Comprehensive Genome-Wide Approaches to Activity-Dependent Translational Control in Neurons
Abstract
:1. The Emerging Importance of Translational Regulation in Neuronal Gene Expression
2. Translational Control Bridging Neuronal Activation and Genetic Program
3. Mechanisms Underlying Activity-Dependent Translational Control
3.1. Kinase Pathway Modifying Translation Factors
3.2. mTOR Signaling Pathway
3.3. Local Translation
4. Translational Control in Neurodevelopmental and Psychiatric Disorders
4.1. Autism Spectrum Disorder
4.2. Depression
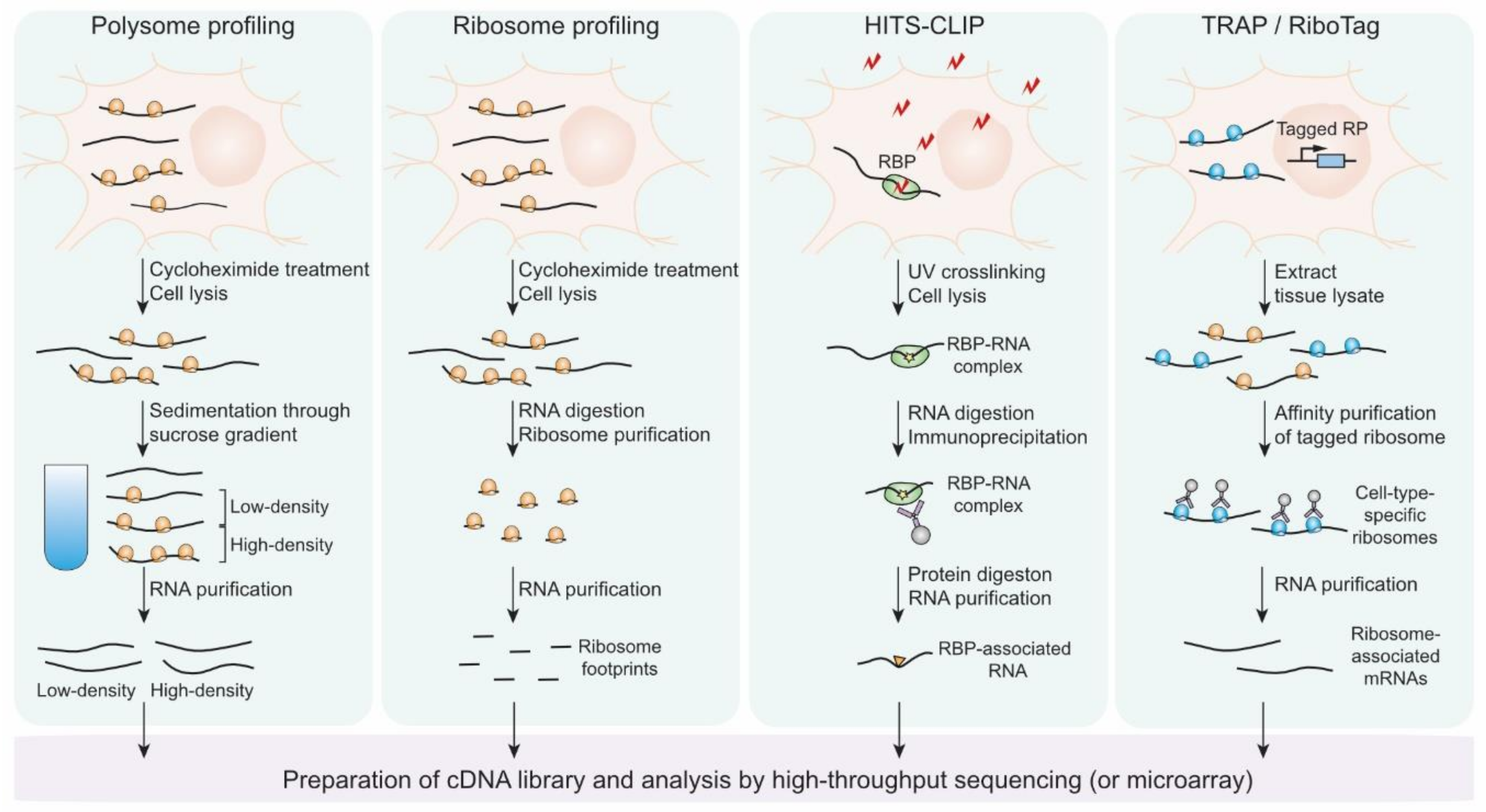
5. Genome-Wide Approaches to Study Translational Controls in Neurons
5.1. Polysome Profiling
5.2. Ribosome Profiling
5.3. Targeting Ribonucleoprotein Complexes
5.4. Targeting Ribosomes
6. Unbiased Translatome-Wide Approach to Bridge Translation and Activity: PhosphoTRAP and IEG-RiboTag
7. Conclusion and Outlook
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Russell, J.B.; Cook, G.M. Energetics of bacterial growth: Balance of anabolic and catabolic reactions. Microbiol. Rev. 1995, 59, 48–62. [Google Scholar] [CrossRef] [Green Version]
- Cleary, J.D.; Ranum, L.P. Repeat-associated non-ATG (RAN) translation in neurological disease. Hum. Mol. Genet. 2013, 22, R45–R51. [Google Scholar] [CrossRef]
- Bolze, A.; Mahlaoui, N.; Byun, M.; Turner, B.; Trede, N.; Ellis, S.R.; Abhyankar, A.; Itan, Y.; Patin, E.; Brebner, S.; et al. Ribosomal protein SA haploinsufficiency in humans with isolated congenital asplenia. Science 2013, 340, 976–978. [Google Scholar] [CrossRef] [Green Version]
- Kapur, M.; Ackerman, S.L. mRNA Translation Gone Awry: Translation Fidelity and Neurological Disease. Trends Genet. 2018, 34, 218–231. [Google Scholar] [CrossRef]
- Mills, E.W.; Green, R. Ribosomopathies: There’s strength in numbers. Science 2017, 358. [Google Scholar] [CrossRef] [Green Version]
- McCann, K.L.; Baserga, S.J. Genetics. Mysterious ribosomopathies. Science 2013, 341, 849–850. [Google Scholar] [CrossRef] [Green Version]
- Mirabello, L.; Khincha, P.P.; Ellis, S.R.; Giri, N.; Brodie, S.; Chandrasekharappa, S.C.; Donovan, F.X.; Zhou, W.; Hicks, B.D.; Boland, J.F.; et al. Novel and known ribosomal causes of Diamond-Blackfan anaemia identified through comprehensive genomic characterisation. J. Med. Genet. 2017, 54, 417–425. [Google Scholar] [CrossRef]
- Khajuria, R.K.; Munschauer, M.; Ulirsch, J.C.; Fiorini, C.; Ludwig, L.S.; McFarland, S.K.; Abdulhay, N.J.; Specht, H.; Keshishian, H.; Mani, D.R.; et al. Ribosome Levels Selectively Regulate Translation and Lineage Commitment in Human Hematopoiesis. Cell 2018, 173, 90–103. [Google Scholar] [CrossRef] [Green Version]
- Delepine, M.; Nicolino, M.; Barrett, T.; Golamaully, M.; Lathrop, G.M.; Julier, C. EIF2AK3, encoding translation initiation factor 2-alpha kinase 3, is mutated in patients with Wolcott-Rallison syndrome. Nat. Genet. 2000, 25, 406–409. [Google Scholar] [CrossRef]
- Calkhoven, C.F.; Muller, C.; Leutz, A. Translational control of gene expression and disease. Trends Mol. Med. 2002, 8, 577–583. [Google Scholar] [CrossRef]
- Kapur, M.; Monaghan, C.E.; Ackerman, S.L. Regulation of mRNA Translation in Neurons-A Matter of Life and Death. Neuron 2017, 96, 616–637. [Google Scholar] [CrossRef] [Green Version]
- Chesnokova, E.; Bal, N.; Kolosov, P. Kinases of eIF2a Switch Translation of mRNA Subset during Neuronal Plasticity. Int. J. Mol. Sci. 2017, 18, 2213. [Google Scholar] [CrossRef]
- Li, W.; Wang, X.; Van Der Knaap, M.S.; Proud, C.G. Mutations linked to leukoencephalopathy with vanishing white matter impair the function of the eukaryotic initiation factor 2B complex in diverse ways. Mol. Cell Biol. 2004, 24, 3295–3306. [Google Scholar] [CrossRef] [Green Version]
- Brown, V.; Jin, P.; Ceman, S.; Darnell, J.C.; O’Donnell, W.T.; Tenenbaum, S.A.; Jin, X.; Feng, Y.; Wilkinson, K.D.; Keene, J.D.; et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell 2001, 107, 477–487. [Google Scholar] [CrossRef] [Green Version]
- Darnell, J.C.; Van Driesche, S.J.; Zhang, C.; Hung, K.Y.; Mele, A.; Fraser, C.E.; Stone, E.F.; Chen, C.; Fak, J.J.; Chi, S.W.; et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 2011, 146, 247–261. [Google Scholar] [CrossRef] [Green Version]
