Arabidopsis RETICULON-LIKE4 (RTNLB4) Protein Participates in Agrobacterium Infection and VirB2 Peptide-Induced Plant Defense Response
Abstract
1. Introduction
2. Results
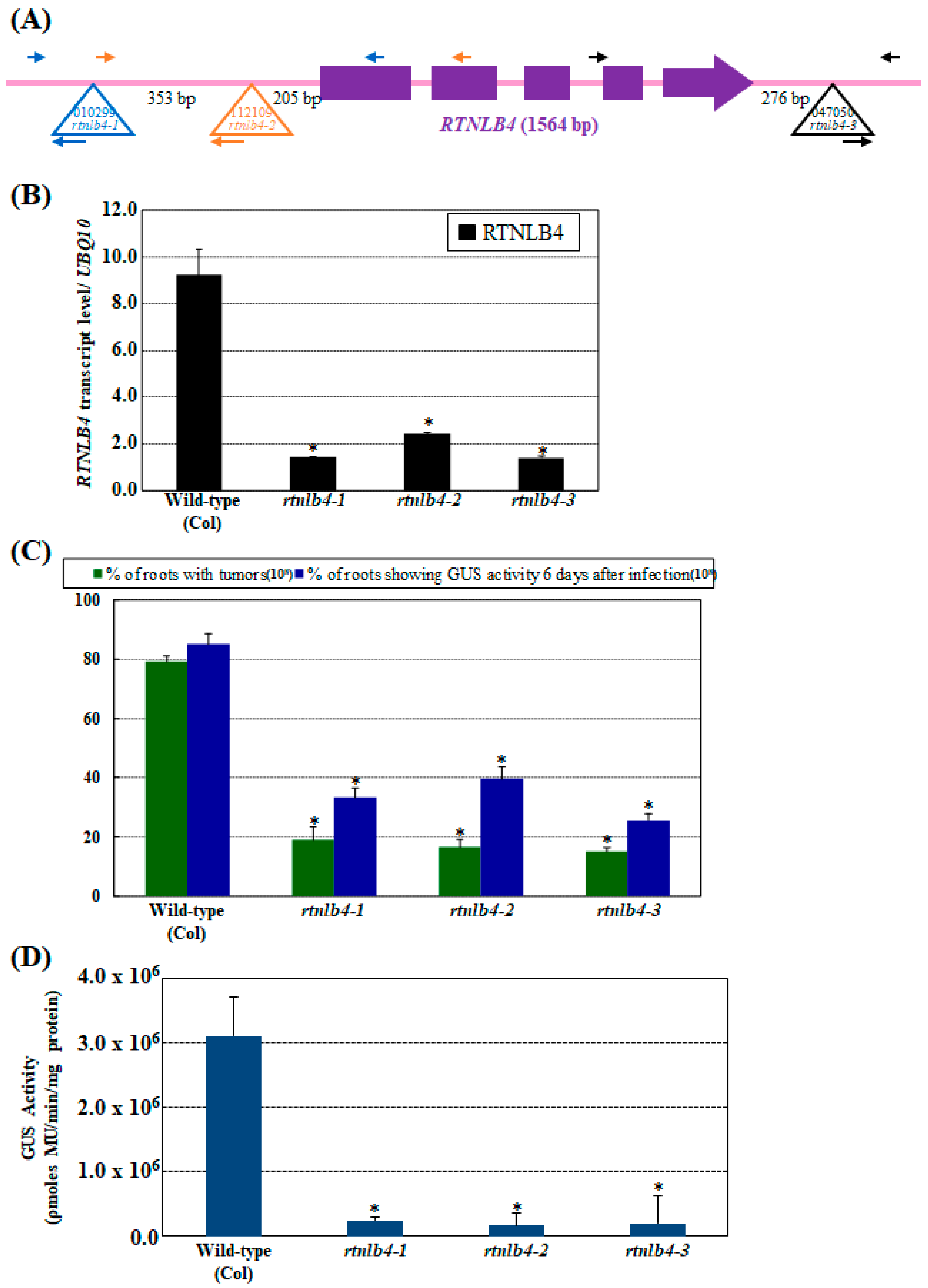
2.1. rtnlb4 Mutants Were Recalcitrant to Agrobacterium-Mediated Transformation in Roots and Seedlings
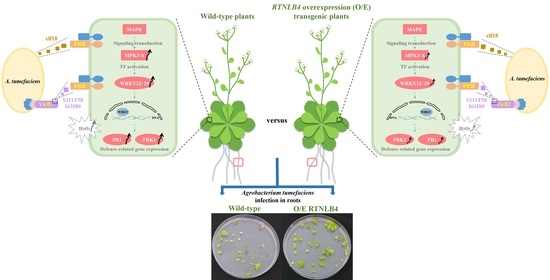
2.2. Overexpression of RTNLB4 in Transgenic Plants Enhanced A. tumefaciens Infection Rates in Roots and Seedlings
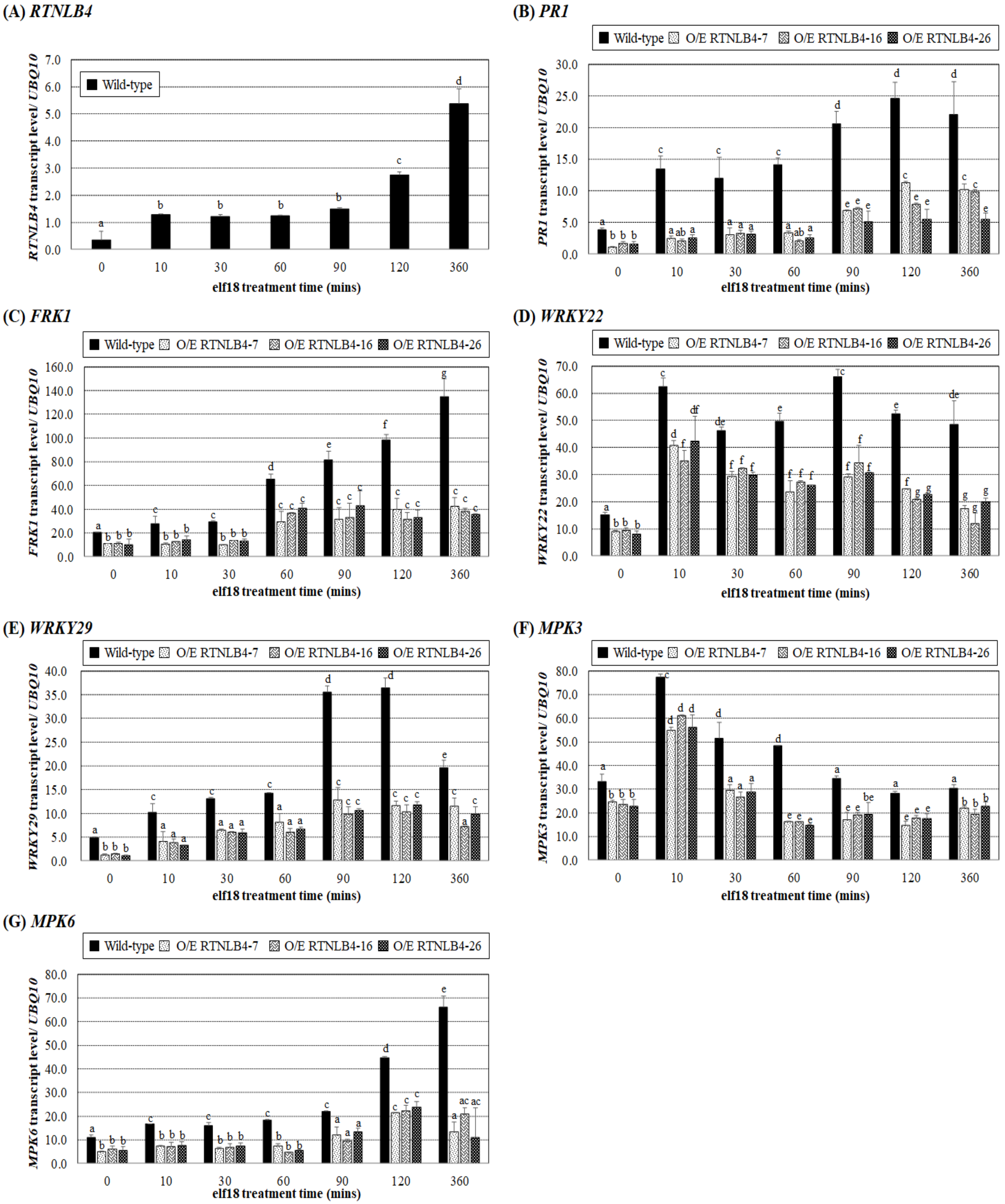
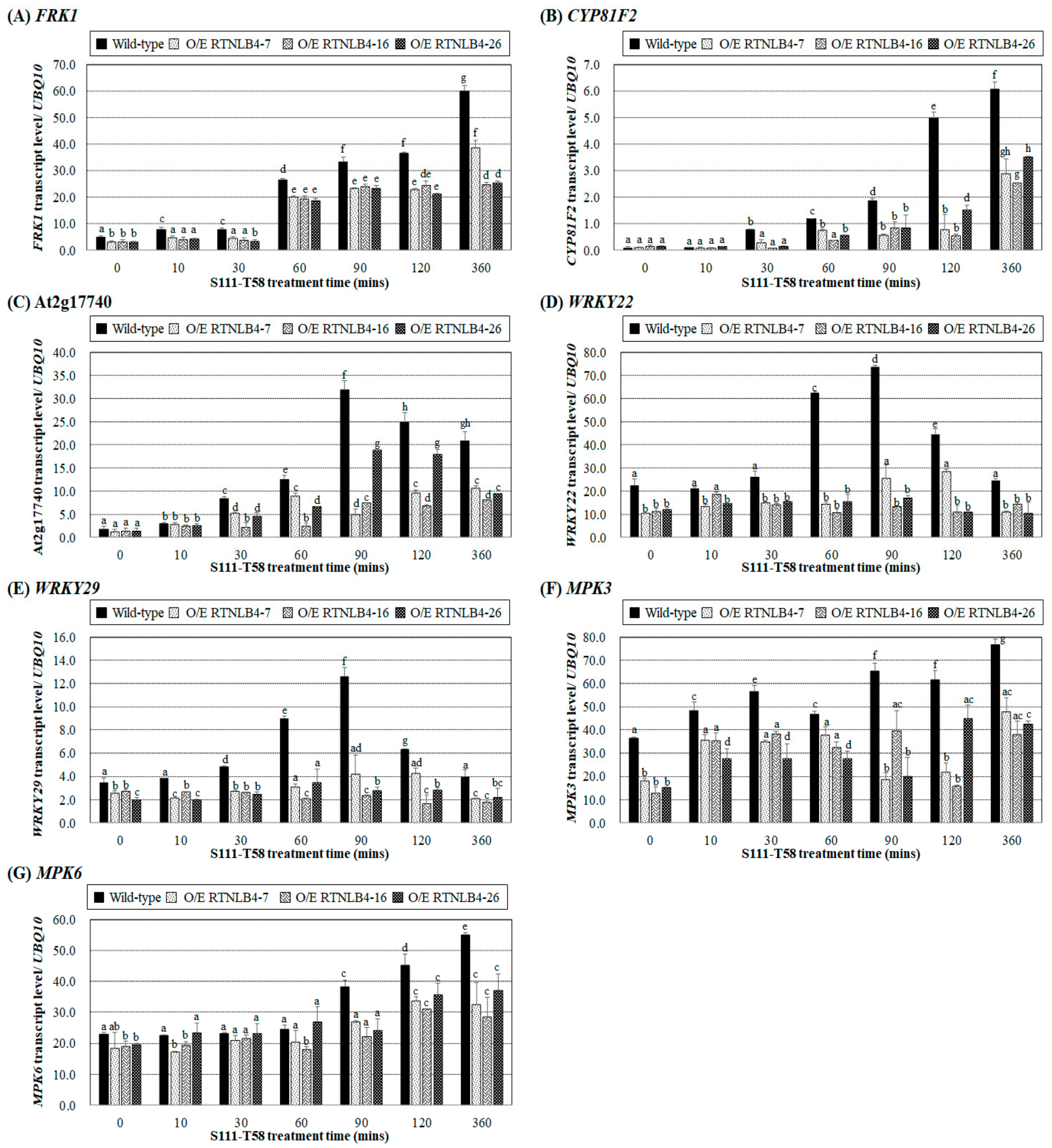
2.3. Induced Expression of Defense-Related Genes Was Affected in both RTNLB4 O/E Transgenic Plants and rtnlb4 Mutants after elf18 Treatment
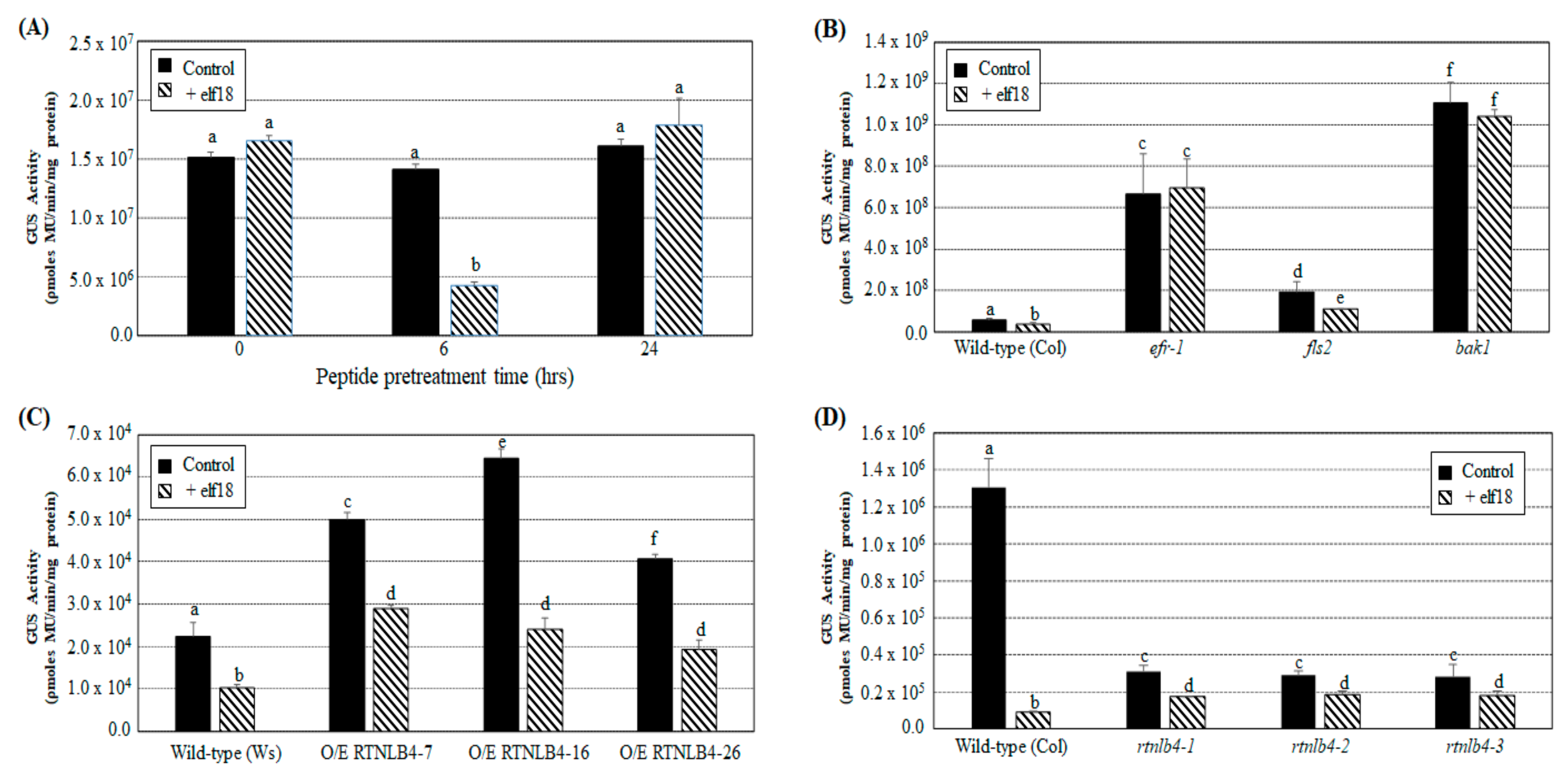
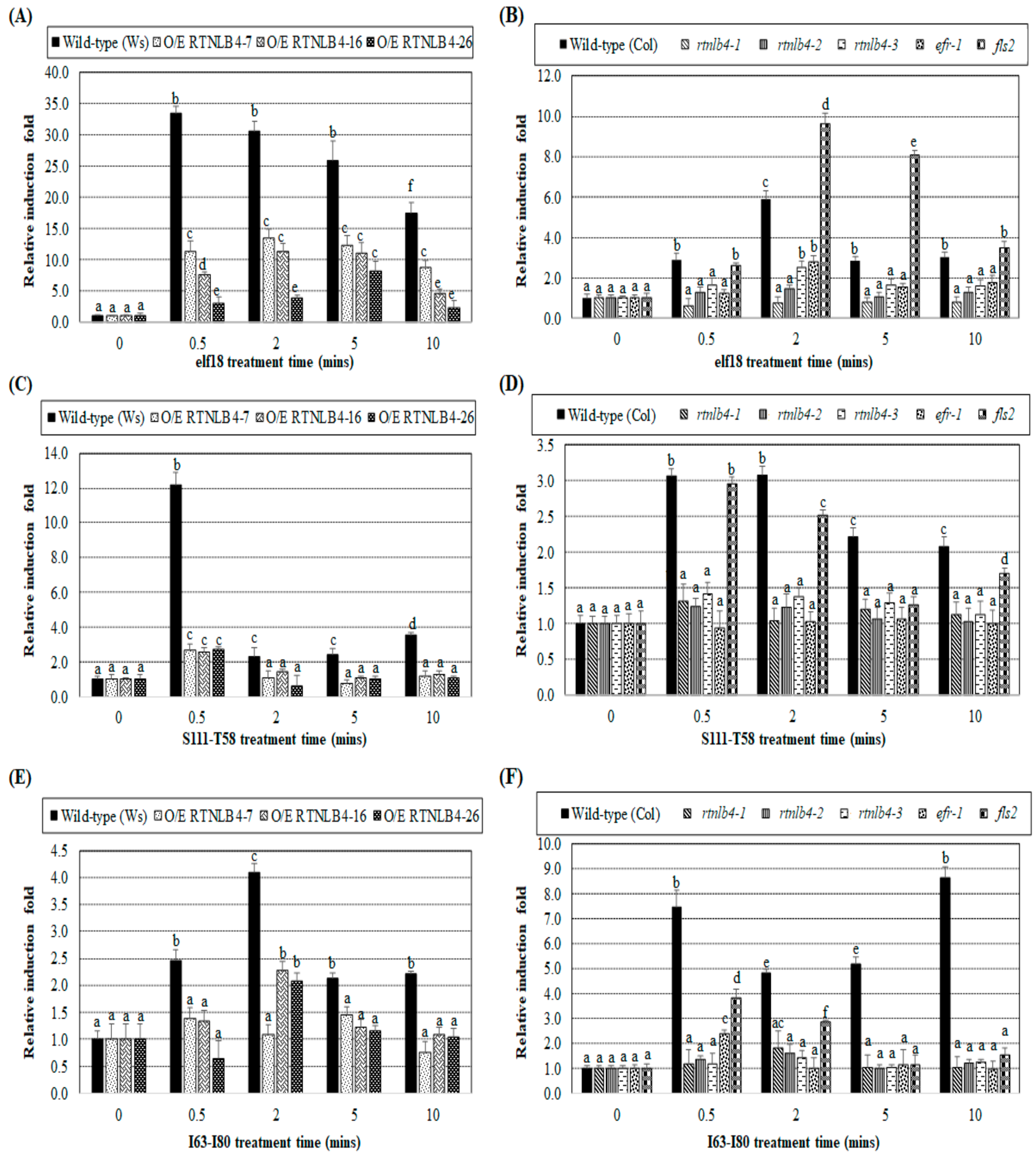
2.4. Agrobacterium-Mediated Transient Expression Efficiency Is Decreased in Wild-Type Plants and in RTNLB4 O/E Transgenic Plants to a Lesser Extent after elf18 Peptide Pretreatment
2.5. VirB2 Peptide Pretreatment Affected Transient T-DNA Expression in Wild-Type, RTNLB4 O/E Transgenic, and rtnlb4 Mutant Plants
2.6. Levels of Defense-Related Genes in RTNLB4 O/E Transgenic and rtnlb4 Mutant Plants Were Less Induced after VirB2 Peptide Treatment
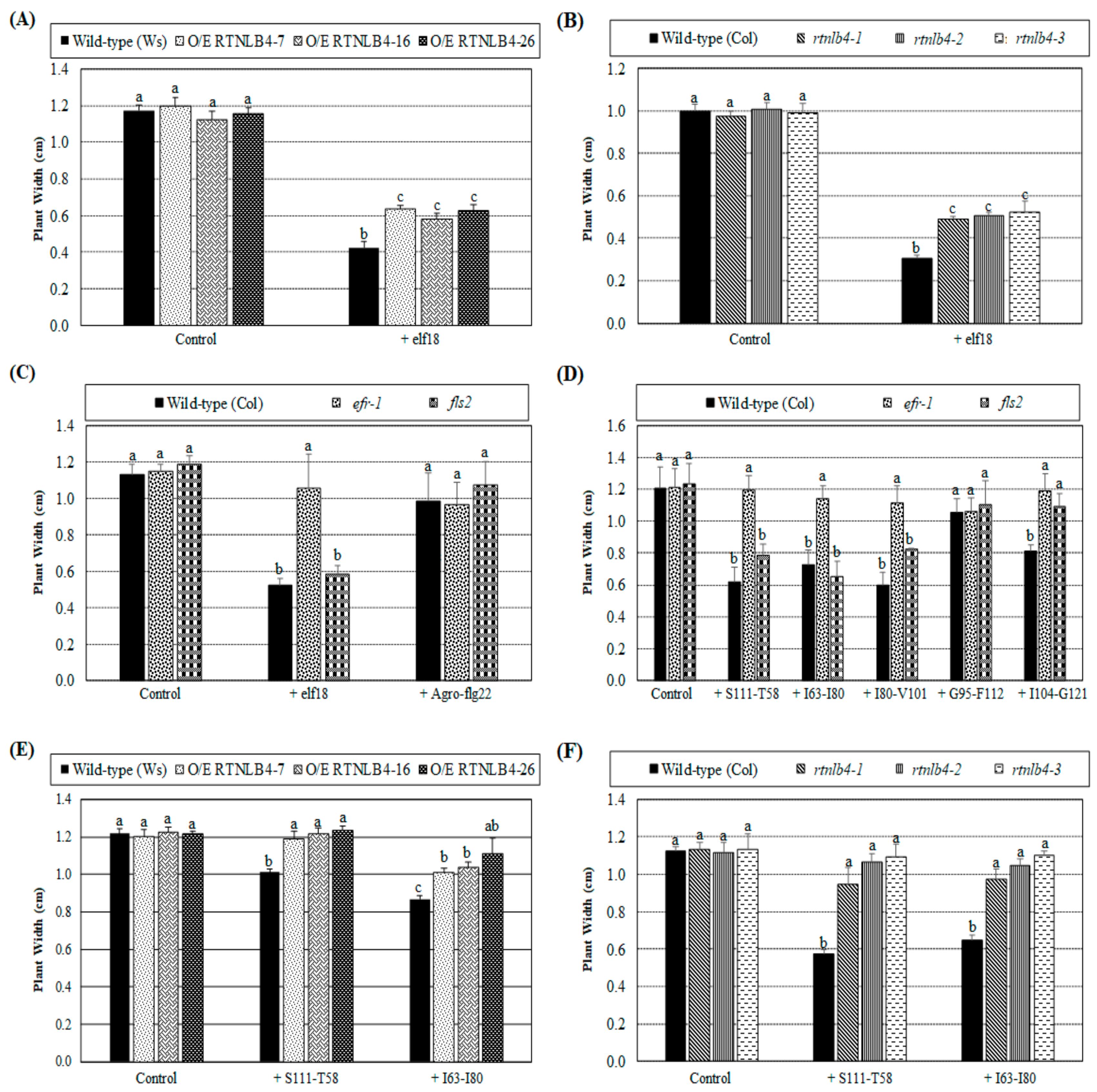
2.7. Arabidopsis Seedling Growth Was Less Inhibited in RTNLB4 O/E Transgenic and rtnlb4 Mutant Plants after elf18 and VirB2 Peptide Treatments
2.8. H2O2 Accumulation Was Lower in RTNLB4 O/E Transgenic and rtnlb4 Mutant Plants after elf18 and VirB2 Peptide Treatment
3. Discussion
3.1. RTNLB4 Plays a Role in Plant Defense Responses and Affects A. tumefaciens Infection
3.2. Elf18 and VirB2 Peptides May Induce a Common Set of Plant Defense Responses
4. Materials and Methods
4.1. Generation of RTNLB4 and T7-tagged-RTNLB4 Overexpression (O/E) Arabidopsis Thaliana Transgenic Plants
4.2. DNA Isolation from Arabidopsis Plants and Genomic DNA PCR Analysis
4.3. RNA Isolation from Arabidopsis Plants and Quantitative Real-Time PCR (qPCR) Analysis
4.4. Protein Extraction from Arabidopsis Plants and Protein Gel Blot Analysis
4.5. Agrobacterium Tumefaciens-Mediated Stable, Transient Root and Seedling Transformation Assays of rtnlb4 Mutant Plants and RTNLB4 O/E Arabidopsis Transgenic Plants
4.6. Isolation of Pili of A. tumefaciens
