Intra-Molecular Homologous Recombination of Scarless Plasmid
Abstract
:1. Introduction
2. Results
2.1. Intra-HR into Plasmid
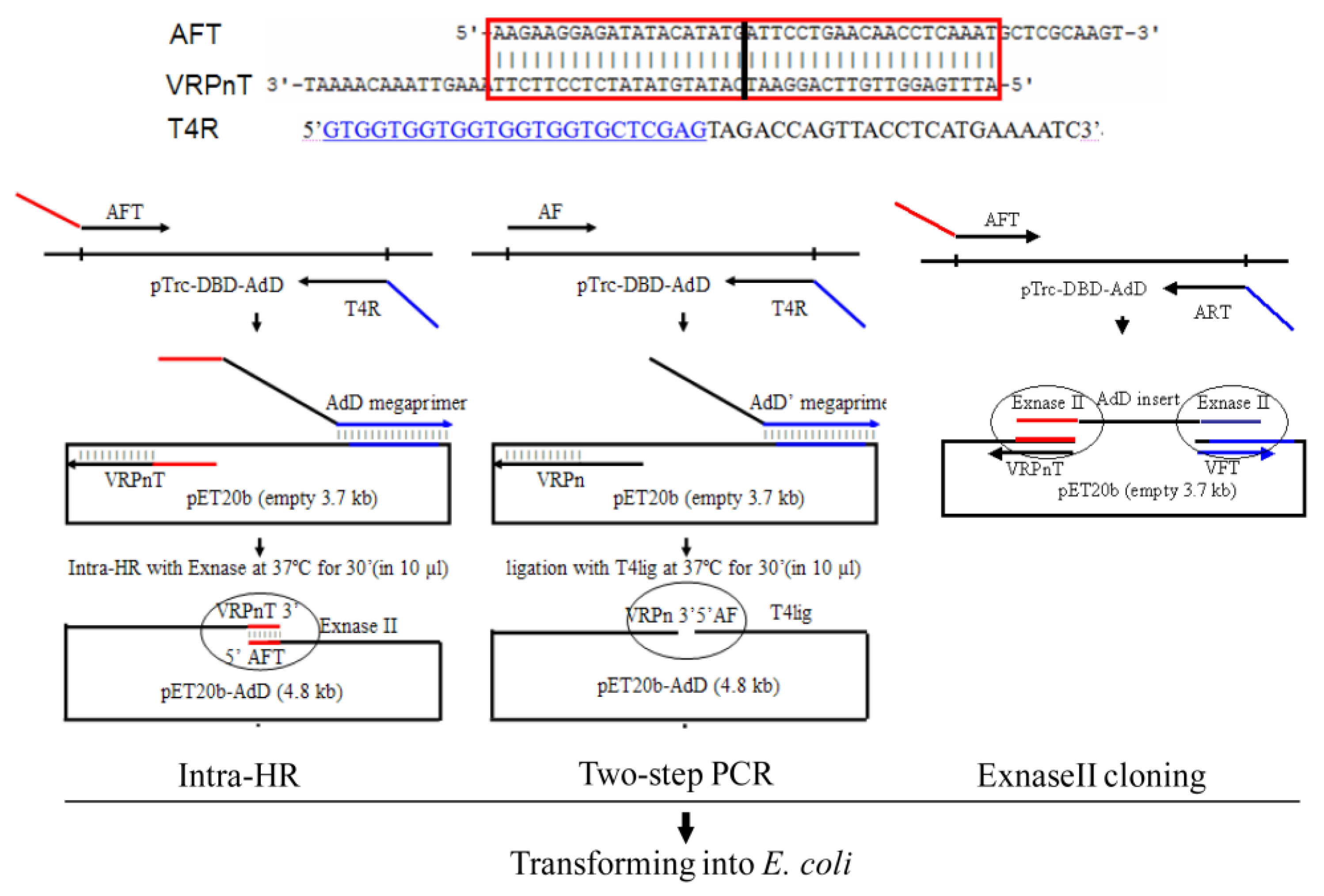
2.1.1. Protocol Overview
2.1.2. Primer Design Rule
2.1.3. Plasmid Amplification, Recombination, and Transformation
2.1.4. Transformant Efficiency and Plasmid Accuracy
2.1.5. The Intra-HR Generality
2.2. Comparing with the Two-Step PCR
2.3. Comparing with the ExnaseII Cloning
2.4. Optimal Length of Homology Arm
2.5. Origin of Transformant
3. Discussion
3.1. Advantages
3.2. Limitations
3.3. Scar-Producing Mechanism
4. Materials and Methods
4.1. Materials
4.2. Intra-HR of Plasmid pET20b-AdD
4.2.1. Megaprimer Amplification
4.2.2. Plasmid Amplification
4.2.3. Intra-Molecular HR and Transformation
4.3. Comparing with the Two-Step PCR
4.4. Comparing with the ExnaseII Cloning
4.4.1. Inter-Molecular HR
4.4.2. Intra-Molecular HR
4.5. Optimal Length of Homology Arm
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Liu, L.; Zhang, G.; Zhang, Z.; Wang, S.; Chen, H. Terminal amino-acids disturb xylanase thermostability and activity. J. Biol. Chem. 2011, 286, 44710–44715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, A.; Cheng, J.; Liu, M.; Shangguan, Y.; Liu, L. Sandwich fusion of CBM9_2 to enhance xylanase thermostability and activity. Int. J. Biol. Macromol. 2018, 117, 586–591. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001. [Google Scholar]
- Miyazaki, K.; Takenouchi, M. Creating random mutagenesis libraries using megaprimer PCR of whole plasmid. BioTechniques 2002, 33, 1036–1038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, B.; Cai, G.; Hall, E. In-fusion assembly: Seamless engineering of multidomain fusion proteins, modular vectors, and mutations. BioTechniques 2007, 43, 354–359. [Google Scholar] [CrossRef]
- Bryksin, A.; Matsumura, I. Overlap extension PCR cloning: A simple and reliable way to create recombinant plasmids. BioTechniques 2010, 48, 463–465. [Google Scholar] [CrossRef]
- Spiliotis, M. Inverse fusion PCR cloning. PLoS ONE 2012, 7, e35407. [Google Scholar] [CrossRef] [Green Version]
- Ulrich, A.; Andersen, K.; Schwartz, T. Exponential megapriming PCR (EMP) cloning--seamless DNA insertion into any target plasmid without sequence constraints. PLoS ONE 2012, 7, e53360. [Google Scholar] [CrossRef] [Green Version]
- Aslanidis, C.; de Jong, P. Ligation-independent cloning of PCR products (LIC-PCR). Nucleic Acids Res. 1990, 18, 6009–6074. [Google Scholar] [CrossRef]