- Buffington, S.A.; Huang, W.; Costa-Mattioli, M. Translational control in synaptic plasticity and cognitive dysfunction. Annu. Rev. Neurosci. 2014, 37, 17–38. [Google Scholar] [CrossRef] [Green Version]
- Cho, J.; Yu, N.K.; Choi, J.H.; Sim, S.E.; Kang, S.J.; Kwak, C.; Lee, S.W.; Kim, J.I.; Choi, D.I.; Kim, V.N.; et al. Multiple repressive mechanisms in the hippocampus during memory formation. Science 2015, 350, 82–87. [Google Scholar] [CrossRef]
- Kandel, E.R. The molecular biology of memory storage: A dialogue between genes and synapses. Science 2001, 294, 1030–1038. [Google Scholar] [CrossRef] [Green Version]
- Sutton, M.A.; Ito, H.T.; Cressy, P.; Kempf, C.; Woo, J.C.; Schuman, E.M. Miniature neurotransmission stabilizes synaptic function via tonic suppression of local dendritic protein synthesis. Cell 2006, 125, 785–799. [Google Scholar] [CrossRef] [Green Version]
- Sutton, M.A.; Wall, N.R.; Aakalu, G.N.; Schuman, E.M. Regulation of dendritic protein synthesis by miniature synaptic events. Science 2004, 304, 1979–1983. [Google Scholar] [CrossRef] [Green Version]
- Yap, E.L.; Greenberg, M.E. Activity-Regulated Transcription: Bridging the Gap between Neural Activity and Behavior. Neuron 2018, 100, 330–348. [Google Scholar] [CrossRef] [Green Version]
- Flavell, S.W.; Greenberg, M.E. Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu. Rev. Neurosci. 2008, 31, 563–590. [Google Scholar] [CrossRef] [Green Version]
- Hernandez, P.J.; Abel, T. The role of protein synthesis in memory consolidation: Progress amid decades of debate. Neurobiol. Learn. Mem. 2008, 89, 293–311. [Google Scholar] [CrossRef] [Green Version]
- Buxbaum, A.R.; Wu, B.; Singer, R.H. Single beta-actin mRNA detection in neurons reveals a mechanism for regulating its translatability. Science 2014, 343, 419–422. [Google Scholar] [CrossRef] [Green Version]
- Taniguchi, Y.; Choi, P.J.; Li, G.W.; Chen, H.; Babu, M.; Hearn, J.; Emili, A.; Xie, X.S. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 2010, 329, 533–538. [Google Scholar] [CrossRef] [Green Version]
- Tebaldi, T.; Re, A.; Viero, G.; Pegoretti, I.; Passerini, A.; Blanzieri, E.; Quattrone, A. Widespread uncoupling between transcriptome and translatome variations after a stimulus in mammalian cells. BMC Genom. 2012, 13, 220. [Google Scholar] [CrossRef] [Green Version]
- Moritz, C.P.; Muhlhaus, T.; Tenzer, S.; Schulenborg, T.; Friauf, E. Poor transcript-protein correlation in the brain: Negatively correlating gene products reveal neuronal polarity as a potential cause. J. Neurochem. 2019, 149, 582–604. [Google Scholar] [CrossRef] [Green Version]
- Hershey, J.W.B.; Sonenberg, N.; Mathews, M.B. Principles of Translational Control. Csh. Perspect. Biol. 2019, 11, a032607. [Google Scholar] [CrossRef]
- Costa-Mattioli, M.; Sossin, W.S.; Klann, E.; Sonenberg, N. Translational Control of Long-Lasting Synaptic Plasticity and Memory. Neuron 2009, 61, 10–26. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Z.; Belforte, J.E.; Lu, Y.; Yabe, Y.; Pickel, J.; Smith, C.B.; Je, H.S.; Lu, B.; Nakazawa, K. eIF2 alpha Phosphorylation-Dependent Translation in CA1 Pyramidal Cells Impairs Hippocampal Memory Consolidation without Affecting General Translation. J. Neurosci. 2010, 30, 2582–2594. [Google Scholar] [CrossRef] [Green Version]
- Taha, E.; Gildish, I.; Gal-Ben-Ari, S.; Rosenblum, K. The role of eEF2 pathway in learning and synaptic plasticity. Neurobiol. Learn. Mem. 2013, 105, 100–106. [Google Scholar] [CrossRef] [Green Version]
- Park, S.; Park, J.M.; Kim, S.; Kim, J.A.; Shepherd, J.D.; Smith-Hicks, C.L.; Chowdhury, S.; Kaufmann, W.; Kuhl, D.; Ryazanov, A.G.; et al. Elongation factor 2 and fragile X mental retardation protein control the dynamic translation of Arc/Arg3.1 essential for mGluR-LTD. Neuron 2008, 59, 70–83. [Google Scholar] [CrossRef] [Green Version]
- Crino, P.B. The mTOR signalling cascade: Paving new roads to cure neurological disease. Nat. Rev. Neurol. 2016, 12, 379–392. [Google Scholar] [CrossRef]