4.7. Seedling Growth Inhibition Assays
4.8. Hydrogen Peroxide (H2O2) Detection by Ferrous Oxidation Xylenol Orange (XO) Assays
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Hwang, H.H.; Yu, M.; Lai, E.M. Agrobacterium-mediated plant transformation: Biology and applications. Arab. Book 2017, 15, e0186. [Google Scholar] [CrossRef] [PubMed]
- Lacroix, B.; Citovsky, V. The roles of bacterial and host plant factors in Agrobacterium-mediated genetic transformation. Int. J. Dev. Biol. 2013, 57, 467–481. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.H.; Gao, R.; Binns, A.N.; Lynn, D.G. Capturing the VirA/VirG TCS of Agrobacterium tumefaciens. Adv. Exp. Med. Biol. 2008, 631, 161–177. [Google Scholar] [CrossRef] [PubMed]
- Christie, P.J. The mosaic type IV secretion systems. EcoSal Plus 2016, 7. [Google Scholar] [CrossRef]
- Li, Y.G.; Christie, P.J. The Agrobacterium VirB/VirD4 T4SS: Mechanism and architecture defined through in vivo mutagenesis and chimeric systems. Curr. Top. Microbiol. Immunol. 2018, 418, 233–260. [Google Scholar] [CrossRef]
- Vergunst, A.C.; Schrammeijer, B.; den Dulk-Ras, A.; de Vlaam, C.M.; Regensburg-Tuink, T.J.; Hooykaas, P.J. VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science 2000, 290, 979–982. [Google Scholar] [CrossRef]
- Duckely, M.; Hohn, B. The VirE2 protein of Agrobacterium tumefaciens: The Yin and Yang of T-DNA transfer. FEMS Lett. 2003, 223, 1–6. [Google Scholar] [CrossRef]
- Li, X.; Pan, S.Q. Agrobacterium delivers VirE2 protein into host cells via clathrin-mediated endocytosis. Sci. Adv. 2017, 3, e1601528. [Google Scholar] [CrossRef]
- Tu, H.; Li, X.; Yang, Q.; Peng, L.; Pan, S.Q. Real-time trafficking of Agrobacterium virulence protein VirE2 inside host cells. Curr. Top. Microbiol. Immunol. 2018, 418, 261–286. [Google Scholar] [CrossRef]
- Yang, Q.; Li, X.; Tu, H.; Pan, S.Q. Agrobacterium-delivered virulence protein VirE2 is trafficked inside host cells via a myosin XI-K-powered ER/actin network. Proc. Natl. Acad. Sci. USA 2017, 114, 2982–2987. [Google Scholar] [CrossRef]