- Chen, G.; Qiu, N.; Karrer, C.; Karrer, C. Restriction site-free insertion of PCR products directionally into vectors. BioTechniques 2000, 28, 498–500. [Google Scholar] [CrossRef]
- Geiser, M.; Cebe, R.; Drewello, D. Integration of PCR fragments at any specific site within cloning vectors without the use of restriction enzymes and DNA ligase. BioTechniques 2001, 31, 88–90. [Google Scholar] [CrossRef] [Green Version]
- Benoit, R.; Wilhelm, R.; Scherer-Becker, D.; Ostermeier, C. An improved method for fast, robust, and seamless integration of DNA fragments into multiple plasmids. Protein Expr. Purif. 2006, 45, 66–71. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Elledge, S. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat. Methods 2007, 4, 251–256. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Evans, R. Ligation independent cloning irrespective of restriction site compatibility. Nucleic Acids Res. 1997, 25, 4165–4166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Buchholz, F.; Muyrers, J.; Stewart, A. A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet. 1998, 20, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Bian, X.; Hu, S.; Wang, H.; Huang, F.; Seibert, P.; Plaza, A.; Xia, L.; Muller, R.; Stewart, A. Full-length RecE enhances linear-linear homologous recombination and facilitates direct cloning for bioprospecting. Nat. Biotechnol. 2012, 30, 440–446. [Google Scholar] [CrossRef]
- Rossi, R.; Montecucco, A.; Ciarrocchi, G.; Biamonti, G. Functional characterization of the T4 DNA ligase: A new insight into the mechanism of action. Nucleic Acids Res. 1997, 25, 2016–2113. [Google Scholar] [CrossRef]
- Lehman, I. DNA ligase: Structure, mechanism, and function. Science 1974, 186, 790–797. [Google Scholar] [CrossRef]
- Shuman, S. DNA ligases: Progress and prospects. J. Biol. Chem. 2009, 284, 17365–17369. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, K.; Nakazawa, M.; Ishizaki, Y.; Hiraoka, N.; Obayashi, A. Regulation of inter- and intramolecular ligation with T4 DNA ligase in the presence of polyethylene glycol. Nucleic Acids Res. 1986, 14, 7617–7631. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, M.; Uchida, T. Thermophilic HB8 DNA Ligase: Effects of polyethylene glycols and polyamines on blunt-end ligation of DNA. J. Biochem. 1986, 100, 123–131. [Google Scholar] [CrossRef]
- Lu, Q. Seamless cloning and gene fusion. Trends Biotechnol. 2005, 23, 199–202. [Google Scholar] [CrossRef] [PubMed]