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Guan, K.L. mTOR as a central hub of nutrient signalling and cell growth. Nat. Cell Biol. 2019, 21, 63–71. [Google Scholar] [CrossRef]
- Ryskalin, L.; Limanaqi, F.; Frati, A.; Busceti, C.L.; Fornai, F. mTOR-Related Brain Dysfunctions in Neuropsychiatric Disorders. Int. J. Mol. Sci. 2018, 19, 2226. [Google Scholar] [CrossRef] [Green Version]
- Lipton, J.O.; Sahin, M. The neurology of mTOR. Neuron 2014, 84, 275–291. [Google Scholar] [CrossRef] [Green Version]
- Bockaert, J.; Marin, P. mTOR in Brain Physiology and Pathologies. Physiol. Rev. 2015, 95, 1157–1187. [Google Scholar] [CrossRef]
- Tang, S.J.; Reis, G.; Kang, H.; Gingras, A.C.; Sonenberg, N.; Schuman, E.M. A rapamycin-sensitive signaling pathway contributes to long-term synaptic plasticity in the hippocampus. Proc. Natl. Acad. Sci. USA 2002, 99, 467–472. [Google Scholar] [CrossRef] [Green Version]
- Cammalleri, M.; Lutjens, R.; Berton, F.; King, A.R.; Simpson, C.; Francesconi, W.; Sanna, P.P. Time-restricted role for dendritic activation of the mTOR-p70S6K pathway in the induction of late-phase long-term potentiation in the CA1. Proc. Natl. Acad. Sci. USA 2003, 100, 14368–14373. [Google Scholar] [CrossRef] [Green Version]
- Dash, P.K.; Orsi, S.A.; Moore, A.N. Spatial memory formation and memory-enhancing effect of glucose involves activation of the tuberous sclerosis complex-Mammalian target of rapamycin pathway. J. Neurosci. 2006, 26, 8048–8056. [Google Scholar] [CrossRef] [Green Version]
- Caccamo, A.; Branca, C.; Talboom, J.S.; Shaw, D.M.; Turner, D.; Ma, L.; Messina, A.; Huang, Z.; Wu, J.; Oddo, S. Reducing Ribosomal Protein S6 Kinase 1 Expression Improves Spatial Memory and Synaptic Plasticity in a Mouse Model of Alzheimer’s Disease. J. Neurosci. 2015, 35, 14042–14056. [Google Scholar] [CrossRef] [Green Version]
- Rogan, M.T.; Staubli, U.V.; LeDoux, J.E. Fear conditioning induces associative long-term potentiation in the amygdala. Nature 1997, 390, 604–607. [Google Scholar] [CrossRef]
- Sigurdsson, T.; Doyere, V.; Cain, C.K.; LeDoux, J.E. Long-term potentiation in the amygdala: A cellular mechanism of fear learning and memory. Neuropharmacology 2007, 52, 215–227. [Google Scholar] [CrossRef]
- Parsons, R.G.; Gafford, G.M.; Helmstetter, F.J. Translational control via the mammalian target of rapamycin pathway is critical for the formation and stability of long-term fear memory in amygdala neurons. J. Neurosci. 2006, 26, 12977–12983. [Google Scholar] [CrossRef]
- Luscher, C.; Malenka, R.C. Drug-evoked synaptic plasticity in addiction: From molecular changes to circuit remodeling. Neuron 2011, 69, 650–663. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Luo, Y.X.; He, Y.Y.; Li, F.Q.; Shi, H.S.; Xue, L.F.; Xue, Y.X.; Lu, L. Nucleus accumbens core mammalian target of rapamycin signaling pathway is critical for cue-induced reinstatement of cocaine seeking in rats. J. Neurosci. 2010, 30, 12632–12641. [Google Scholar] [CrossRef] [Green Version]
- Barak, S.; Liu, F.; Ben Hamida, S.; Yowell, Q.V.; Neasta, J.; Kharazia, V.; Janak, P.H.; Ron, D. Disruption of alcohol-related memories by mTORC1 inhibition prevents relapse. Nat. Neurosci. 2013, 16, 1111–1117. [Google Scholar] [CrossRef]
- Ma, X.M.; Blenis, J. Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell Biol. 2009, 10, 307–318. [Google Scholar] [CrossRef]
- Wang, Z.G.; Li, H.; Huang, Y.; Li, R.; Wang, X.F.; Yu, L.X.; Guang, X.Q.; Li, L.; Zhang, H.Y.; Zhao, Y.Z.; et al. Nerve growth factor-induced Akt/mTOR activation protects the ischemic heart via restoring autophagic flux and attenuating ubiquitinated protein accumulation. Oncotarget 2017, 8, 5400–5413. [Google Scholar] [CrossRef] [Green Version]
- Shaw, R.J.; Cantley, L.C. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature 2006, 441, 424–430. [Google Scholar] [CrossRef] [PubMed]
- Ronesi, J.A.; Collins, K.A.; Hays, S.A.; Tsai, N.P.; Guo, W.; Birnbaum, S.G.; Hu, J.H.; Worley, P.F.; Gibson, J.R.; Huber, K.M. Disrupted Homer scaffolds mediate abnormal mGluR5 function in a mouse model of fragile X syndrome. Nat. Neurosci. 2012, 15, 431–440. [Google Scholar] [CrossRef] [PubMed]