- Ballas, N.; Citovsky, V. Nuclear localization signal binding protein from Arabidopsis mediates nuclear import of Agrobacterium VirD2 protein. Proc. Natl. Acad. Sci. USA 1997, 94, 10723–10728. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, S.; Lee, L.Y.; Oltmanns, H.; Cao, H.; Veena Cuperus, J.; Gelvin, S.B. IMPa-4, an Arabidopsis importin alpha isoform, is preferentially involved in Agrobacterium-mediated plant transformation. Plant Cell 2008, 20, 2661–2680. [Google Scholar] [CrossRef]
- Gelvin, S.B. Integration of Agrobacterium T-DNA into the plant genome. Annu. Rev. Genet. 2017, 51, 195–217. [Google Scholar] [CrossRef] [PubMed]
- Djamei, A.; Pitzschke, A.; Nakagami, H.; Rajh, I.; Hirt, H. Trojan horse strategy in Agrobacterium transformation: Abusing MAPK defense signaling. Science 2007, 318, 453–456. [Google Scholar] [CrossRef] [PubMed]
- Pitzschke, A. Agrobacterium infection and plant defense-transformation success hangs by a thread. Front. Plant Sci. 2013, 4, 519. [Google Scholar] [CrossRef]
- Pitzschke, A.; Djamei, A.; Teige, M.; Hirt, H. VIP1 response elements mediate mitogen-activated protein kinase 3-induced stress gene expression. Proc. Natl. Acad. Sci. USA 2009, 106, 18414–18419. [Google Scholar] [CrossRef]
- Garcia-Cano, E.; Hak, H.; Magori, S.; Lazarowitz, S.G.; Citovsky, V. The Agrobacterium F-Box protein effector VirF destabilizes the Arabidopsis GLABROUS1 enhancer/binding protein-like transcription factor VFP4, a transcriptional activator of defense response genes. Mol. Plant Microbe Interact. 2018, 31, 576–586. [Google Scholar] [CrossRef]
- Tzfira, T.; Vaidya, M.; Citovsky, V. VIP1, an Arabidopsis protein that interacts with Agrobacterium VirE2, is involved in VirE2 nuclear import and Agrobacterium infectivity. EMBO J. 2001, 20, 3596–3607. [Google Scholar] [CrossRef]
- Zaltsman, A.; Krichevsky, A.; Loyter, A.; Citovsky, V. Agrobacterium induces expression of a host F-box protein required for tumorigenicity. Cell Host Microbe 2010, 7, 197–209. [Google Scholar] [CrossRef]
- Zaltsman, A.; Lacroix, B.; Gafni, Y.; Citovsky, V. Disassembly of synthetic Agrobacterium T-DNA-protein complexes via the host SCF (VBF) ubiquitin-ligase complex pathway. Proc. Natl. Acad. Sci. USA 2013, 110, 169–174. [Google Scholar] [CrossRef]