- Genebridges. Direct cloning-proficient E. coli strain GB05-dir. Technical Protocol 2014, 1, 4. [Google Scholar]
- Muyrers, J.; Zhang, Y.; Buchholz, F.; Stewart, A. RecE/RecT and Redalpha/Redbeta initiate double-stranded break repair by specifically interacting with their respective partners. Genes Dev. 2000, 14, 1971–1982. [Google Scholar] [PubMed]
- Zhang, Y.; Werling, Y.; Edelmann, W. SLiCE: A novel bacterial cell extract-based DNA cloning method. Nucleic Acids Res. 2012, 40, e55. [Google Scholar] [CrossRef] [Green Version]
- Pascal, J.; Brien, P.; Tomkinson, A.; Ellenberger, T. Human DNA ligase I completely encircles and partially unwinds nicked DNA. Nature 2004, 432, 473–478. [Google Scholar] [CrossRef]
- Gibson, D.; Young, L.; Chuang, R.; Venter, J.; Hutchison, C.r.; Smith, H. Integration of PCR fragments at any specific site within cloning vectors without the use of restriction enzymes and DNA ligase. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef]
- Bologna, S.; Altmannova, V.; Valtorta, E.; Koenig, C.; Liberali, P.; Gentili, C.; Anrather, D.; Ammerer, G.; Pelkmans, L.; Krejci, L.; et al. Sumoylation regulates EXO1 stability and processing of DNA damage. Cell Cycle 2015, 142, 2439–2450. [Google Scholar] [CrossRef] [Green Version]
- Ahel, I.; Rass, U.; El-Khamisy, S.; Katyal, S.; Clements, P.; McKinnon, P.; Caldecott, K.; West, S. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature 2006, 443, 713–716. [Google Scholar] [CrossRef]
- Zahn, K.E.; Averill, A.; Aller, P.; Wood, R.; Doublie, S. Human DNA polymerase theta grasps the primer terminus to mediate DNA repair. Nat. Struct. Mol. Biol. 2015, 22, 304–311. [Google Scholar] [CrossRef] [Green Version]
- Khodaverdian, V.; Hanscom, T.; Yu, A.; Yu, T.; Mak, V.; Brown, A.; Roberts, S.; McVey, M. Secondary structure forming sequences drive SD-MMEJ repair of DNA double-strand breaks. Nucleic Acids Res. 2017, 45, 12848–12861. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Yang, A.; Shangguan, Y.; He, T.; Xu, W.; Liu, L. Amplification of DNA with double annealing temperature PCR. Acta Microbiol. Sin. 2017, 57, 1262–1269. [Google Scholar]





| Method | Enzyme/Reaction | Plasmids | Efficiency (Transformants/ng) | Nicked DNA (ng) | Accuracy (%)/Scarless Plasmids |
|---|---|---|---|---|---|
| Intra-HR in parallel with the two-step PCR | ExanseII 37°C, 30 min/intra-molecular HR | pET20b-AdD (1′ in Figure 3) | 6 ± 0.5 | 7.2 | 50/1′L1-2,4-5,7 |
| pET21a-AdD | 9.8 ± 1.5 | 14.1 | 80/L1-4/7-10 | ||
| pET22b-AdD | 37.4 ± 6.8 | 41.3 | 90/L1-4/6-10 | ||