- Huber, K.M.; Klann, E.; Costa-Mattioli, M.; Zukin, R.S. Dysregulation of Mammalian Target of Rapamycin Signaling in Mouse Models of Autism. J. Neurosci. 2015, 35, 13836–13842. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Lipton, J.O.; Boyle, L.M.; Madsen, J.R.; Goldenberg, M.C.; Pascual-Leone, A.; Sahin, M.; Rotenberg, A. Direct current stimulation induces mGluR5-dependent neocortical plasticity. Ann. Neurol. 2016, 80, 233–246. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.; Gkogkas, C.G.; Sonenberg, N.; Holt, C.E. Remote control of gene function by local translation. Cell 2014, 157, 26–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lecuyer, E.; Yoshida, H.; Parthasarathy, N.; Alm, C.; Babak, T.; Cerovina, T.; Hughes, T.R.; Tomancak, P.; Krause, H.M. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 2007, 131, 174–187. [Google Scholar] [CrossRef] [Green Version]
- Thomas, M.G.; Pascual, M.L.; Maschi, D.; Luchelli, L.; Boccaccio, G.L. Synaptic control of local translation: The plot thickens with new characters. Cell Mol. Life Sci. 2014, 71, 2219–2239. [Google Scholar] [CrossRef]
- Sambandan, S.; Akbalik, G.; Kochen, L.; Rinne, J.; Kahlstatt, J.; Glock, C.; Tushev, G.; Alvarez-Castelao, B.; Heckel, A.; Schuman, E.M. Activity-dependent spatially localized miRNA maturation in neuronal dendrites. Science 2017, 355, 634–637. [Google Scholar] [CrossRef]
- Ellis-Davies, G.C. Caged compounds: Photorelease technology for control of cellular chemistry and physiology. Nat. Methods 2007, 4, 619–628. [Google Scholar] [CrossRef] [Green Version]
- Bartley, C.M.; O’Keefe, R.A.; Blice-Baum, A.; Mihailescu, M.R.; Gong, X.; Miyares, L.; Karaca, E.; Bordey, A. Mammalian FMRP S499 Is Phosphorylated by CK2 and Promotes Secondary Phosphorylation of FMRP. eNeuro 2016, 3. [Google Scholar] [CrossRef] [Green Version]
- Bartley, C.M.; O’Keefe, R.A.; Bordey, A. FMRP S499 is phosphorylated independent of mTORC1-S6K1 activity. PLoS ONE 2014, 9, e96956. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, U.; Nalavadi, V.; Nakamoto, M.; Pallas, D.C.; Ceman, S.; Bassell, G.J.; Warren, S.T. FMRP phosphorylation reveals an immediate-early signaling pathway triggered by group I mGluR and mediated by PP2A. J. Neurosci. 2007, 27, 14349–14357. [Google Scholar] [CrossRef] [PubMed]
- Tsang, B.; Arsenault, J.; Vernon, R.M.; Lin, H.; Sonenberg, N.; Wang, L.Y.; Bah, A.; Forman-Kay, J.D. Phosphoregulated FMRP phase separation models activity-dependent translation through bidirectional control of mRNA granule formation. Proc. Natl. Acad. Sci. USA 2019, 116, 4218–4227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beaulieu-Laroche, L.; Toloza, E.H.S.; van der Goes, M.S.; Lafourcade, M.; Barnagian, D.; Williams, Z.M.; Eskandar, E.N.; Frosch, M.P.; Cash, S.S.; Harnett, M.T. Enhanced Dendritic Compartmentalization in Human Cortical Neurons. Cell 2018, 175, 643–651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- American Psychiatric Association. DSM-5 Task Force. Diagnostic and statistical manual of mental disorders: DSM-5, 5th ed.; American Psychiatric Association: Washington, DC, USA, 2013; p. 947. [Google Scholar]
- Ebert, D.H.; Greenberg, M.E. Activity-dependent neuronal signalling and autism spectrum disorder. Nature 2013, 493, 327–337. [Google Scholar] [CrossRef] [Green Version]
- Richter, J.D.; Bassell, G.J.; Klann, E. Dysregulation and restoration of translational homeostasis in fragile X syndrome. Nat. Rev. Neurosci. 2015, 16, 595–605. [Google Scholar] [CrossRef] [Green Version]
- Santoro, M.R.; Bray, S.M.; Warren, S.T. Molecular mechanisms of fragile X syndrome: A twenty-year perspective. Annu. Rev. Pathol. 2012, 7, 219–245. [Google Scholar] [CrossRef] [Green Version]
- Waung, M.W.; Pfeiffer, B.E.; Nosyreva, E.D.; Ronesi, J.A.; Huber, K.M. Rapid translation of Arc/Arg3.1 selectively mediates mGluR-dependent LTD through persistent increases in AMPAR endocytosis rate. Neuron 2008, 59, 84–97. [Google Scholar] [CrossRef] [Green Version]
- Weiler, I.J.; Irwin, S.A.; Klintsova, A.Y.; Spencer, C.M.; Brazelton, A.D.; Miyashiro, K.; Comery, T.A.; Patel, B.; Eberwine, J.; Greenough, W.T. Fragile X mental retardation protein is translated near synapses in response to neurotransmitter activation. Proc. Natl. Acad. Sci. USA 1997, 94, 5395–5400. [Google Scholar] [CrossRef] [Green Version]
