proteins in leaves and roots in two cultivars of creeping bentgrass differing in salinity tolerance. Plant Cell Rep. 2010, 29, 595–615. [Google Scholar] [CrossRef] [PubMed]
- Yamori, W.; Takahashi, S.; Makino, A.; Price, G.D.; Badger, M.R.; von Caemmerer, S. The roles of ATP synthase and the cytochrome b6/f complexes in limiting chloroplast electron transport and determining photosynthetic capacity. Plant Physiol. 2011, 155, 956–962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lv, G.Y.; Guo, X.G.; Xie, L.P.; Xie, C.G.; Zhang, X.H.; Yang, Y.; Xiao, L.; Tang, Y.Y.; Pan, X.L.; Guo, A.G.; et al. Molecular characterization, gene evolution, and expression analysis of the fructose-1, 6-bisphosphate aldolase (FBA) gene family in wheat (Triticum aestivum L.). Front. Plant Sci. 2017, 8, 1030. [Google Scholar] [CrossRef] [Green Version]
- Fan, W.; Zhang, Z.; Zhang, Y. Cloning and molecular characterization of fructose-1, 6-bisphosphate aldolase gene regulated by high-salinity and drought in Sesuvium portulacastrum. Plant Cell Rep. 2009, 28, 975–984. [Google Scholar] [CrossRef]
- Uematsu, K.; Suzuki, N.; Iwamae, T.; Inui, M.; Yukawa, H. Increased fructose 1, 6-bisphosphate aldolase in plastids enhances growth and photosynthesis of tobacco plants. J. Exp. Bot. 2012, 63, 3001–3009. [Google Scholar] [CrossRef]
- Munns, R.; Passioura, J.B.; Colmer, T.D.; Byrt, C.S. Osmotic adjustment and energy limitations to plant growth in saline soil. New Phytol. 2019, 225, 1091–1096. [Google Scholar] [CrossRef] [Green Version]
- Liska, A.J.; Shevchenko, A.; Pick, U.; Katz, A. Enhanced photosynthesis and redox energy production contribute to salinity tolerance in Dunaliella as revealed by homology-based proteomics. Plant Physiol. 2004, 136, 2806–2817. [Google Scholar] [CrossRef] [Green Version]
- Neto, M.L.; Lobo, A.K.M.; Martins, M.O.; Fontenele, A.V.; Silveira, J.A.G. Dissipation of excess photosynthetic energy contributes to salinity tolerance: A comparative study of salt-tolerant Ricinus communis and salt-sensitive Jatropha curcas. J. Plant Physiol. 2014, 171, 23–30. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Fu, J.; Yu, C.; Wang, X.; Jiang, Q.; Hong, J.; Lu, K.; Xue, G.; Yan, C.; James, A.; et al. Increasing cyclic electron flow is related to Na+ sequestration into vacuoles for salt tolerance in soybean. J. Exp. Bot. 2015, 66, 6877–6889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gniazdowska, A.; Krasuska, U.; Bogatek, R. Dormancy removal in apple embryos by nitric oxide or cyanide involves modifications in ethylene biosynthetic pathway. Planta 2010, 232, 1397–1407. [Google Scholar] [CrossRef] [PubMed]
- Dionisio-Sese, M.L.; Tobita, S. Antioxidant responses of rice seedlings to salinity stress. Plant Sci. 1998, 135, 1–9. [Google Scholar] [CrossRef]
- Bates, L.S.; Waldren, R.P.; Teare, I.D. Rapid determination of free proline for water-stress studies. Plant Soil 1973, 39, 205–207. [Google Scholar] [CrossRef]
- Jaipal Reddy, P.; Anand Rao, A.; Malhotra, D.; Sharma, S.; Kumar, R.; Jain, R.; Gollapalli, K.; Pendharkar, N.; Rapole, S.; Srivastava, S. A simple protein extraction method for proteomic analysis of diverse biological specimens. Curr. Proteom. 2013, 10, 298–311. [Google Scholar] [CrossRef]
- Guo, B.; Chen, Y.; Li, C.; Wang, T.; Wang, R.; Wang, B.; Hu, S.; Du, X.; Xing, H.; Song, X.; et al. Maize (Zea mays L.) seedling leaf nuclear proteome and differentially expressed proteins between a hybrid and its parental lines. Proteomics 2014, 14, 1071–1087. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, J.; Fan, Y.; Shabala, S.; Li, C.; Lv, C.; Guo, B.; Xu, R.; Zhou, M. Understanding Mechanisms of Salinity Tolerance in Barley by Proteomic and Biochemical Analysis of Near-Isogenic Lines. Int. J. Mol. Sci. 2020, 21, 1516. https://doi.org/10.3390/ijms21041516
Zhu J, Fan Y, Shabala S, Li C, Lv C, Guo B, Xu R, Zhou M. Understanding Mechanisms of Salinity Tolerance in Barley by Proteomic and Biochemical Analysis of Near-Isogenic Lines. International Journal of Molecular Sciences. 2020; 21(4):1516. https://doi.org/10.3390/ijms21041516
Chicago/Turabian StyleZhu, Juan, Yun Fan, Sergey Shabala, Chengdao Li, Chao Lv, Baojian Guo, Rugen Xu, and Meixue Zhou. 2020. "Understanding Mechanisms of Salinity Tolerance in Barley by Proteomic and Biochemical Analysis of Near-Isogenic Lines" International Journal of Molecular Sciences 21, no. 4: 1516. https://doi.org/10.3390/ijms21041516