| Two-step PCR | T4 DNA ligase 16°C, 16 h/intra-molecular ligation | pET20b-AdD (2′ in Figure 3) | 2.1 ± 0.5 | 16.2 | 0/ |
| pET21a-AdD | 25 ± 0.1 | 30.5 | 50/L2,4,6-7,9 | ||
| pET22b-AdD | 32.2 ± 5.7 | 27.9 | 70/L1-4,8-10 | ||
| ExnaseII cloning | ExanseII 37°C, 30 min/inter-molecular HR | pET20b-AdD with inter-20bp | 5.7 ± 1.1 | nd | 80 |
| pET20b-AdD with inter-40bp | 13.5 ± 2.7 | nd | 30 | ||
| Intra-HR in parallel with the ExnaseII cloning | ExanseII 37°C, 30 min/intra-molecular HR | pET20b-AdD with intra-20bp | 9.7 ± 1.2 | nd | 80 |
| pET20b-AdD with intra-40bp | 13.7 ± 2.3 | nd | 90 | ||
| Intra-HR for optimal arm length | ExanseII 37°C, 30 min/intra-molecular HR | pET20b-AdD with intra-10bp | 12.7 ± 0.4 | nd | 90 |
| pET20b-AdD with intra-30bp | 14.2 ± 0.2 | nd | 60 | ||
| pET20b-AdD with intra-50bp | 11.4 ± 0.9 | nd | 80 |
| Primer | Sequence | Tm (°C) |
|---|---|---|
| AFT | AAGAAGGAGATATACATATG‖ ATTCCTGAACAACCTCAAATGCTCGCAAGT | 72 |
| T4R | GTGGTGGTGGTGGTGGTGCTCGAGTAGACCAGTTACCTCATGAAAATC | 61 |
| VRPnT | ATTTGAGGTTGTTCAGGAAT‖ CATATGTATATCTCCTTCTTAAAGTTAAACAAAAT | 61 |
| AF | p-ATTCCTGAACAACCTCAAATG | 72 |
| VRPn | p-CATATGTATATCTCCTTCTTAAAGTTAAACAAAAT | 62 |
| AF-L | ATTCCTGAACAACCTCAAATGCTCGCAAGT | 72 |
| AR-L | TTCTTTAAATTTATAAAGATTTTTTGAACG | 57 |
| ART | TGGTGGTGGTGGTGCTCGAGTTCTTTAAATTTATAAAGATTTTTTGAACG | 57 |
| VF | CTCGAGCACCACCACCACC | 65.2 |
| VFT | ATCTTTATAAATTTAAAGAACTCGAGCACCACCACCACCA | 73 |
| ARP1 | GTTCAGGAATCATATGTATATCTCCTTCTTAAAGTTAAACAAAATTATTTCTAGA | 68 |
| ARP3 | ACTTGCGAGCATTTGAGGTTGTTCAGGAATCATATGTATATCTCCTTCTTAAAGT | 75 |
| ARP5 | ATGCCTTTTTCATCATAAGAACTTGCGAGCATTTGAGGTTGTTCAGGAATCATAT | 76 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, Y.; Zhang, Y.; Liu, L. Intra-Molecular Homologous Recombination of Scarless Plasmid. Int. J. Mol. Sci. 2020, 21, 1697. https://doi.org/10.3390/ijms21051697
Liang Y, Zhang Y, Liu L. Intra-Molecular Homologous Recombination of Scarless Plasmid. International Journal of Molecular Sciences. 2020; 21(5):1697. https://doi.org/10.3390/ijms21051697
Chicago/Turabian StyleLiang, Yaping, Yu Zhang, and Liangwei Liu. 2020. "Intra-Molecular Homologous Recombination of Scarless Plasmid" International Journal of Molecular Sciences 21, no. 5: 1697. https://doi.org/10.3390/ijms21051697
APA StyleLiang, Y., Zhang, Y., & Liu, L. (2020). Intra-Molecular Homologous Recombination of Scarless Plasmid. International Journal of Molecular Sciences, 21(5), 1697. https://doi.org/10.3390/ijms21051697