- Tzfira, T.; Vaidya, M.; Citovsky, V. Involvement of targeted proteolysis in plant genetic transformation by Agrobacterium. Nature 2004, 431, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Boutrot, F.; Zipfel, C. Function, discovery, and exploitation of plant pattern recognition receptors for broad-spectrum disease resistance. Annu. Rev. Phytopathol. 2017, 55, 257–286. [Google Scholar] [CrossRef] [PubMed]
- Saijo, Y.; Loo, E.P.; Yasuda, S. Pattern recognition receptors and signaling in plant-microbe interactions. Plant J. 2018, 93, 592–613. [Google Scholar] [CrossRef] [PubMed]
- Felix, G.; Duran, J.D.; Volko, S.; Boller, T. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 1999, 18, 265–276. [Google Scholar] [CrossRef]
- Chinchilla, D.; Bauer, Z.; Regenass, M.; Boller, T.; Felix, G. The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 2006, 18, 465–476. [Google Scholar] [CrossRef]
- Ali, G.S.; Reddy, A. PAMP-triggered immunity: Early events in the activation of FLAGELLIN SENSITIVE2. Plant Signal. Behav. 2008, 3, 423–426. [Google Scholar] [CrossRef]
- Asai, T.; Tena, G.; Plotnikova, J.; Willmann, M.R.; Chiu, W.L.; Gomez-Gomez, L.; Boller, T.; Ausubel, F.M.; Sheen, J. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 2002, 415, 977–983. [Google Scholar] [CrossRef]
- Heese, A.; Hann, D.R.; Gimenez-Ibanez, S.; Jones, A.M.; He, K.; Li, J.; Schroeder, J.I.; Peck, S.C.; Rathjen, J.P. The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. USA 2007, 104, 12217–12222. [Google Scholar] [CrossRef]
- Ishihama, N.; Yoshioka, H. Post-translational regulation of WRKY transcription factors in plant immunity. Curr. Opin. Plant Biol. 2012, 15, 431–437. [Google Scholar] [CrossRef]
- Meng, X.; Zhang, S. MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol. 2013, 51, 245–266. [Google Scholar] [CrossRef]
- Rasmussen, M.W.; Roux, M.; Petersen, M.; Mundy, J. MAP kinase cascades in Arabidopsis innate immunity. Front. Plant Sci. 2012, 3, 169. [Google Scholar] [CrossRef] [PubMed]
- Zipfel, C.; Robatzek, S.; Navarro, L.; Oakeley, E.J.; Jones, J.D.; Felix, G.; Boller, T. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 2004, 428, 764–767. [Google Scholar] [CrossRef] [PubMed]
- Zipfel, C.; Kunze, G.; Chinchilla, D.; Caniard, A.; Jones, J.D.; Boller, T.; Felix, G. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 2006, 125, 749–760. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Zavaliev, R.; Dong, X. Membrane trafficking in plant immunity. Mol. Plant 2017, 10, 1026–1034. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.M.; Liu, P.Q.; Xu, Y.J.; Xiao, S. Protein trafficking during plant innate immunity. J. Integr. Plant Biol. 2016, 58, 284–298. [Google Scholar] [CrossRef]