- Michalon, A.; Sidorov, M.; Ballard, T.M.; Ozmen, L.; Spooren, W.; Wettstein, J.G.; Jaeschke, G.; Bear, M.F.; Lindemann, L. Chronic pharmacological mGlu5 inhibition corrects fragile X in adult mice. Neuron 2012, 74, 49–56. [Google Scholar] [CrossRef] [Green Version]
- Dolen, G.; Osterweil, E.; Rao, B.S.; Smith, G.B.; Auerbach, B.D.; Chattarji, S.; Bear, M.F. Correction of fragile X syndrome in mice. Neuron 2007, 56, 955–962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, J.M.; Sahin, M. TSC1/TSC2 signaling in the CNS. FEBS Lett. 2011, 585, 973–980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Auerbach, B.D.; Osterweil, E.K.; Bear, M.F. Mutations causing syndromic autism define an axis of synaptic pathophysiology. Nature 2011, 480, 63–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kilincaslan, A.; Kok, B.E.; Tekturk, P.; Yalcinkaya, C.; Ozkara, C.; Yapici, Z. Beneficial Effects of Everolimus on Autism and Attention-Deficit/Hyperactivity Disorder Symptoms in a Group of Patients with Tuberous Sclerosis Complex. J. Child. Adolesc. Psychopharmacol. 2017, 27, 383–388. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.L.; Luo, L.; Mu, R.H.; Liu, B.B.; Geng, D.; Liu, Q.; Yi, L.T. Fluoxetine regulates mTOR signalling in a region-dependent manner in depression-like mice. Sci. Rep. 2015, 5, 16024. [Google Scholar] [CrossRef] [PubMed]
- Fond, G.; Loundou, A.; Rabu, C.; Macgregor, A.; Lancon, C.; Brittner, M.; Micoulaud-Franchi, J.A.; Richieri, R.; Courtet, P.; Abbar, M.; et al. Ketamine administration in depressive disorders: A systematic review and meta-analysis. Psychopharmacology 2014, 231, 3663–3676. [Google Scholar] [CrossRef] [PubMed]
- Berman, R.M.; Cappiello, A.; Anand, A.; Oren, D.A.; Heninger, G.R.; Charney, D.S.; Krystal, J.H. Antidepressant effects of ketamine in depressed patients. Biol. Psychiatr. 2000, 47, 351–354. [Google Scholar] [CrossRef]
- Autry, A.E.; Adachi, M.; Nosyreva, E.; Na, E.S.; Los, M.F.; Cheng, P.F.; Kavalali, E.T.; Monteggia, L.M. NMDA receptor blockade at rest triggers rapid behavioural antidepressant responses. Nature 2011, 475, 91–95. [Google Scholar] [CrossRef] [Green Version]
- Fuchs, T.; Jefferson, S.J.; Hooper, A.; Yee, P.H.; Maguire, J.; Luscher, B. Disinhibition of somatostatin-positive GABAergic interneurons results in an anxiolytic and antidepressant-like brain state. Mol. Psychiatry 2017, 22, 920–930. [Google Scholar] [CrossRef]
- Cavalleri, L.; Merlo Pich, E.; Millan, M.J.; Chiamulera, C.; Kunath, T.; Spano, P.F.; Collo, G. Ketamine enhances structural plasticity in mouse mesencephalic and human iPSC-derived dopaminergic neurons via AMPAR-driven BDNF and mTOR signaling. Mol. Psychiatry 2018, 23, 812–823. [Google Scholar] [CrossRef]
- Schena, M.; Shalon, D.; Davis, R.W.; Brown, P.O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 1995, 270, 467–470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heller, M.J. DNA microarray technology: Devices, systems, and applications. Annu. Rev. Biomed. Eng. 2002, 4, 129–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zong, Q.; Schummer, M.; Hood, L.; Morris, D.R. Messenger RNA translation state: The second dimension of high-throughput expression screening. Proc. Natl. Acad. Sci. USA 1999, 96, 10632–10636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schratt, G.M.; Nigh, E.A.; Chen, W.G.; Hu, L.; Greenberg, M.E. BDNF regulates the translation of a select group of mRNAs by a mammalian target of rapamycin-phosphatidylinositol 3-kinase-dependent pathway during neuronal development. J. Neurosci. 2004, 24, 7366–7377. [Google Scholar] [CrossRef] [PubMed]
- Sivan, G.; Kedersha, N.; Elroy-Stein, O. Ribosomal slowdown mediates translational arrest during cellular division. Mol. Cell Biol. 2007, 27, 6639–6646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reuter, J.A.; Spacek, D.V.; Snyder, M.P. High-throughput sequencing technologies. Mol. Cell 2015, 58, 586–597. [Google Scholar] [CrossRef] [Green Version]