- Yun, H.S.; Kwon, C. Vesicle trafficking in plant immunity. Curr. Opin. Plant Biol. 2017, 40, 34–42. [Google Scholar] [CrossRef]
- Nziengui, H.; Schoefs, B. Functions of reticulons in plants: What we can learn from animals and yeasts. Cell Mol. Life Sci. 2009, 66, 584–595. [Google Scholar] [CrossRef]
- Oertle, T.; Schwab, M.E. Nogo and its paRTNers. Trends Cell Biol. 2003, 13, 187–194. [Google Scholar] [CrossRef]
- Nziengui, H.; Bouhidel, K.; Pillon, D.; Der, C.; Marty, F.; Schoefs, B. Reticulon-like proteins in Arabidopsis thaliana: Structural organization and ER localization. FEBS Lett. 2007, 581, 3356–3362. [Google Scholar] [CrossRef]
- Sparkes, I.; Tolley, N.; Aller, I.; Svozil, J.; Osterrieder, A.; Botchway, S.; Mueller, C.; Frigerio, L.; Hawes, C. Five Arabidopsis reticulon isoforms share endoplasmic reticulum location, topology, and membrane-shaping properties. Plant Cell 2010, 22, 1333–1343. [Google Scholar] [CrossRef]
- Tolley, N.; Sparkes, I.A.; Hunter, P.R.; Craddock, C.P.; Nuttall, J.; Roberts, L.M.; Hawes, C.; Pedrazzini, E.; Frigerio, L. Overexpression of a plant reticulon remodels the lumen of the cortical endoplasmic reticulum but does not perturb protein transport. Traffic 2008, 9, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Hwang, H.H.; Gelvin, S.B. Plant proteins that interact with VirB2, the Agrobacterium pilin protein, mediate plant transformation. Plant Cell 2004, 16, 3148–3167. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.Y.; Bowen, C.H.; Popescu, G.V.; Kang, H.G.; Kato, N.; Ma, S.; Dinesh-Kumar, S.; Snyder, M.; Popescu, S.C. Arabidopsis RTNLB1 and RTNLB2 reticulon-like proteins regulate intracellular trafficking and activity of the FLS2 immune receptor. Plant Cell 2011, 23, 3374–3391. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.C.; Fu, B.J.; Liu, Y.T.; Chang, Y.R.; Chi, S.F.; Chien, P.R.; Huang, S.C.; Hwang, H.H. Arabidopsis RETICULON-LIKE3 (RTNLB3) and RTNLB8 participate in Agrobacterium-mediated plant transformation. Int. J. Mol. Sci. 2018, 19, 638. [Google Scholar] [CrossRef]