- Ingolia, N.T.; Ghaemmaghami, S.; Newman, J.R.; Weissman, J.S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 2009, 324, 218–223. [Google Scholar] [CrossRef] [Green Version]
- Ingolia, N.T.; Brar, G.A.; Rouskin, S.; McGeachy, A.M.; Weissman, J.S. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc. 2012, 7, 1534–1550. [Google Scholar] [CrossRef]
- Andreev, D.E.; O’Connor, P.B.; Fahey, C.; Kenny, E.M.; Terenin, I.M.; Dmitriev, S.E.; Cormican, P.; Morris, D.W.; Shatsky, I.N.; Baranov, P.V. Translation of 5’ leaders is pervasive in genes resistant to eIF2 repression. Elife 2015, 4, e03971. [Google Scholar] [CrossRef]
- Sidrauski, C.; McGeachy, A.M.; Ingolia, N.T.; Walter, P. The small molecule ISRIB reverses the effects of eIF2alpha phosphorylation on translation and stress granule assembly. Elife 2015, 4, e05033. [Google Scholar] [CrossRef]
- Brubaker, S.W.; Gauthier, A.E.; Mills, E.W.; Ingolia, N.T.; Kagan, J.C. A bicistronic MAVS transcript highlights a class of truncated variants in antiviral immunity. Cell 2014, 156, 800–811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, H.; Ingolia, N.T.; Weissman, J.S.; Bartel, D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 2010, 466, 835–840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ingolia, N.T.; Lareau, L.F.; Weissman, J.S. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 2011, 147, 789–802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oh, E.; Becker, A.H.; Sandikci, A.; Huber, D.; Chaba, R.; Gloge, F.; Nichols, R.J.; Typas, A.; Gross, C.A.; Kramer, G.; et al. Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell 2011, 147, 1295–1308. [Google Scholar] [CrossRef] [Green Version]
- Hsieh, A.C.; Liu, Y.; Edlind, M.P.; Ingolia, N.T.; Janes, M.R.; Sher, A.; Shi, E.Y.; Stumpf, C.R.; Christensen, C.; Bonham, M.J.; et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 2012, 485, 55–61. [Google Scholar] [CrossRef] [Green Version]
- Li, G.W.; Burkhardt, D.; Gross, C.; Weissman, J.S. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 2014, 157, 624–635. [Google Scholar] [CrossRef] [Green Version]
- Chotewutmontri, P.; Barkan, A. Dynamics of Chloroplast Translation during Chloroplast Differentiation in Maize. PLoS Genet. 2016, 12, e1006106. [Google Scholar] [CrossRef] [Green Version]
- Blair, J.D.; Hockemeyer, D.; Doudna, J.A.; Bateup, H.S.; Floor, S.N. Widespread Translational Remodeling during Human Neuronal Differentiation. Cell Rep. 2017, 21, 2005–2016. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Li, Y.; Stackpole, E.E.; Novak, A.; Gao, Y.; Zhao, Y.; Zhao, X.; Richter, J.D. Regulatory discrimination of mRNAs by FMRP controls mouse adult neural stem cell differentiation. Proc. Natl. Acad. Sci. USA 2018, 115, E11397–E11405. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.K.; Cho, J.; Kim, S.H.; Kang, H.C.; Kim, D.S.; Kim, V.N.; Lee, J.H. Brain somatic mutations in MTOR reveal translational dysregulations underlying intractable focal epilepsy. J. Clin. Invest. 2019, 129, 4207–4223. [Google Scholar] [CrossRef]
- Cho, J.; Yu, N.K.; Kim, V.N.; Kaang, B.K. Response to Comment on “Multiple repressive mechanisms in the hippocampus during memory formation”. Science 2016, 353, 453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathew, R.S.; Mullan, H.; Blusztajn, J.K.; Lehtinen, M.K. Comment on “Multiple repressive mechanisms in the hippocampus during memory formation”. Science 2016, 353, 453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hornstein, N.; Torres, D.; Das Sharma, S.; Tang, G.; Canoll, P.; Sims, P.A. Ligation-free ribosome profiling of cell type-specific translation in the brain. Genome Biol. 2016, 17, 149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biever, A.; Glock, C.; Tushev, G.; Ciirdaeva, E.; Dalmay, T.; Langer, J.D.; Schuman, E.M. Monosomes actively translate synaptic mRNAs in neuronal processes. Science 2020, 367. [Google Scholar] [CrossRef] [PubMed]
- Darnell, R.B. RNA protein interaction in neurons. Annu. Rev. Neurosci. 2013, 36, 243–270. [Google Scholar] [CrossRef] [Green Version]