- Knox, K.; Wang, P.; Kriechbaumer, V.; Tilsner, J.; Frigerio, L.; Sparkes, I.; Hawes, C.; Oparka, K. Putting the squeeze on plasmodesmata: A Role for reticulons in primary plasmodesmata formation. Plant Physiol. 2015, 168, 1563–1572. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.H.; Lee, S.C.; Wang, C.W. Viral protein targeting to the cortical endoplasmic reticulum is required for cell-cell spreading in plants. J. Cell Biol. 2011, 193, 521–535. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.Y.; Liu, K.H.; Wang, Y.C.; Wu, J.F.; Chiu, W.L.; Chen, C.Y.; Wu, S.H.; Sheen, J.; Lai, E.M. AGROBEST: An efficient Agrobacterium-mediated transient expression method for versatile gene function analyses in Arabidopsis seedlings. Plant Methods 2014, 10, 19. [Google Scholar] [CrossRef]
- Eisenbrandt, R.; Kalkum, M.; Lai, E.M.; Lurz, R.; Kado, C.I.; Lanka, E. Conjugative pili of IncP plasmids, and the Ti plasmid T pilus are composed of cyclic subunits. J. Biol. Chem. 1999, 274, 22548–22555. [Google Scholar] [CrossRef]
- Lai, E.M.; Chesnokova, O.; Banta, L.M.; Kado, C.I. Genetic and environmental factors affecting T-pilin export and T-pilus biogenesis in relation to flagellation of Agrobacterium tumefaciens. J. Bacteriol. 2000, 182, 3705–3716. [Google Scholar] [CrossRef]
- Lai, E.M.; Eisenbrandt, R.; Kalkum, M.; Lanka, E.; Kado, C.I. Biogenesis of T pili in Agrobacterium tumefaciens requires precise VirB2 propilin cleavage and cyclization. J. Bacteriol. 2002, 184, 327–330. [Google Scholar] [CrossRef]
- Kunze, G.; Zipfel, C.; Robatzek, S.; Niehaus, K.; Boller, T.; Felix, G. The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 2004, 16, 3496–3507. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.Y.; Chen, C.Y.; Lai, E.M. Expression and functional characterization of the Agrobacterium VirB2 amino acid substitution variants in T-pilus biogenesis, virulence, and transient transformation efficiency. PLoS ONE 2014, 9, e101142. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: New York, NY, USA, 2005; pp. 571–607. [Google Scholar]
- Peptide Hydrophobicity/Hydrophilicity Analysis Home Page. Available online: http://https://www.peptide2.com/N_peptide_hydrophobicity_hydrophilicity.php (accessed on 12 August 2018).
- Po-Wen, C.; Singh, P.; Zimmerli, L. Priming of the Arabidopsis pattern-triggered immunity response upon infection by necrotrophic Pectobacterium carotovorum bacteria. Mol. Plant Pathol. 2013, 14, 58–70. [Google Scholar] [CrossRef]
- Schwessinger, B.; Roux, M.; Kadota, Y.; Ntoukakis, V.; Sklenar, J.; Jones, A.; Zipfel, C. Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS Genet. 2011, 7, e1002046. [Google Scholar] [CrossRef] [PubMed]
- Roux, M.; Schwessinger, B.; Albrecht, C.; Chinchilla, D.; Jones, A.; Holton, N.; Malinovsky, F.G.; Tor, M.; de Vries, S.; Zipfel, C. The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 2011, 23, 2440–2455. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Gómez, L.; Boller, T. FLS2: An LRR receptor–like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 2000, 5, 1003–1011. [Google Scholar] [CrossRef]
- Gómez-Gómez, L.; Felix, G.; Boller, T. A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J. 1999, 18, 277–284. [Google Scholar] [CrossRef]
- Cui, X.; Lu, L.; Wang, Y.; Yuan, X.; Chen, X. The interaction of soybean reticulon homology domain protein (GmRHP) with Soybean mosaic virus encoded P3 contributes to the viral infection. Biochem. Biophys. Res. Commun. 2018, 495, 2105–2110. [Google Scholar] [CrossRef]
- Marmagne, A.; Rouet, M.A.; Ferro, M.; Rolland, N.; Alcon, C.; Joyard, J.; Garin, J.; Barbier-Brygoo, H.; Ephritikhine, G. Identification of new intrinsic proteins in Arabidopsis plasma membrane proteome. Mol. Cell Proteom. 2004, 3, 675–691. [Google Scholar] [CrossRef]
- Ben Khaled, S.; Postma, J.; Robatzek, S. A moving view: Subcellular trafficking processes in pattern recognition receptor-triggered plant immunity. Annu. Rev. Phytopathol. 2015, 53, 379–402. [Google Scholar] [CrossRef]
- Faulkner, C.; Petutschnig, E.; Benitez-Alfonso, Y.; Beck, M.; Robatzek, S.; Lipka, V.; Maule, A.J. LYM2-dependent chitin perception limits molecular flux via plasmodesmata. Proc. Natl. Acad. Sci. USA 2013, 110, 9166–9170. [Google Scholar] [CrossRef] [PubMed]
- Korner, C.J.; Du, X.; Vollmer, M.E.; Pajerowska-Mukhtar, K.M. Endoplasmic reticulum stress signaling in plant immunity--at the crossroad of life and death. Int. J. Mol. Sci. 2015, 16, 26582–26598. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Zhang, J.; Xiao, W.; Lee, K.; Li, Z.; Wen, J.; He, L.; Gui, D.; Xue, R.; Jian, G.; et al. Rtn1a-mediated endoplasmic reticulum stress in podocyte injury and diabetic nephropathy. Sci. Rep. 2017, 7, 323. [Google Scholar] [CrossRef] [PubMed]
- Pina, F.J.; Fleming, T.; Pogliano, K.; Niwa, M. Reticulons regulate the ER inheritance block during ER stress. Develop. Cell 2016, 37, 279–288. [Google Scholar] [CrossRef]
- Stachel, S.E.; Nester, E.W. The genetic and transcriptional organization of the vir region of the A6 Ti plasmid of Agrobacterium tumefaciens. EMBO J. 1986, 5, 1445–1454. [Google Scholar] [CrossRef]
- Jakubowski, S.J.; Cascales, E.; Krishnamoorthy, V.; Christie, P.J. Agrobacterium tumefaciens VirB9, an outer-membrane-associated component of a type IV secretion system, regulates substrate selection and T-pilus biogenesis. J. Bacteriol. 2005, 187, 3486–3495. [Google Scholar] [CrossRef]
- Sagulenko, E.; Sagulenko, V.; Chen, J.; Christie, P.J. Role of Agrobacterium VirB11 ATPase in T-pilus assembly and substrate selection. J. Bacteriol. 2001, 183, 5813–5825. [Google Scholar] [CrossRef]
- Zhou, X.R.; Christie, P.J. Suppression of mutant phenotypes of the Agrobacterium tumefaciens VirB11 ATPase by overproduction of VirB proteins. J. Bacteriol. 1997, 179, 5835–5842. [Google Scholar] [CrossRef]
- Ahlfors, R.; Macioszek, V.; Rudd, J.; Brosche, M.; Schlichting, R.; Scheel, D.; Kangasjarvi, J. Stress hormone-independent activation and nuclear translocation of mitogen-activated protein kinases in Arabidopsis thaliana during ozone exposure. Plant J. 2004, 40, 512–522. [Google Scholar] [CrossRef]
- Beckers, G.J.; Jaskiewicz, M.; Liu, Y.; Underwood, W.R.; He, S.Y.; Zhang, S.; Conrath, U. Mitogen-activated protein kinases 3 and 6 are required for full priming of stress responses in Arabidopsis thaliana. Plant Cell 2009, 21, 944–953. [Google Scholar] [CrossRef]
- Ma, Y.; Zhao, Y.; Walker, R.K.; Berkowitz, G.A. Molecular steps in the immune signaling pathway evoked by plant elicitor peptides: Ca2+-dependent protein kinases, nitric oxide, and reactive oxygen species are downstream from the early Ca2+ signal. Plant Physiol. 2013, 163, 1459–1471. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zheng, Y.; Gu, C.; He, C.; Yang, M.; Zhang, X.; Guo, J.; Zhao, H.; Niu, D. Bacillus cereus AR156 activates defense responses to Pseudomonas syringae pv. tomato in Arabidopsis thaliana similarly to flg22. Mol. Plant Microbe Interact. 2018, 31, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Meng, J.; Meng, X.; Zhao, Y.; Liu, J.; Sun, T.; Liu, Y.; Wang, Q.; Zhang, S. Pathogen-responsive MPK3 and MPK6 reprogram the biosynthesis of indole glucosinolates and their derivatives in Arabidopsis immunity. Plant Cell 2016, 28, 1144–1162. [Google Scholar] [CrossRef] [PubMed]