- Licatalosi, D.D.; Mele, A.; Fak, J.J.; Ule, J.; Kayikci, M.; Chi, S.W.; Clark, T.A.; Schweitzer, A.C.; Blume, J.E.; Wang, X.; et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 2008, 456, 464–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, J.; Chang, H.; Kwon, S.C.; Kim, B.; Kim, Y.; Choe, J.; Ha, M.; Kim, Y.K.; Kim, V.N. LIN28A is a suppressor of ER-associated translation in embryonic stem cells. Cell 2012, 151, 765–777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polymenidou, M.; Lagier-Tourenne, C.; Hutt, K.R.; Huelga, S.C.; Moran, J.; Liang, T.Y.; Ling, S.C.; Sun, E.; Wancewicz, E.; Mazur, C.; et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat. Neurosci. 2011, 14, 459–468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tollervey, J.R.; Curk, T.; Rogelj, B.; Briese, M.; Cereda, M.; Kayikci, M.; Konig, J.; Hortobagyi, T.; Nishimura, A.L.; Zupunski, V.; et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat. Neurosci. 2011, 14, 452–458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katz, Y.; Wang, E.T.; Airoldi, E.M.; Burge, C.B. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods 2010, 7, 1009–1015. [Google Scholar] [CrossRef]
- Konig, J.; Zarnack, K.; Rot, G.; Curk, T.; Kayikci, M.; Zupan, B.; Turner, D.J.; Luscombe, N.M.; Ule, J. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat. Struct. Mol. Biol. 2010, 17, 909–915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugimoto, Y.; Chakrabarti, A.M.; Luscombe, N.M.; Ule, J. Using hiCLIP to identify RNA duplexes that interact with a specific RNA-binding protein. Nat. Protoc. 2017, 12, 611–637. [Google Scholar] [CrossRef] [PubMed]
- Eberwine, J.; Sul, J.Y.; Bartfai, T.; Kim, J. The promise of single-cell sequencing. Nat. Methods 2014, 11, 25–27. [Google Scholar] [CrossRef] [PubMed]
- Nawy, T. Single-cell sequencing. Nat. Methods 2014, 11, 18. [Google Scholar] [CrossRef] [PubMed]
- Heiman, M.; Schaefer, A.; Gong, S.; Peterson, J.D.; Day, M.; Ramsey, K.E.; Suarez-Farinas, M.; Schwarz, C.; Stephan, D.A.; Surmeier, D.J.; et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell 2008, 135, 738–748. [Google Scholar] [CrossRef] [Green Version]
- Sanz, E.; Yang, L.; Su, T.; Morris, D.R.; McKnight, G.S.; Amieux, P.S. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc. Natl. Acad. Sci. USA 2009, 106, 13939–13944. [Google Scholar] [CrossRef] [Green Version]
- Knight, Z.A.; Tan, K.; Birsoy, K.; Schmidt, S.; Garrison, J.L.; Wysocki, R.W.; Emiliano, A.; Ekstrand, M.I.; Friedman, J.M. Molecular profiling of activated neurons by phosphorylated ribosome capture. Cell 2012, 151, 1126–1137. [Google Scholar] [CrossRef] [Green Version]
- Ye, L.; Allen, W.E.; Thompson, K.R.; Tian, Q.; Hsueh, B.; Ramakrishnan, C.; Wang, A.C.; Jennings, J.H.; Adhikari, A.; Halpern, C.H.; et al. Wiring and Molecular Features of Prefrontal Ensembles Representing Distinct Experiences. Cell 2016, 165, 1776–1788. [Google Scholar] [CrossRef] [Green Version]
- Simsek, D.; Tiu, G.C.; Flynn, R.A.; Byeon, G.W.; Leppek, K.; Xu, A.F.; Chang, H.Y.; Barna, M. The Mammalian Ribo-interactome Reveals Ribosome Functional Diversity and Heterogeneity. Cell 2017, 169, 1051–1065. [Google Scholar] [CrossRef] [Green Version]
- Luo, L.; Callaway, E.M.; Svoboda, K. Genetic Dissection of Neural Circuits: A Decade of Progress. Neuron 2018, 98, 256–281. [Google Scholar] [CrossRef] [Green Version]
- Le, T.T.; Savitz, J.; Suzuki, H.; Misaki, M.; Teague, T.K.; White, B.C.; Marino, J.H.; Wiley, G.; Gaffney, P.M.; Drevets, W.C.; et al. Identification and replication of RNA-Seq gene network modules associated with depression severity. Transl. Psychiatry 2018, 8, 180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathys, H.; Davila-Velderrain, J.; Peng, Z.; Gao, F.; Mohammadi, S.; Young, J.Z.; Menon, M.; He, L.; Abdurrob, F.; Jiang, X.; et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 2019, 570, 332–337. [Google Scholar] [CrossRef] [PubMed]
- Jordao, M.J.C.; Sankowski, R.; Brendecke, S.M.; Sagar Locatelli, G.; Tai, Y.H.; Tay, T.L.; Schramm, E.; Armbruster, S.; Hagemeyer, N.; Gross, O.; et al. Single-cell profiling identifies myeloid cell subsets with distinct fates during neuroinflammation. Science 2019, 363, eaat7554. [Google Scholar] [CrossRef] [PubMed]