- Shih, P.Y.; Chou, S.J.; Mueller, C.; Halkier, B.A.; Deeken, R.; Lai, E.M. Differential roles of glucosinolates and camalexin at different stages of Agrobacterium-mediated transformation. Mol. Plant Pathol. 2018, 19, 1956–1970. [Google Scholar] [CrossRef]
- Koncz, C.; Schell, J. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 1986, 204, 383–396. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Alonso, J.M.; Stepanova, A.N.; Leisse, T.J.; Kim, C.J.; Chen, H.; Shinn, P.; Stevenson, D.K.; Zimmerman, J.; Barajas, P.; Cheuk, R.; et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 2003, 301, 653–657. [Google Scholar] [CrossRef]
- Dellaporta, S.L.; Wood, J.; Hicks, J.B. A plant DNA minipreperation: Version 2. Plant Mol. Biol. Rep. 1983, 1, 19–22. [Google Scholar] [CrossRef]
- Nam, K.H.; Li, J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 2002, 110, 203–212. [Google Scholar] [CrossRef]
- Hwang, H.H.; Wang, M.H.; Lee, Y.L.; Tsai, Y.L.; Li, Y.H.; Yang, F.J.; Liao, Y.C.; Lin, S.K.; Lai, E.M. Agrobacterium-produced and exogenous cytokinin-modulated Agrobacterium-mediated plant transformation. Mol. Plant Pathol. 2010, 11, 677–690. [Google Scholar] [CrossRef]
- Lai, E.M.; Kado, C.I. Processed VirB2 is the major subunit of the promiscuous pilus of Agrobacterium tumefaciens. J. Bacteriol. 1998, 180, 2711–2717. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.Y.; Woollard, A.C.; Wolff, S.P. Hydrogen peroxide production during experimental protein glycation. FEBS Lett. 1990, 268, 69–71. [Google Scholar] [CrossRef]
- Journot-Catalino, N.; Somssich, I.E.; Roby, D.; Kroj, T. The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana. Plant Cell 2006, 18, 3289–3302. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Shen, W.; Chan, Z.; Wu, Y. Endogenous cytokinin overproduction modulates ROS homeostasis and decreases salt stress resistance in Arabidopsis thaliana. Front. Plant Sci. 2015, 6, 1004. [Google Scholar] [CrossRef]
- Singh, P.; Chien, C.C.; Mishra, S.; Tsai, C.H.; Zimmerli, L. The Arabidopsis LECTIN RECEPTOR KINASE-VI.2 is a functional protein kinase and is dispensable for basal resistance to Botrytis cinerea. Plant Signal. Bevhav. 2013, 8, e22611. [Google Scholar] [CrossRef]
- He, P.; Shan, L.; Lin, N.C.; Martin, G.B.; Kemmerling, B.; Nurnberger, T.; Sheen, J. Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell 2006, 125, 563–575. [Google Scholar] [CrossRef]
- Nam, J.; Matthysse, A.G.; Gelvin, S.B. Differences in susceptibility of Arabidopsis ecotypes to crown gall disease may result from a deficiency in T-DNA integration. Plant Cell 1997, 9, 317–333. [Google Scholar] [CrossRef]
- Deblaere, R.; Bytebier, B.; De Greve, H.; Deboeck, F.; Schell, J.; Van Montagu, M.; Leemans, J. Efficient octopine Ti plasmid-derived vectors for Agrobacterium-mediated gene transfer to plants. Nucleic. Acids Res. 1985, 13, 4777–4788. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.C.; Kado, C.I. Studies on Agrobacterium tumefaciens VIII avirulence induced by temperature and ethidium bromide. Can. J. Microbiol. 1977, 23, 1554–1561. [Google Scholar] [CrossRef]


| Peptide Names | Peptide Position of the VirB2 Protein | Peptide Sequences | Length (a. a.) | pI | Hydrophobicity | Mw (Da) | References |
|---|---|---|---|---|---|---|---|
| VirB2-S111-T58 | C-terminus connected with N-terminus | S111thFLGKTLTGGGQSAGGGTDPAT58th | 22 | 5.55 | 27.3 % | 1980.12 | This study |
| VirB2-I63-I80 | Transmembrane domain (TM) 1 | I63thCTFILGPFGQSLAVLGI80th | 18 | 5.52 | 61.1 % | 1849.26 | This study |
| VirB2-I80-V101 | Part of TM1, region between 2 TM domains, part of TM2 | I80thVAIGISWMFGRASLGLVAGVV101th | 22 | 9.75 | 68.2 % | 2216.71 | This study |
| VirB2-G95-F112 | Transmembrane domain (TM) 2 | G95thLVAGVVGGIVIMFGASF112th | 18 | 5.52 | 66.7 % | 1694.06 | This study |
| VirB2-I104-G121 | C-terminal region | I104thVIMFGASFLGKTLTGGG121th | 18 | 8.75 | 50.0 % | 1769.13 | This study |
| Elf18 | N-terminal region | M1stSKEKFERTKPHVNVGTI18th | 18 | 9.70 | 33.3 % | 2101.45 | [51] |
| Agro-Flg22 | N-terminal region | S19thRVSSGLRVKSASDNAAYWSIA40th | 22 | 9.98 | 36.4 % | 2325.57 | [24] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, F.-C.; Hwang, H.-H. Arabidopsis RETICULON-LIKE4 (RTNLB4) Protein Participates in Agrobacterium Infection and VirB2 Peptide-Induced Plant Defense Response. Int. J. Mol. Sci. 2020, 21, 1722. https://doi.org/10.3390/ijms21051722
Huang F-C, Hwang H-H. Arabidopsis RETICULON-LIKE4 (RTNLB4) Protein Participates in Agrobacterium Infection and VirB2 Peptide-Induced Plant Defense Response. International Journal of Molecular Sciences. 2020; 21(5):1722. https://doi.org/10.3390/ijms21051722
Chicago/Turabian StyleHuang, Fan-Chen, and Hau-Hsuan Hwang. 2020. "Arabidopsis RETICULON-LIKE4 (RTNLB4) Protein Participates in Agrobacterium Infection and VirB2 Peptide-Induced Plant Defense Response" International Journal of Molecular Sciences 21, no. 5: 1722. https://doi.org/10.3390/ijms21051722
APA StyleHuang, F.-C., & Hwang, H.-H. (2020). Arabidopsis RETICULON-LIKE4 (RTNLB4) Protein Participates in Agrobacterium Infection and VirB2 Peptide-Induced Plant Defense Response. International Journal of Molecular Sciences, 21(5), 1722. https://doi.org/10.3390/ijms21051722