- Velmeshev, D.; Schirmer, L.; Jung, D.; Haeussler, M.; Perez, Y.; Mayer, S.; Bhaduri, A.; Goyal, N.; Rowitch, D.H.; Kriegstein, A.R. Single-cell genomics identifies cell type-specific molecular changes in autism. Science 2019, 364, 685–689. [Google Scholar] [CrossRef]
- Meyuhas, O. Ribosomal Protein S6 Phosphorylation: Four Decades of Research. Int. Rev. Cell Mol. Biol. 2015, 320, 41–73. [Google Scholar]
- Biever, A.; Valjent, E.; Puighermanal, E. Ribosomal Protein S6 Phosphorylation in the Nervous System: From Regulation to Function. Front. Mol. Neurosci. 2015, 8, 75. [Google Scholar] [CrossRef] [Green Version]
- Gressner, A.M.; Wool, I.G. The phosphorylation of liver ribosomal proteins in vivo. Evidence that only a single small subunit protein (S6) is phosphorylated. J. Biol. Chem. 1974, 249, 6917–6925. [Google Scholar]
- Kelleher, R.J., 3rd; Govindarajan, A.; Jung, H.Y.; Kang, H.; Tonegawa, S. Translational control by MAPK signaling in long-term synaptic plasticity and memory. Cell 2004, 116, 467–479. [Google Scholar] [CrossRef] [Green Version]
- Tan, C.L.; Cooke, E.K.; Leib, D.E.; Lin, Y.C.; Daly, G.E.; Zimmerman, C.A.; Knight, Z.A. Warm-Sensitive Neurons that Control Body Temperature. Cell 2016, 167, 47–59. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Z.D.; Yang, W.Z.; Gao, C.; Fu, X.; Zhang, W.; Zhou, Q.; Chen, W.; Ni, X.; Lin, J.K.; Yang, J.; et al. A hypothalamic circuit that controls body temperature. Proc. Natl. Acad. Sci. USA 2017, 114, 2042–2047. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Gong, N.N.; Hu, X.S.; Ni, M.J.; Pasi, R.; Matsunami, H. Molecular profiling of activated olfactory neurons identifies odorant receptors for odors in vivo. Nat. Neurosci. 2015, 18, 1446–1454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Isogai, Y.; Wu, Z.; Love, M.I.; Ahn, M.H.; Bambah-Mukku, D.; Hua, V.; Farrell, K.; Dulac, C. Multisensory Logic of Infant-Directed Aggression by Males. Cell 2018, 175, 1827–1841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruvinsky, I.; Sharon, N.; Lerer, T.; Cohen, H.; Stolovich-Rain, M.; Nir, T.; Dor, Y.; Zisman, P.; Meyuhas, O. Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes Dev. 2005, 19, 2199–2211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guenthner, C.J.; Miyamichi, K.; Yang, H.H.; Heller, H.C.; Luo, L. Permanent genetic access to transiently active neurons via TRAP: Targeted recombination in active populations. Neuron 2013, 78, 773–784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeNardo, L.; Luo, L. Genetic strategies to access activated neurons. Curr. Opin. Neurobiol. 2017, 45, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Romanov, R.A.; Zeisel, A.; Bakker, J.; Girach, F.; Hellysaz, A.; Tomer, R.; Alpar, A.; Mulder, J.; Clotman, F.; Keimpema, E.; et al. Molecular interrogation of hypothalamic organization reveals distinct dopamine neuronal subtypes. Nat. Neurosci. 2017, 20, 176–188. [Google Scholar] [CrossRef]
- Haring, M.; Zeisel, A.; Hochgerner, H.; Rinwa, P.; Jakobsson, J.E.T.; Lonnerberg, P.; La Manno, G.; Sharma, N.; Borgius, L.; Kiehn, O.; et al. Neuronal atlas of the dorsal horn defines its architecture and links sensory input to transcriptional cell types. Nat. Neurosci. 2018, 21, 869–880. [Google Scholar] [CrossRef]
- Kim, D.W.; Yao, Z.; Graybuck, L.T.; Kim, T.K.; Nguyen, T.N.; Smith, K.A.; Fong, O.; Yi, L.; Koulena, N.; Pierson, N.; et al. Multimodal Analysis of Cell Types in a Hypothalamic Node Controlling Social Behavior. Cell 2019, 179, 713–728. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choe, H.K.; Cho, J. Comprehensive Genome-Wide Approaches to Activity-Dependent Translational Control in Neurons. Int. J. Mol. Sci. 2020, 21, 1592. https://doi.org/10.3390/ijms21051592
Choe HK, Cho J. Comprehensive Genome-Wide Approaches to Activity-Dependent Translational Control in Neurons. International Journal of Molecular Sciences. 2020; 21(5):1592. https://doi.org/10.3390/ijms21051592
Chicago/Turabian StyleChoe, Han Kyoung, and Jun Cho. 2020. "Comprehensive Genome-Wide Approaches to Activity-Dependent Translational Control in Neurons" International Journal of Molecular Sciences 21, no. 5: 1592. https://doi.org/10.3390/ijms21051592




